Abstract
New Delhi metallo-β-lactamase (NDM-1) was initially identified in various Enterobacteriaceae and recently in Acinetobacter baumannii. This study described the clonal dissemination of an NDM-2-producing A. baumannii isolate in an Israeli rehabilitation ward and the genetic surroundings of the gene. The blaNDM-2 gene was surrounded by the ble and trpF genes downstream and two copies of the ISAba125 on both sides. These are the first NDM-producing A. baumannii strains in Israel from patients with no previous travel or hospitalization on the Indian subcontinent.
TEXT
Carbapenem resistance in Gram-negative bacteria is an important worldwide problem, particularly because of the production of class A, D, and B metallo-β-lactamase enzymes (MBLs) as a resistance mechanism and the facility to spread by mobile genetic elements (12). The new MBL, New Delhi metallo-β-lactamase 1 (NDM-1), initially reported in Klebsiella pneumoniae and Escherichia coli recovered from a Swedish patient who was previously hospitalized in India (23), has disseminated to several countries and other Enterobacteriaceae (4, 9, 13, 15–18, 22). Recently, cases of NDM-producing Acinetobacter baumannii have been described in India, Egypt, and China (1, 6, 8).
Five carbapenem-resistant A. baumannii isolates were recovered from female patients at the TA-Sourasky-MA Rehabilitation hospital in Tel Aviv, Israel (Table 1). The five elderly patients (mean age, 81) were hospitalized in the same geriatric rehabilitation ward. The cultures were taken as a point prevalence study from 70 patients hospitalized in two wards in the rehabilitation center. Surveillance skin cultures were taken from six body sites (armpit, thigh, and groin, bilaterally). Four of the five patients were admitted to rehabilitation after orthopedic surgery in two different orthopedic wards located in the same hospital, adjacent to the rehabilitation center. Three of the patients shared a room with each other at a point during their hospital stay, and others shared with them common facilities. None of the patients had any clinical culture that grew Acinetobacter spp., and no signs of infection due to Acinetobacter were evident. There was no specific history taken regarding travel (Table 1). Isolates were initially identified using the Vitek-2 automatic system (bioMérieux, Marcy, France) and confirmed by amplified rRNA gene restriction analysis (ARDRA) (20). The epidemiological relationship was corroborated by pulsed-field gel electrophoresis (PFGE) under conditions described elsewhere (10). PFGE results showed an identical pattern for all the strains. Multiplex PCR to identify clonal lineages (19) showed that the strains did not belong to pan-European clone I, II, or III. Multilocus sequence typing (MLST) indicated that the strain corresponded to sequence type (ST) 103 according to the Pasteur system (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html), which is in agreement with the ST found in the NDM-2-producing A. baumannii isolate reported from Egypt (6).
Table 1.
Epidemiological information on Acinetobacter baumannii strainsa
| Strain | Dates of hospitalization | Sex | Age (yr) | Source of isolate | Days of hospitalization | Comorbidityb | Surgery site(s) | Invasive devicesc | Treatment | Screening CRAd | Date of screening CRA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| I-15 | 14/06/2009–01/07/2009 | Female | 79 | Skin | 18 | CVD | Limbs | UC, ID | Cephalosporins | Positive | 09/07/2009 |
| I-1 | 02/07/2009–26/07/2009 | Female | 84 | Skin | 24 | CVD | Limbs, joints | UC, ID | Cephalosporins | Positive | 09/07/2009 |
| I-16 | 08/07/2009–20/08/2009 | Female | 85 | Skin | 43 | CVD, CLD | Limbs, joints | UC, ID | Cephalosporins | Positive | 12/07/2009 |
| I-2 | 09/07/2009–07/08/2009 | Female | 81 | Skin | 29 | CVD, DM | Head-neck | UC, ID, Tr | No | Positive | 09/07/2009 |
| I-17 | 09/07/2009–01/10/2009 | Female | 75 | Skin | 84 | CVD, DM | Limbs, joints | UC, ID | Cephalosporins | Positive | 12/07/2009 |
Dates are given as day/month/year.
CVD, cardiovascular disease; CLD, chronic lung disease; DM, diabetes mellitus.
UC, urinary catheter; ID, intravascular device; Tr, tracheostomy.
CRA, carbapenem-resistant Acinetobacter baumannii.
Antibiotic susceptibility was performed by MicroScan (Siemens, CA), and the results were interpreted according to CLSI guidelines (2). The strains were resistant to aztreonam, cefepime, ceftazidime, and amikacin (MIC, ≥64 mg/liter), ampicillin-sulbactam (>16/8 mg/liter), ciprofloxacin (≥4 mg/liter), gentamicin (≥16 mg/liter), imipenem and meropenem (≥16 mg/liter), piperacillin (32 to ≥128 mg/liter), piperacillin-tazobactam (32 to ≥128 mg/liter), and ticarcillin (≥128 mg/liter). MICs of tigecycline and colistin were 2 mg/liter and ≤0.5 mg/liter, respectively. MBL production was confirmed by Etest strips (AB Biodisk, Sweden). The MIC of imipenem was ≥256 mg/liter, and that of imipenem/EDTA was ≤1 mg/liter.
Multiplex PCR for class D β-lactamases (blaOXA-51, blaOXA-23, blaOXA-24, and blaOXA-58) (21) was positive only for blaOXA-51 in all the strains. PCRs for class B β-lactamases blaIMP, blaVIM, blaSIM, blaSPM, blaGIM (11), and blaNDM (NDM-1F, 5′-CCAATATTATGCACCCGGTCG; NDM-1R, 5′-ATGCGGGCCGTATGAGTGATTG) were performed with specific primers. All strains were positive for blaNDM. Sequence analysis of the PCR products showed 99% identity with the blaNDM-1 previously reported (23). The sequence of the blaNDM gene detected in our study showed a double nucleotide substitution from C to G at position 82 and A to G at position 468 from the start codon. Only the first change resulted in an amino acid substitution from P (proline) to A (alanine) at position 28, as was already described and named blaNDM-2 (6), and the other was a silent mutation. Although the armA gene has been associated with blaNDM-1 in A. baumannii (8), PCRs to detect the 16S rRNA methylase-encoding genes rmtA, rmtB, rmtC, rmtD, armA (3), and npmA (npmA-F, 5′-CTCAAAGGAACAAAGACGGTTG-3′; npmA-R, 5′-GTTTCTGGCCATGTTCAAAAC-3′) were negative in our strains, in agreement with the report by Kaase et al. (6).
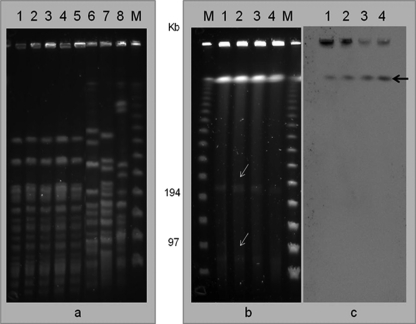
Plasmid identification by the Kado and Liu method (7) and conjugation experiments using a ciprofloxacin-resistant, imipenem-susceptible A. baumannii isolate as a recipient were unsuccessful. Southern blot analysis was performed by digestion with the S1 nuclease. Digested genomic DNA was first separated by PFGE and then hybridized with the blaNDM-1 probe marked with the PCR DIG probe synthesis kit (Roche, Barcelona, Spain). Detection was performed with antidigoxigenin antibody conjugated to alkaline phosphatase and CDP-Star chemiluminescence substrate (Roche). The data showed two plasmids of approximately 70 and 200 kb with no signal hybridization with the probe, but it was clearly demonstrated that blaNDM-2 is located in the chromosome (Fig. 1).
Fig. 1.
(a) PFGE analysis of Acinetobacter baumannii strains. (b) Plasmid identification by digestion with S1 nuclease. (c) Hybridization with blaNDM-1 probe. Lanes: 1, A. baumannii AB-I1; 2, AB-I2; 3, AB-I3; 4, AB-I4; 5, AB-I5. Lanes 6 to 8, A. baumannii European clones EC-I (strain RUH-875), EC-II (strain RUH-134), and EC-III (strain RUH-5875), respectively. Bands with white arrows indicate the presence of plasmids without signal hybridization with the blaNDM-1 probe; black arrow indicates the chromosomal position with positive hybridization with the blaNDM-1 probe.
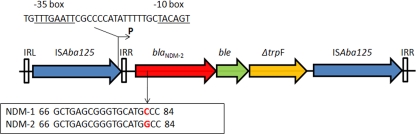
In order to determine the genetic structure surrounding the blaNDM-2 gene, DNA from strain AB-I1 was digested with RsaI (Promega). The fragments obtained were autoligated at 16°C with T4 DNA ligase (Promega). The fragment of DNA containing the blaNDM-2 gene was used as a template for an inverse PCR with primers designed from the blaNDM-1 gene sequence (NDM-inv-F, 5′-TGCCGACACTGAGCACTAC-3′; NDM-inv-R, 5′-GGTCGCCAGTTTCCATTTGC-3′). Analysis of the genetic surroundings showed that the blaNDM-2 gene was similar to that described for plasmid pNDM-HK (5) with the ble (bleomycin resistance) and trpF [N-(5′-phosphoribosyl) anthranilate isomerase] genes downstream; however, two copies of the insertion sequence ISAba125, one upstream close to the promoter region and the second at the 3′end of the truncated trpF gene, with the respective left (IRL) and right inverted repeats (IRR), were observed in our strain (Fig. 2). A promoter made of −35 (TTGAAT) and −10 (TACAGT) sequences separated by a distance of 17 bp was found at 104 bp from the blaNDM-2 start codon. A similar position of the promoter has been reported (5), suggesting that NDM enzymes are under the control of the promoter upstream of the gene.
Fig. 2.
Genetic surroundings of the blaNDM-2 in A. baumannii strain AB-I1. IRL, left inverted repeat; IRR, right inverted repeat. P, promoter; ble, bleomycin resistance gene; trpF, N-(5′-phosphoribosyl) anthranilate isomerase.
Sequence alignment of our strain AB-I1 with the previously reported arrays of E. coli 271, pNDM-HK, and pkpANDM-1 (5, 14) showed a high degree of homology in the region corresponding to the IRR of the ISAba125 gene, suggesting the presence of truncated structures by the participation of mobile genetic elements that can move from one site to another by transposition. With this, blaNDM-1 or blaNDM-2 flanked by IS can also be shuttled between plasmids (1) and the chromosome. Therefore, we can hypothesize that the ISAba125 specific element from A. baumannii could be the origin of the dissemination among plasmids from the Enterobacteriaceae.
Despite the epidemiological evidence that travel to the Indian subcontinent is related to infection caused by blaNDM-1 (9), the occurrence of sporadic colonizers and their clonal dissemination in the same unit, as observed in the present study, may be possible without any association with previous travels or hospitalization on the Indian subcontinent.
In conclusion, we report for the first time a clonal dissemination of an NDM-2-producing A. baumannii isolate in an Israeli rehabilitation ward and the genetic surroundings of the gene. Epidemiological control and adequate identification of NDM-producing A. baumannii will prevent an increase in resistance and better applications of therapeutic measures.
Nucleotide sequence accession number.
The GenBank accession number for the strain AB-I1 is JF821215.
Acknowledgments
This study was supported by the Spanish Ministry of Health (FIS 08/00195), by grant 2009SGR1256, and by Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III-FEDER, Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008). This work was supported in part by the European Commission Grant (FP6): European Network for Mastering Hospital Antimicrobial Resistance and its Spread into the Community (MOSAR; LSHPCT-2007-037941).
Footnotes
Published ahead of print on 8 August 2011.
REFERENCES
- 1. Chen Y., Zhou Z., Jiang Y., Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66:1255–1259 [DOI] [PubMed] [Google Scholar]
- 2. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 3. Doi Y., Arakawa Y. 21 May 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88–94 [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 4. Gottig S., et al. 2010. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect. Dis. 10:828–829 [DOI] [PubMed] [Google Scholar]
- 5. Ho P. L., et al. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaase M., et al. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262 [DOI] [PubMed] [Google Scholar]
- 7. Kado C. I., Liu S. T. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karthikeyan K., Thirunarayan M. A., Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65:2253–2254 [DOI] [PubMed] [Google Scholar]
- 9. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marcos M. A., Jimenez de Anta M. T., Vila J. 1995. Correlation of six methods for typing nosocomial isolates of Acinetobacter baumannii. J. Med. Microbiol. 42:328–335 [DOI] [PubMed] [Google Scholar]
- 11. Mendes R. E., et al. 2007. Rapid detection and identification of metallo-beta-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J. Clin. Microbiol. 45:544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miriagou V., et al. 2010. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16:112–122 [DOI] [PubMed] [Google Scholar]
- 13. Poirel L., Al Maskari Z., Al Rashdi F., Bernabeu S., Nordmann P. 2011. NDM-1-producing Klebsiella pneumoniae isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 66:304–306 [DOI] [PubMed] [Google Scholar]
- 14. Poirel L., Lagrutta E., Taylor P., Pham J., Nordmann P. 2010. Emergence of metallo-beta-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poirel L., Revathi G., Bernabeu S., Nordmann P. 2011. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 55:934–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poirel L., et al. 2011. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob. Agents Chemother. 55:447–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rolain J. M., Parola P., Cornaglia G. 2010. New Delhi metallo-beta-lactamase (NDM-1): towards a new pandemia?. Clin. Microbiol. Infect. 16:1699–1701 [DOI] [PubMed] [Google Scholar]
- 18. Struelens M. J., Monnet D. L., Magiorakos A. P., Santos O'Connor F., Giesecke J. 2010. New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill. 15(46):pii=19716. [DOI] [PubMed] [Google Scholar]
- 19. Turton J. F., Gabriel S. N., Valderrey C., Kaufmann M. E., Pitt T. L. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815 [DOI] [PubMed] [Google Scholar]
- 20. Vaneechoutte M., et al. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Woodford N. 2010. Rapid characterization of beta-lactamases by multiplex PCR. Methods Mol. Biol. 642:181–192 [DOI] [PubMed] [Google Scholar]
- 22. Wu H. S., et al. 2010. First identification of a patient colonized with Klebsiella pneumoniae carrying blaNDM-1 in Taiwan. J. Chin. Med. Assoc. 73:596–598 [DOI] [PubMed] [Google Scholar]
- 23. Yong D., et al. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]