Abstract
(S)-1-[3-hydroxy-2-(phosphonomethoxy)propyl]cytosine (HPMPC [cidofovir]) and (S)-9-[3-hydroxy-2-(phosphonomethoxy)propyl]adenine (HPMPA) are potent inhibitors of a variety of DNA viruses. These drugs possess a 3′-hydroxyl equivalent which could support chain extension from an incorporated drug molecule. HPMPC and HPMPA were initially reported to lack activity against human immunodeficiency virus type 1 (HIV-1); more recent results have shown that the octadecyloxyethyl (ODE) and hexadecyloxypropyl (HDP) esters of HPMPA are potent inhibitors of the virus. We have synthesized the ODE esters of a series of (S)-[3-hydroxy-2-(phosphonomethoxy)propyl] (HPMP) nucleosides, including HPMPC, HPMP-guanine (HPMPG), HPMP-thymine (HPMPT), and HPMP-diaminopurine (HPMPDAP), as well as the ODE ester of the obligate chain terminator (S)-9-[3-methoxy-2-(phosphonomethoxy)-propyl]adenine (MPMPA). All compounds except ODE-HPMPT were inhibitors of HIV-1 replication at low nanomolar concentrations. These compounds were also inhibitors of the replication of HIV-1 variants that are resistant to various nucleoside reverse transcriptase (RT) inhibitors at concentrations several times lower than would be expected to be achieved in vivo. To investigate the mechanism of the antiviral activity, the active metabolites of HPMPC and HPMPA were studied for their effects on reactions catalyzed by HIV-1 RT. Incorporation of HPMPC and HPMPA into a DNA primer strand resulted in multiple inhibitory effects exerted on the enzyme and showed that neither compound acts as an absolute chain terminator. Further, inhibition of HIV-1 RT also occurred when these drugs were located in the template strand. These results indicate that HPMPC and HPMPA inhibit HIV-1 by a complex mechanism and suggest that this class of drugs has a broader spectrum of activity than previously shown.
INTRODUCTION
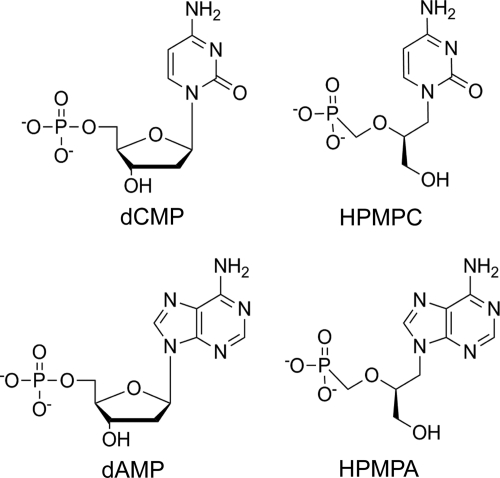
The nucleoside phosphonates (S)-1-[3-hydroxy-2-(phosphonomethoxypropyl)]cytosine (HPMPC), also known as cidofovir, and (S)-9-[3-hydroxy-(2-phosphonomethoxy)propyl]adenine (HPMPA) are acyclic analogs of dCMP and dAMP, respectively (Fig. 1). These drugs are inhibitors of a wide range of double-stranded DNA (dsDNA) viruses, including herpes simplex virus, cytomegalovirus, varicella zoster virus, adenovirus, and the poxviruses vaccinia, monkeypox, and variola virus (3, 10; reviewed in reference 9). We have previously used the orthopoxvirus vaccinia virus in a model system to investigate the mechanism of action of these compounds (23, 24). Our results have shown that the active intracellular metabolites of these drugs, HPMPC diphosphate (HPMPCpp) and HPMPA diphosphate (HPMPApp), respectively, are substrates for vaccinia virus DNA polymerase and, once incorporated into the penultimate 3′ end of the primer strand, inhibit the enzyme's polymerase and 3′-to-5′ proofreading exonuclease activities. Further, we found that, when these drugs are incorporated into the template strand, they profoundly inhibit replication across the drug lesion. These results suggest that this class of antiviral agents has a complex mechanism of action, inhibiting DNA synthesis when present in both the primer and template strands and blocking 3′-to-5′ exonuclease activity when located in the primer strand. We have also shown that drug-resistant vaccinia viruses that encode independently acting mutations in the 3′-to-5′ exonuclease and 5′-to-3′ DNA polymerase domains can be isolated (2, 21). This pattern of mutation is consistent with a multifaceted mode of drug action.
Fig. 1.
Structures of key nucleoside monophosphates and the corresponding antiviral acyclic nucleoside phosphonate analogs.
Although effective against several DNA viruses, HPMPA does not have activity against HIV-1 in its original form (4, 27). Interestingly, unmodified HPMPC was able to inhibit HIV-1 in epithelioid HeLa-CD4 cells but not in the T-lymphocyte cell line MT-2, a result partially explained by differences between the metabolism of HPMPC and that of HPMPCpp in the two cell lines (29). The idea of the importance of effective cellular metabolism for the efficacy of these drugs was strengthened by our previous data showing that alkoxyalkyl ester derivatives of HPMPA are inhibitory to HIV-1 in MT-2 cells (17). The octadecyloxyethyl (ODE) and hexadecyloxypropyl (HDP) esters of HPMPA are potent inhibitors of HIV-1 replication, with low or subnamomolar 50% effective concentration (EC50) values (17). These prodrugs of HPMPC and HPMPA are taken up by cells and metabolized to the active HPMPCpp and HPMPApp metabolites to a much greater extent than are the underivatized forms (1, 23). ODE-HPMPA and ODE-HPMPC are also powerful inhibitors of a variety of dsDNA viruses, and HDP-HPMPC (CMX001) is in phase II clinical trials for cytomegalovirus and other dsDNA virus infections (16).
Based on these data, we were interested in extending our studies on the mechanism of action of HPMPC and HPMPA to HIV-1 reverse transcriptase (RT) to determine whether a pattern of inhibition similar to that seen with vaccinia virus DNA polymerase would be observed. The antiviral nucleosides currently approved for treating HIV-1 infections are all obligatory chain terminators (14). However, HPMPC and HPMPA both have a hydroxyl group in the acyclic chain, which could support continued DNA synthesis after drug incorporation into DNA, raising the possibility of alternative mechanisms of action against HIV-1. For this report, we synthesized the ODE esters of HPMPC, HPMP-diaminopurine (HPMPDAP), HPMP-guanine (HPMPG), and HPMP-thymine (HPMPT) and found that, with the exception of HPMPT, they are all potent inhibitors of HIV-1 replication in vitro. We also used steady-state primer extension analyses to determine the effects of HPMPCpp and HPMPApp on HIV-1 RT. Our data indicate that HIV-1 RT can use HPMPCpp and HPMPApp as substrates and incorporate them into newly synthesized DNA. This incorporation of HPMPC or HPMPA into DNA inhibits further DNA synthesis; the pattern and extent of inhibition are dependent on whether the template is RNA or DNA and whether a single analog molecule or two consecutive molecules are incorporated. Further, the enzyme is inhibited by the drugs when they are incorporated into the template strand. Overall, these data suggest that HPMPC and HPMPA have a complex mechanism of action against HIV-1, which is consistent with previous results showing a multifaceted mechanism of action against the dsDNA of vaccinia virus.
MATERIALS AND METHODS
Synthesis of ODE esters of nucleoside phosphonates.
Briefly, 4-O-methylthymine was prepared by hydrolysis of 2,4-dimethoxy-5-methylpyrimidine (Reddy Chemtech, Inc., Kennesaw, GA) (33). Nucleoside bases 4-O-methylthymine, N4-benzoylcytosine (Aldrich, St. Louis, MO), 6-O-benzylguanine (AK Scientific, Mountain View, CA), and 2,6-diaminopurine (TCI America, Portland, OR) reacted with (S)-trityl glycidyl ether (Sanyo Fine Co., Nishi-ku, Osaka, Japan) to give the corresponding 3-trityloxy-2-hydroxypropyl nucleosides, which were then alkylated with the octadecyloxyethyl p-toluenesulfonyloxy-methylphosphonate as previously described (31). The 2-methoxy analog of HPMPA was prepared by substituting (S)-methyl glycidyl ether (TCI America, Portland, OR) for (S)-trityl glycidyl ether in the reaction scheme; details of this synthesis have been reported elsewhere (32). After acidic deprotection, the desired octadecyloxyethyl esters of (S)-[3-hydroxy-2-(phosphonomethoxy)propyl] nucleosides and (S)-9-[3-methoxy-2-(phosphonomethoxy)propyl]adenine were obtained. Compound purity was assessed to be >95%. Adefovir and tenofovir were obtained from Moravek Biochemicals (Brea, CA).
HIV-1 inhibition assays.
To assess the antiviral activity of the ODE-HPMP nucleosides, the inhibition of HIV-1 by each compound was first examined using MT-2 cells (D. D. Richman, personal stocks). Cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 10 mM HEPES buffer, 50 IU of penicillin/ml, and 50 μg of streptomycin/ml (Invitrogen). HIV-1LAI and nucleoside RT inhibitor (NRTI)-resistant isolates were obtained from personal stocks (D. D. Richman) or from the AIDS Research and Reference Reagent Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The antiviral activity of each compound was determined by inoculating MT-2 cells with HIV-1LAI at a multiplicity of infection (MOI) of 0.001 50% tissue culture infectious dose (TCID50)/cell, followed by incubation in the presence of 3-fold serial drug dilutions (three wells per dilution) as previously described (15). Four days after infection, culture supernatants were harvested, lysed with 0.5% Triton X-100, and assayed for p24 antigen concentrations using a commercial enzyme-linked immunosorbent assay (ELISA) (PerkinElmer Life Sciences). The antiviral activity of each compound was determined and is expressed as the 50% effective concentration (EC50), which is the concentration required for inhibition of p24 antigen production by 50%.
Inhibition of replication of wild-type and NRTI-resistant HIV-1 by the compounds was determined in donor peripheral blood mononuclear cells (PBMCs). PBMCs were prepared by Ficoll-Hypaque (Histopaque 1077; Sigma, St. Louis, MO) density gradient centrifugation of heparinized blood from normal volunteers. PBMCs were suspended to 2 × 106/ml in R-3 medium (RPMI 1640 medium supplemented with 15 to 20% heat-inactivated fetal bovine serum, 5% purified human interleukin-2, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM l-glutamine) and were stimulated with phytohemagglutinin (PHA) (2 to 3 μg/ml) for 2 to 4 days. Stocks of the test compounds were made in dimethyl sulfoxide (DMSO) and diluted with complete media to obtain the desired drug concentrations. The standardized drug susceptibility assay was done as previously described (19). Briefly, a total of 4,000 TCID50 of HIV-1 virus stock (NL43 [wild type], Y071 [69 insert], G871-2 [151 complex], G910-6 [TAMS], or U463-1 [K65R]) was added to 4 × 106 PHA-stimulated normal donor PBMCs. Cells and virus were incubated for 1 h at 37°C. Cells were then washed and resuspended in 2.0 ml of fresh medium to achieve a final concentration of 2 × 106 cells per ml. The infected cells were seeded at 2 × 105 cells per well into triplicate wells of a 96-well microtiter plate, and drug-containing medium was added to achieve final drug concentrations ranging from 0.1 to 10,000 nM. Plates were incubated at 37°C in humidified air with 5% CO2. On day 4, the cells were resuspended and two-thirds of the medium was removed and replaced with fresh medium containing the appropriate concentration of drug. On day 7, the supernatant from each well was diluted approximately 1:750 to 1:3,000 for quantitative p24 antigen determinations. The EC50 of the drugs was determined by comparing p24 antigen values for drug-containing wells with those for control wells as previously described (19). The 50% cytotoxic concentration (CC50) values were determined as previously described (26).
Cytotoxicity assay.
To assess cytotoxicity, MT-2 cells were incubated with the compounds for 4 days and harvested. Flow count beads (Beckman Coulter) were added to the cell suspension followed by propidium iodide staining and analysis using an Epics Elite flow cytometer (Beckman Coulter). The CC50 was calculated from the cell counts and viability (17).
HIV-1 RT assay reagents.
HPMPCpp and HPMPApp were prepared by custom synthesis by TriLink Biotechnologies (San Diego, CA). Unlabeled deoxynucleoside triphosphates (dNTPs) and dideoxynucleoside triphosphates (ddNTPs) were from Fermentas (Burlington, Ontario, Canada), and radiolabeled [γ-32P]ATP and cordycepin triphosphate ([α-32P]3′-deoxyATP) were purchased from PerkinElmer (Woodbridge, Ontario, Canada). DNA and RNA oligonucleotides were purchased from Sigma-Genosys (Oakville, Ontario, Canada) or Integrated DNA Technologies (Coralville, IA). A DNA oligonucleotide labeled on its 5′ end with FAM (6-carboxyfluorescein) was obtained from Integrated DNA Technologies. Wild-type clade B HIV-1 RT produced using Escherichia coli was a generous gift from M. Götte (McGill University). T4 polynucleotide kinase, a Klenow fragment of DNA polymerase I, uracil-DNA glycosylase (UDG), and terminal deoxynucleotidyl transferase (TdT) were obtained from Fermentas. Moloney murine leukemia virus RT was purchased from Invitrogen (Burlington, Ontario, Canada).
HPMPCpp and HPMPApp incorporation assays.
Oligonucleotide primer-template pairs (Fig. 2) were used as substrates to examine the ability of HIV-1 RT to use HPMPCpp or HPMPApp and to incorporate them into DNA. Both RNA and DNA templates were used to represent reverse transcription and DNA polymerase activities, respectively. Various combinations of dNTPs and HPMPCpp or HPMPApp were added as indicated. In some experiments, ddNTPs were also added to reaction mixtures. HIV-1 RT was used at a final concentration of 50 nM in a solution containing 50 mM Tris·HCl (pH 7.8), 50 mM NaCl, and 6 mM MgCl2. After incubation at 37°C, reactions were stopped by the addition of an equal volume of gel loading buffer (95% [vol/vol] formamide, 20 mM EDTA [pH 8.0]). Reaction products were resolved on 15% denaturing polyacrylamide gels (38 by 30 cm) processed at 45 W for 1.5 h in half-strength Tris-borate-EDTA. Gels were transferred to a Typhoon Trio variable mode imager (GE Healthcare) using plastic sheeting (Protect All; Studio) and scanned using a 488-nm enhanced chemiluminescence-positive (ECL+) excitation setting, a 520-nm band-pass emission filter, and normal sensitivity and 100-μm-resolution settings. Images were analyzed using ImageQuant TL image analysis version 7.0 software (GE Healthcare).
Fig. 2.
Oligonucleotide primer-template pairs used in this study. Unlabeled primer P1 was originally described by Xiong et al. (34). Templates T9 and T2 were originally described by Magee et al. (24), and primers P9, P10, P11, P13, P14, P17, P18, and P19 and templates T11, T12, T13, T14, T17, and T18 were previously described by Magee et al. (23). The RNA oligonucleotides T9RNA, T11RNA, T2RNA, and T12RNA have sequences corresponding to those of the DNA oligonucleotides T9, T11, T2, and T12, respectively.
Preparation of HPMPC- and HPMPA-containing templates.
HPMPC- and HPMPA-containing templates were prepared by enzymatic synthesis as described previously (23) using the primer-template pairs shown in Fig. 2. Control templates containing dCMP and dAMP, respectively, were also prepared as described previously (23). Aliquots of the synthesized DNA were subjected to 3′-end labeling using [α-32P]3′-deoxyATP and TdT to monitor complete extension of each template (data not shown).
Template assays.
DNA polymerase reactions using the HPMPC- or HPMPA-containing templates were performed as previously described (23), except that HIV-1 RT (50 nM) and its buffer (50 mM Tris·HCl [pH 7.8], 50 mM NaCl, 6 mM MgCl2) were used in place of vaccinia virus DNA polymerase. The primers (Fig. 2) were first subjected to end labeling by using [γ-32P]ATP and T4 polynucleotide kinase prior to annealing to the template strand. Size standards were generated by using the Klenow fragment and dideoxy sequencing reactions (28). After incubation at 37°C, reaction mixtures were stopped by the addition of one-half volume of gel loading buffer (80% [vol/vol] formamide, 10 mM EDTA [pH 8.0], 1 mg of xylene cyanol FF/ml (sigma), 1 mg of bromophenol blue/ml). Reaction products were resolved on 10% denaturing polyacrylamide gels (38 by 30 cm) at 70 W for 35 min in half-strength Tris-borate-EDTA and subjected to phosphorimager analysis as previously described (24) using a Typhoon 9400 phosphorimager (GE Healthcare).
RESULTS
Antiviral evaluations.
In HIV-1LAI-infected MT-2 cells, all compounds were highly active in the low nanomolar concentration range, with the exception of ODE-HPMPT. ODE-HPMPC and the previously described ODE-HPMPA (17) were the most active, with identical EC50s of 0.1 ± 0.1 nM (Table 1). ODE-HPMPDAP and ODE-HPMPG were less active, with EC50s of 1.0 ± 2.0 nM and 9.0 ± 6.0 nM, respectively. Blocking the 3-hydroxyl of ODE-HPMPA by converting it to a 3-methoxy residue, the ODE ester of the obligate chain terminator (S)-9-[3-methoxy-2-(phosphonomethoxy)-propyl]adenine (MPMPA) (ODE-MPMPA), reduced but did not abolish activity against HIV-1 (EC50 of 30 ± 15 nM). Two conventional nucleoside phosphonates, adefovir and tenofovir, showed much higher EC50s (1,300 ± 700 nM and 680 ± 460 nM, respectively) than the ODE-HPMP nucleosides and ODE-MPMPA. Cytotoxicity in rapidly dividing MT-2 cells was assessed by flow cytometry with propidium iodide staining. The CC50 values ranged from 20 nM to 25,000 nM for the ODE-HPMP nucleosides. In human foreskin fibroblast cells, however, the CC50 values for these compounds are much higher and range from 1,400 to 70,000 nM (31). ODE-MPMPA was the least cytotoxic ODE ester tested, with a CC50 of 15,000 ± 3,100 nM. Selectivity index (SI) values ranged from <2.5 to 1,900; ODE-HPMPDAP (SI of 1,900) and ODE-MPMPA (SI of 500) were the most selective compounds. ODE-HPMPT was essentially inactive.
Table 1.
Antiviral activity of ODE-HPMP nucleosides and ODE-MPMPA in HIV-1-infected MT-2 cellsa
| Compound | Mean EC50 (nM) ± SD | Mean CC50 (nM) ± SD | Selectivity indexb |
|---|---|---|---|
| Reference compounds | |||
| Adefovir | 1,300 ± 700 (7) | 157,000 ± 54,000 (3) | 121 |
| Tenofovir | 680 ± 460 (6) | >100,000 (7) | >147 |
| 3′-Hydroxyl compounds | |||
| ODE-(S)-HPMPA | 0.1 ± 0.1 (6) | 30 ± 20 (3) | 300 |
| ODE-(S)-HPMPC | 0.1 ± 0.1 (4) | 20 ± 3 (3) | 200 |
| ODE-(S)-HPMPDAP | 1.0 ± 2.0 (3) | 1,900 ± 800 (3) | 1,900 |
| ODE-(S)-HPMPG | 9.0 ± 6.0 (3) | 240 ± 30 (3) | 27 |
| ODE-(S)-HPMPT | >10,000 (3) | 25,000 (3) | <2.5 |
| 3-Methoxy compound | |||
| ODE-(S)-MPMPA | 30 ± 15 (3) | 15,000 ± 3,100 (3) | 500 |
Values in parentheses represent the numbers of independent experiments.
Selectivity index values were determined as follows: mean CC50/mean EC50.
In human PBMC, the ODE esters of the HPMP nucleosides were active against wild-type HIV-1NL43, with EC50s in the 3 to 14 nM range, while their CC50s ranged from 6,000 to 21,700 (Table 2). In general, all compounds in a small resistance panel of HIV-1 variants were highly active, with EC50s ranging from 5 to 27 nM. Several compounds showed lower activity against some NRTI-resistant HIV-1 variants. For example, in PBMCs infected with the 69 insert variant, ODE-HPMPC had an EC50 of 83 nM (6.4× the wild-type value), ODE-HPMPG exhibited an EC50 of 61 nM against the 151 complex HIV-1 variant (20× the wild-type value), and ODE-HPMPDAP had an EC50 of 57 nM against the TAMS HIV-1 variant (4.1× the wild-type value). In PBMCs infected with the K65R HIV-1 variant, the adenosine, cytidine, and guanosine analogs exhibited only modest increases in EC50 that ranged from 22 to 27 nM compared with 3 to 13 nM in wild-type HIV-1. ODE-HPMPDAP showed a 5-fold higher EC50 against K65R (5.3× the wild-type value). Nevertheless, all compounds would be expected to be active against these NRTI-resistant strains in vivo based on their EC50s of 3 to 74 nM and the known ability of related compounds of a similar design to produce human drug concentrations > 1,000 nM after oral administration (22, 26).
Table 2.
Antiviral activity of octadexyloxyethyl esters of HPMP nucleosides in wild-type and NRTI-resistanta HIV-1-infected human PBMCs
| Compound | CC50 (nM) | EC50 (nM) |
||||
|---|---|---|---|---|---|---|
| Wild type | 69 insert | 151 complex | TAMS | K65R | ||
| ODE-(S)-HPMPA | 16,500 ± 6,400 (4) | 10 ± 6 (6) | 16 ± 4 | 14 ± 4 | 5 ± 9 | 24 ± 3 |
| ODE-(S)-HPMPC | 7,500 ± 200 | 13 ± 5 (6) | 83 ± 21 | 27 ± 3 | 12 ± 7 | 27 ± 11 |
| ODE-(S)-HPMPDAP | 6,000 ± 4,900 (5) | 14 ± 8 (6) | 8 ± 4 | 16 ± 3 | 57 ± 39 | 74 ± 14 |
| ODE-(S)-HPMPG | 21,700 ± 11,700 | 3 ± 2 (6) | 11 ± 5 | 61 ± 15 | 5 ± 5 | 22 ± 12 |
Mutations present: M41L, T69SS, L74V, M184V, and L210W (69 insert); V75I, F77L, F116Y, and Q151M (151 complex); M41L, D67N, K70R, T215Y, and K219Q (TAMS); and K65R. Results represent the means ± standard deviations of the results of 3 replicate experiments unless otherwise indicated (values in parentheses represent the numbers of independent experiments).
The addition of HPMPCpp or HPMPApp to HIV-1 RT primer extension assays results in the inhibition of DNA synthesis.
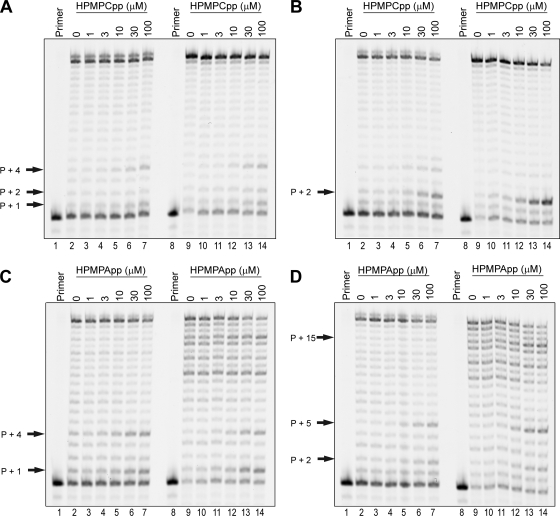
To begin to determine the mechanism of inhibition of HPMPCpp and HPMPApp, we examined the influence of increasing concentrations of each compound on HIV-1 RT activity in the presence of all four dNTPs. Because retroviruses use both RNA and DNA templates to synthesize DNA, we performed these assays using both types of templates to represent first-strand (reverse transcription) and second-strand syntheses, respectively. Further, we utilized templates that directed the incorporation of one drug molecule (T9RNA, T9, T11RNA, T11) or two consecutive drug molecules (T2RNA, T2, T12RNA, T12) immediately after the primer terminus (Fig. 2). The results of these experiments are shown in Fig. 3. When HIV-1 RT was used to catalyze primer extension opposite templates carrying a single (d)GMP (T9RNA and T9) and in the presence of 10 μM dATP, 5 μM dCTP, 10 μM dGTP, 10 μM dTTP, the addition of increasing concentrations of HPMPCpp resulted in little inhibition of DNA synthesis compared to the results seen with reaction mixtures incubated in the absence of drug (Fig. 3A). In both reaction mixtures, at HPMPCpp concentrations ≥ 10 μM, weak pause sites are observed at positions primer + 1 and primer + 4. There is also a weak pause site observed at position primer + 2 when 30 or 100 μM HPMPCpp was added to a P1-T9 reaction mixture (Fig. 3A, lanes 13 and 14); this pause was not observed when an RNA template of the same sequence was used (Fig. 3A, lanes 6 and 7). When templates T2RNA and T2 (directing the incorporation of two consecutive drug molecules) were used, greater inhibition of DNA synthesis by HPMPCpp was observed (Fig. 3B). In both reaction mixtures, the addition of ≥10 μM HPMPCpp resulted in pausing at a position corresponding to primer + 2. This pausing was more pronounced when the DNA template was used compared to the RNA template (compare lanes 13 and 14 of Fig. 3B with lanes 6 and 7).
Fig. 3.
Effects of increasing concentrations of HPMPCpp or HPMPApp on HIV-1 RT activity. (A) Primer extension reactions using RNA or DNA templates directing the incorporation of a single HPMPC molecule. Each 10-μl reaction mixture contained FAM-labeled primer P1 annealed to template T9RNA (lanes 2 to 7) or T9 (lanes 9 to 14), 5 μM dCTP, 10 μM dATP, 10 μM dGTP, 10 μM dTTP, and 50 nM HIV-1 RT. Reactions were further supplemented with HPMPCpp at 0, 1, 3, 10, 30, or 100 μM as indicated. After incubation at 37°C for 5 min, each reaction was stopped by adding gel loading buffer and reaction products were separated on a 15% polyacrylamide gel and then analyzed by fluorescent imaging. Lanes marked “Primer” show the position of the labeled primer, and the positions of pause sites are indicated by arrows. (B) Primer extension reactions using RNA or DNA templates directing the incorporation of two consecutive HPMPC molecules. Reaction conditions were as described for panel A, except that templates T2RNA (lanes 2 to 7) and T2 (lanes 9 to 14) were used. (C) Primer extension reactions using RNA or DNA templates directing the incorporation of a single HPMPA molecule. The reaction mixtures contained a FAM-labeled primer P1 annealed to template T11RNA (lanes 2 to 7) or template T11 (lanes 9 to 14), 5 μM dATP, 10 μM dCTP, 10 μM dGTP, 10 μM dTTP, and 50 nM HIV-1 RT. HPMPApp was also added at the concentrations indicated. The reactions were processed and imaged as described for panel A. (D) Primer extension reactions using RNA or DNA templates directing the incorporation of two consecutive HPMPA molecules. Reaction conditions were as described for panel C, except that templates T12RNA (lanes 2 to 7) and T12 (lanes 9 to 14) were used.
The addition of HPMPApp to HIV-1 RT-catalyzed primer extension reaction mixtures resulted in patterns of inhibition of DNA synthesis that showed similarities to (as well as differences from) HPMPCpp inhibition. As shown in Fig. 3C, when HPMPApp was added at concentrations ≥ 10 μM to reaction mixtures containing template T11RNA or T11 (directing the incorporation of a single drug molecule) and containing all four dNTPs (5 μM dATP, 10 μM dCTP, 10 μM dGTP, and 10 μM dTTP), pause sites were observed at positions primer + 1 and primer + 4. These are the same pause sites seen in the reactions incorporating a single molecule of HPMPC. However, the pause sites observed in the HPMPApp-containing reaction mixtures were stronger than those in the HPMPCpp-containing reactions. Further, the pause at position primer + 2 was not observed in the reaction mixtures incubated with HPMPApp (compare Fig. 3C and 3A). Greater differences between the two compounds were seen when templates directing the incorporation of two consecutive HPMPA molecules (T12RNA or T12) were used (Fig. 3D). When T12RNA was used as the template, the addition of increasing concentrations of HPMPApp to HIV-1 RT reaction mixtures resulted in the appearance of pause sites at positions primer + 2 and primer + 5. Replacement of T12RNA with the DNA template T12 in HIV-1 RT-catalyzed reaction mixtures also resulted in the primer + 2 and primer + 5 pause sites when HPMPApp concentrations ≥ 10 μM were used (Fig. 3D, lanes 12 to 14). An additional pause site at position primer + 15 was also observed in these reaction products. Although use of this template did result in the natural pause sites seen in the absence of drug (Fig. 3D, lane 9), the primer + 15 pause appears to be stronger in the presence of 30 or 100 μM HPMPApp (Fig. 3D, lanes 13 and 14). We have also observed this pause site in preliminary experiments in which a 32P-labeled primer was used (data not shown).
HIV-1 RT can replace dCTP or dATP with HPMPCpp or HPMPApp, respectively, to promote DNA synthesis.
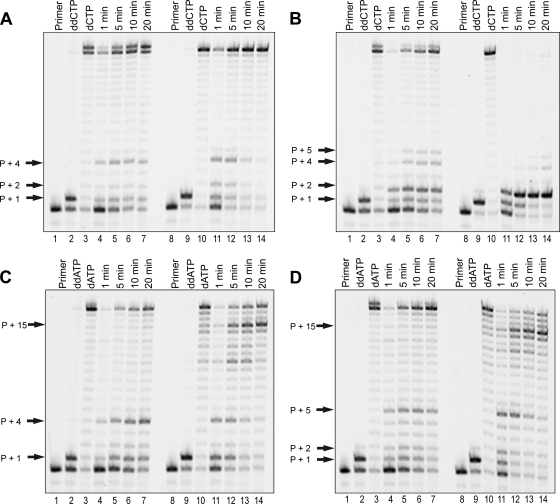
Although the addition of HPMPCpp or HPMPApp to primer extension reactions containing all four dNTPs resulted in various degrees of HIV-1 RT inhibition, it was unclear from these experiments whether each drug could effectively replace its natural nucleotide counterpart. As such, we conducted an experiment that examined HIV-1 RT activity over time: dCTP or dATP was replaced by HPMPCpp or HPMPApp, respectively; the remaining three nucleotides were also added to the reaction mixtures (Fig. 4). Two sets of controls were also used: dCTP or dATP in place of HPMPCpp or HPMPApp, respectively, to show full-length DNA synthesis catalyzed by HIV-1 RT, and ddCTP or ddATP, respectively, to show the effects of obligate chain terminators on enzyme activity. In each case, the concentration of each nucleotide or nucleotide analog was 10 μM. In the reaction mixtures incubated for 20 min with dCTP or dATP in place of HPMPCpp or HPMPApp, respectively, production of full-length DNA molecules was observed (Fig. 4A to D, lanes 3 and 10). In contrast, when ddCTP or ddATP was used as a substrate in the reaction mixtures, there was no significant DNA extension past the site of incorporation of a single molecule of ddCMP or ddAMP, respectively (Fig. 4A to D, lanes 2 and 9). A reaction mixture containing P1-T9RNA and incubated with HPMPCpp over 20 min showed pause sites at positions primer + 1 and primer + 4 (Fig. 4A, lanes 4 to 7). These pause sites were relatively consistent over the 20-min time course and correspond to the pause sites observed in reaction mixtures incubated with all four dNTPs at HPMPCpp concentrations ≥ 10 μM (Fig. 3A). Similarly, a time course reaction experiment examining DNA synthesis in the presence of HPMPCpp and using DNA template T9 showed the same pause sites as previously observed (primer + 1, primer + 2, and primer + 4) (Fig. 4A, lanes 11 to 14). Over time, however, the pause sites become less prominent, suggesting that they are extended into full-length molecules. Figure 4B shows the results obtained when templates directing the incorporation of two consecutive molecules of HPMPC were used. Several pause sites, including primer + 1, primer + 2, primer + 4, and primer + 5 (Fig. 4B, lanes 4 to 7), were observed when T2RNA was used as the template molecule. Full-length DNA molecules were produced in these reactions, and the amount increased with time, indicating that, although the incorporation of two consecutive HPMPC molecules slows DNA extension, it does not terminate it. Very greatly pronounced inhibition was seen when the T2 DNA template was used in place of T2RNA. From these data, it appears that HIV-1 RT has great difficulty extending a DNA primer terminating in two HPMPC molecules. With longer incubation times (10 and 20 min), faint bands corresponding to primer + 4 and primer + 5, as well as full-length molecules, were also seen, suggesting that the incorporation of two HPMPC molecules is not an absolute block to continued DNA synthesis (Fig. 4B, lanes 13 and 14).
Fig. 4.
HIV-1 RT can use HPMPCpp or HPMPApp to support DNA synthesis. (A and B) Primer extension reactions examining the ability of HIV-1 RT to replace dCTP with HPMPCpp. FAM-labeled primer P1 was annealed to template T9RNA or T9 (A) or to template T2RNA or T2 (B) and incubated at 37°C with 50 nM HIV-1 RT in the presence of 10 μM (each) dATP, dGTP, and dTTP. Reactions were further supplemented with 10 μM ddCTP (lanes 2 and 9), 10 μM dCTP (lanes 3 and 10), or 10 μM HPMPCpp (lanes 4 to 7 and 11 to 14). ddCTP- and dCTP-containing reaction mixtures were incubated for 20 min; HPMPCpp-containing reaction mixtures were incubated over 20 min with sampling at 1, 5, 10, and 20 min. (C and D) Primer extension reactions examining the ability of HIV-1 RT to replace dATP with HPMPApp. Reaction mixtures were prepared by annealing FAM-labeled primer P1 to template T11RNA or T11 (C) or to template T12RNA or T12 (D) and incubated at 37°C with 50 nM HIV-1 RT in the presence of 10 μM (each) dCTP, dGTP, and dTTP. ddATP at 10 μM (lanes 2 and 9), dATP at 10 μM (lanes 3 and 10), or HPMPApp at 10 μM (lanes 4 to 7 and 11 to 14) was also added as indicated. ddATP- and dATP-containing reaction mixtures were incubated for 20 min; HPMPApp-containing reaction mixtures were incubated over 20 min, with sampling at 1, 5, 10, and 20 min. Lanes marked “Primer” show labeled primer P1. The locations of pause sites are indicated by arrows.
Products of reaction mixtures containing the P1-T11RNA primer-template pair showed that HPMPApp could substitute for dATP in promoting full-length DNA synthesis (Fig. 4C). Pausing at positions primer + 1 and primer + 4 was also observed in these reactions, indicating that some inhibition of DNA synthesis was taking place. The amount of primer + 1 product was relatively constant throughout the 20-min time course, but the amount of primer + 4 product increased with time (Fig. 4C, lanes 4 to 7). In the reaction using primer-template pair P1-T11, the pause sites at positions primer + 1 and primer + 4 were also observed (Fig. 4C, lanes 11 to 14). In contrast to the results obtained with template T11RNA, however, the amounts of both of these products decreased with time, suggesting that they had extended into full-length molecules. A third pause site, at the primer + 15 location, was also observed in products of that reaction mixture. In contrast to the results seen with the other two pause site products, the amount of that product increased with time. The primer + 1 and primer + 4 pause sites observed with the T11RNA- and T11-containing reaction mixtures were also seen with the reaction mixtures incubated with HPMPApp in the presence of all four dNTPs (Fig. 3A and B). The primer + 15 pause site was not previously observed with the T11-containing reaction mixture. This result was most likely due to the fact that this template causes HIV-1 RT to pause at several sites in the absence of drug (Fig. 3C, lane 9), and yet the enzyme can fully extend these DNA molecules with longer incubation times (Fig. 4C, lane 10). With the longer incubation time (20 min), this drug-dependent primer + 15 pause becomes apparent.
The use of template T12RNA in reactions designed to examine whether HPMPApp can replace dATP showed that, although the incorporation of two consecutive molecules of HPMPA into a DNA primer slows DNA synthesis, further chain elongation is not terminated (Fig. 4D). Pausing at positions primer + 1, primer + 2, and primer + 5 was observed in these reaction products; with increased incubation times, however, these molecules appeared to be extended into full-length molecules (Fig. 4D, lanes 4 to 7). Interestingly, and in contrast to the results obtained with HPMPCpp, the incorporation of two consecutive molecules of HPMPA into DNA opposite a DNA template did not cause a significant inhibition to continued synthesis around the site of drug incorporation (Fig. 4D, lanes 11 to 14). Products of pause sites at positions primer + 1, primer + 2, and primer + 5 were observed at earlier time points in the reaction, but they were nearly negligible at the longest time point of 20 min. In contrast, the amount of a pause site product corresponding to primer + 15 increased with time.
HIV-RT is inhibited by HPMPC and HPMPA in the template strand.
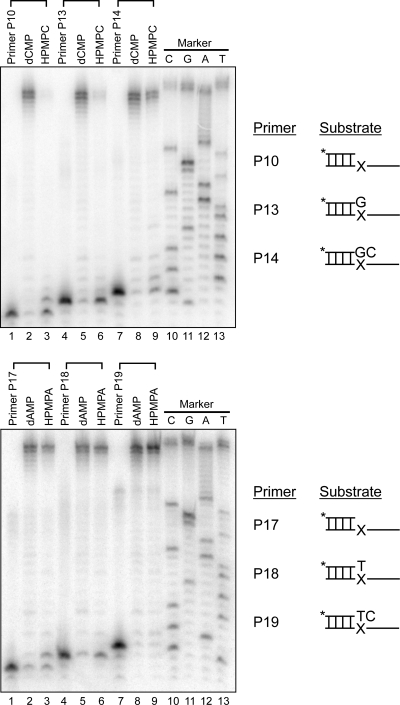
Since HIV-1 RT can use HPMPCpp and HPMPApp to replace dCTP and dATP, respectively, to promote chain elongation, we next examined the ability of the enzyme to copy DNA templates containing each of these drugs. The drug-containing templates were prepared as described previously (23) and annealed to 32P-labeled primers designed to terminate at a position one nucleotide prior to the drug lesion (P10 and P17), at the drug lesion (P13 and P18), or at a position one nucleotide past the drug lesion (P14 and P19) (Fig. 2) (23). The results of these experiments are shown in Fig. 5. In each of the control reaction lanes representing the presence of dCMP or dAMP in place of HPMPC and HPMPA, respectively, HIV-1 RT was able to extend each primer out to the end of the template. There was an array of these fully extended products seen in each of these reaction mixtures; the nature of these products is unknown but may be a result of the biotin tag located at the 5′ end of the template strand. The presence of HPMPC or HPMPA in the template strand resulted in the inhibition of DNA synthesis across the drug lesions. This inhibition was more pronounced with the HPMPC-containing templates than with the HPMPA-containing templates. For the HPMPC-containing template, the use of primers P10 and P13 resulted in faint smearing located at the position of full-length extension products. The strongest products found in each of these reactions, however, were those produced by the primer and primer + 1 positions (P10) and by P13 (Fig. 5, top panel, lanes 3 and 6). In contrast, when primer P14 was annealed to this template, distinct full-length extension products were seen along with two pause sites located at positions corresponding to the primer and primer + 1 (Fig. 5, top panel, lane 9). The presence of HPMPA in the template strand resulted in even less of a block to DNA synthesis. In all three reactions catalyzed by HIV-1 RT, full-length extension products were observed (Fig. 5, bottom panel, lanes 3, 6, and 9). Although pause sites were observed at positions corresponding to primer + 1 and the primer for reactions containing P17 and P18, respectively (and relative to the control reactions), there were no pause sites seen when primer P19 was used. These results suggest that, once the newly synthesized DNA is positioned immediately past the HPMPA drug lesion, HIV-1 RT can continue synthesis unimpeded.
Fig. 5.
Effects of templates bearing HPMPC and HPMPA on DNA synthesis catalyzed by HIV-1 RT. Templates containing dCMP or HPMPC (top panel) or containing dAMP or HPMPA (bottom panel) were prepared and annealed to the indicated 32P-labeled primers. These primers terminate one nucleotide prior to (P10 and P17), at (P13 and P18), or one nucleotide after (P14 and P19) the site of drug incorporation (marked with an “X”). The primer-template pairs were incubated with 200 μM dNTPs and 50 nM HIV-1 RT for 5 min at 37°C. The reaction products were recovered with magnetic beads and analyzed by phosphorimager analysis after separation on a 10% polyacrylamide gel. Lanes marked “Primer” show the position of each primer on the gel, and dideoxy sequencing reaction size markers are shown in lanes 10 to 13 of each gel.
DISCUSSION
Antiviral activity.
HPMPC and HPMPA are broad-spectrum antivirals active against dsDNA viruses and are particularly active in the form of their hexadecyloxypropyl (HDP) or octadecyloxyethyl (ODE) esters (16). One of these compounds, HDP-HPMPC (CMX001), is currently in phase II clinical trials for treatment of dsDNA virus infections. HDP-HPMPA and ODE-HPMPA were recently shown to be inhibitors of HIV-1 replication at low nanomolar concentrations (17). In this report, we show that the ODE esters of HPMPC, HPMPG, and HPMPDAP at low or subnanomolar concentrations are also inhibitors of HIV-1 replication in MT-2 cells. In contrast, ODE-HPMPT was devoid of activity against HIV-1; the latter results are consistent with those of De Clercq et al. (11), who found that HPMPT showed only weak activity against dsDNA viruses. Interestingly, replacing the 3-hydroxyl of ODE-HPMPA with a 3-methoxy group to yield ODE-MPMPA preserves the low nanomolar antiviral activity against HIV-1 but nearly eliminates the antiviral activity against dsDNA viruses characteristic of ODE-HPMPA (32). In PBMCs infected with NRTI-resistant HIV, some moderate (up to 5-fold) base-dependent increases in resistance of some mutant viruses were noted, but the appropriate plasma levels of these compounds are expected to be sufficient to allow successful treatment based on the human plasma drug levels noted in clinical trials performed with HDP-HPMPC (CMX001) and HDP-tenofovir (CMX157).
Mechanism of action.
Primer extension analysis using purified HIV-1 RT showed that HPMPCpp and HPMPApp can replace the natural nucleotides dCTP and dATP, respectively. The incorporation of either compound into the primer strand, however, slows elongation with pausing at specific positions, including the site of drug incorporation and a position three nucleotides away from the site of drug incorporation (Fig. 4). Interestingly, these two pause sites are also observed when entecavir, a 2′-deoxyguanosine analog, is incorporated into DNA by HIV-1 RT (12, 30). There were also some compound-specific pause sites observed in these reactions. For example, the incorporation of two consecutive molecules of HPMPC opposite a DNA template caused a profound block to further elongation (Fig. 4B). In contrast, two consecutive HPMPA incorporations did not result in this profound block; full-length extension was observed in these reactions (Fig. 4D). HPMPA incorporation did result in its own specific pause site, however, at a position 15 nucleotides away from the original primer terminus (Fig. 4C and D). The structural basis for this pausing remains to be determined.
Although the incorporation of HPMPC or HPMPA into the DNA primer strand slows subsequent elongation, absolute chain termination does not occur. It is unclear from these studies, however, how efficiently HIV-1 RT utilizes HPMPCpp and HPMPApp in vivo compared with dCTP and dATP. Cherrington et al. (7) calculated a Ki value of 0.83 μM for HPMPCpp (relative to the Km value of 0.14 μM for dCTP) against HIV-1 RT, and Frangeul et al. (13) used pre-steady-state kinetic analyses to estimate that the Kd (dissociation constant) for HPMPCpp was 23-fold higher than the Kd for dCTP (180 μM versus 7.9 μM, respectively). These observations suggest that these drugs are generally inefficient substrates for HIV-1 RT, with larger Km values relative to those seen with the natural nucleotides. This would explain why the alkoxyalkyl ester prodrugs of HPMPC and HPMPA can inhibit HIV-1 in a T-lymphocyte cell culture (Table 1) whereas HPMPC and HPMPA cannot (4, 17, 27, 29). The alkoxyalkyl ester prodrugs are more effectively taken up by cells and produce higher concentrations of the intracellular active metabolites (1, 23). They can thus produce the levels of HPMPCpp and HPMPApp that are required for use by the HIV-1 RT.
Because incorporation of HPMPC or HPMPA does not cause absolute chain termination and can support continued DNA synthesis, we also examined the effects of incorporation of each compound into the template strand. We observed that HIV-1 RT was inhibited by both drugs in the template strand. When templates containing HPMPC were used, the inhibition was stronger than the inhibition seen with those containing HPMPA. Strong pausing around the drug lesions was observed, although some production of full-length reaction products was still seen in all of the reaction mixtures. Why the RT cannot efficiently transit across a DNA template bearing a drug molecule is unclear, although we have recently described the structure of a duplex DNA containing HPMPC that does exhibit some minor local distortion of the phosphodiester backbone and altered base orientation at the site of the drug (20).
These results lead us to suggest that HPMPC and HPMPA inhibit HIV-1 RT by a complex mechanism of action similar to what we have previously observed with vaccinia virus DNA polymerase (23, 24). HPMPC and HPMPA can be incorporated into DNA, causing a slowing, but not a termination, of DNA synthesis. These compounds can also inhibit DNA replication when incorporated into the template strand. A general observation is that pauses in the chain extension profile (Fig. 3, 4, and 5) tend to occur where an incorporated phosphonate moiety is predicted to form (or break) nonspecific contacts with lysine side chains in the palm of the reverse transcriptase. This is consistent with the hypothesis that, by perturbing the structure of the phosphodiester backbone, these drugs could interfere with DNA translocation through the enzyme's primer-template binding grove. However, more extensively detailed structural and kinetic studies are required to explore this “stripped thread” model for enzyme inhibition (20). This complex mechanism of action may also have implications for the development of resistance to these compounds. It would be of interest to determine the types of resistance mutations that develop in vitro after prolonged passage of HIV-1 in the presence of either drug.
Since it has been reported that HIV-cytomegalovirus (CMV) coinfection may lead to negative outcomes even after successful antiretroviral therapy (8, 18, 25), it is tempting to speculate that the inclusion of a potent inhibitor of HIV-1 replication that also possesses broad-spectrum activity against dsDNA viruses (including CMV) could be very beneficial. For example, ODE-HPMPC and ODE-HPMPA have EC50s against human CMV of 1 to 3 nM (5, 6) and EC50s against HIV-1 of 0.2 to 0.4 nM (Table 1). If one could use either one of those (or similar) agents at a dose effective for treatment of human CMV infection, it appears that an additional benefit of treatment of HIV infection might be achieved. It is important, however, to investigate the impact of such a drug regimen on the development of resistant virus.
ACKNOWLEDGMENTS
We thank M. Götte for the gift of HIV-1 reverse transcriptase and C. Irwin and J. Lin for the preparation of vaccinia virus DNA polymerase used to synthesize the HPMPC- and HPMPA-containing templates and Shahrzad Rostami for technical assistance.
This study was supported in part by operating grant MOP-10923 from the Canadian Institutes of Health Research (D.H.E.), by NIH grants AI071803 and AI074057 (K.Y.H.), and by UCSD (University of California, San Diego) Center for AIDS Research grants AI36214 and AI074621 (D.D.R.).
Footnotes
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Aldern K. A., Ciesla S. L., Winegarden K. L., Hostetler K. Y. 2003. Increased antiviral activity of 1-O-hexadecyloxypropyl-[2-14C]cidofovir in MRC-5 human lung fibroblasts is explained by unique cellular uptake and metabolism. Mol. Pharmacol. 63:678–681 [DOI] [PubMed] [Google Scholar]
- 2. Andrei G., et al. 2006. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 80:9391–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker R. O., Bray M., Huggins J. W. 2003. Potential antiviral therapeutics for smallpox, monkeypox and other orthopoxvirus infections. Antiviral Res. 57:13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balzarini J., et al. 1993. Differential antiherpesvirus and antiretrovirus effects of the (S) and (R) enantiomers of acyclic nucleoside phosphonates: potent and selective in vitro and in vivo antiretrovirus activities of (R)-9-(2-phosphonomethoxypropyl)-2,6-diaminopurine. Antimicrob. Agents Chemother. 37:332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beadle J. R., et al. 2002. Alkoxyalkyl esters of cidofovir and cyclic cidofovir exhibit multiple-log enhancement of antiviral activity against cytomegalovirus and herpesvirus replication in vitro. Antimicrob. Agents Chemother. 46:2381–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beadle J. R., et al. 2006. Synthesis and antiviral evaluation of alkoxyalkyl derivatives of 9-(S)-(3-hydroxy-2-phosphonomethoxypropyl)adenine against cytomegalovirus and orthopoxviruses. J. Med. Chem. 49:2010–2015 [DOI] [PubMed] [Google Scholar]
- 7. Cherrington J. M., et al. 1996. Comparative kinetic analyses of interaction of inhibitors with Rauscher murine leukemia virus and human immunodeficiency virus reverse transcriptases. Antimicrob. Agents Chemother. 40:1270–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deayton J. R., et al. 2004. Importance of cytomegalovirus viraemia in risk of disease progression and death in HIV-infected patients receiving highly active antiretroviral therapy. Lancet 363:2116–2121 [DOI] [PubMed] [Google Scholar]
- 9. De Clercq E., Holý A. 2005. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat. Rev. Drug Discov. 4:928–940 [DOI] [PubMed] [Google Scholar]
- 10. De Clercq E., et al. 1986. A novel selective broad-spectrum anti-DNA virus agent. Nature 323:464–467 [DOI] [PubMed] [Google Scholar]
- 11. De Clercq E., et al. 1987. Antiviral activity of phosphonylmethoxyalkyl derivatives of purine and pyrimidines. Antiviral Res. 8:261–272 [DOI] [PubMed] [Google Scholar]
- 12. Domaoal R. A., et al. 2008. Pre-steady-state kinetic studies establish entecavir 5′-triphosphate as a substrate for HIV-1 reverse transcriptase. J. Biol. Chem. 283:5452–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frangeul A., et al. 2008. Gln151 of HIV-1 reverse transcriptase acts as a steric gate towards clinically relevant acyclic phosphonate nucleotide analogues. Antivir. Ther. 13:115–124 [PubMed] [Google Scholar]
- 14. Gilbert D. N., Moellering R. C., Jr., Eliopoulos G. M., Chambers H. F., Saag M. S. 2010. The Sanford guide to antimicrobial therapy, 40th ed. Antimicrobial Therapy, Inc., Sperryville, VA [Google Scholar]
- 15. Hammond J. L., et al. 2001. Alkylglycerol prodrugs of phosphonoformate are potent in vitro inhibitors of nucleoside-resistant human immunodeficiency virus type 1 and select for resistance mutations that suppress zidovudine resistance. Antimicrob. Agents Chemother. 45:1621–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hostetler K. Y. 2009. Alkoxyalkyl prodrugs of acyclic nucleoside phosphonates enhance oral antiviral activity and reduce toxicity: current state of the art. Antiviral Res. 82:A84–A98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hostetler K. Y., Aldern K. A., Wan W. B., Ciesla S. L., Beadle J. R. 2006. Alkoxyalkyl esters of (S)-9-[3-hydroxy-2-(phosphonomethoxy)propyl]adenine are potent inhibitors of the replication of wild-type and drug-resistant human immunodeficiency virus type 1 in vitro. Antimicrob. Agents Chemother. 50:2857–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jabs D. A., et al. 2005. Risk factors for mortality in patients with AIDS in the era of highly active antiretroviral therapy. Ophthalmology 112:771–779 [DOI] [PubMed] [Google Scholar]
- 19. Japour A. J., et al. 1993. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 37:1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Julien O., et al. 2011. Solution structure of a DNA duplex containing the potent anti-poxvirus agent cidofovir. J. Am. Chem. Soc. 133:2264–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kornbluth R. S., et al. 2006. Mutations in the E9L polymerase gene of cidofovir-resistant vaccinia virus strain WR are associated with the drug resistance phenotype. Antimicrob. Agents Chemother. 50:4038–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lanier R., et al. 2010. Development of CMX001 for the treatment of poxvirus infections. Viruses 2:2740–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Magee W. C., Aldern K. A., Hostetler K. Y., Evans D. H. 2008. Cidofovir and (S)-9-[3-hydroxy-(2-phosphonomethoxy)propyl]adenine are highly effective inhibitors of vaccinia virus DNA polymerase when incorporated into the template strand. Antimicrob. Agents Chemother. 52:586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magee W. C., Hostetler K. Y., Evans D. H. 2005. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob. Agents Chemother. 49:3153–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naeger D. M., et al. 2010. Cytomegalovirus-specific T cells persist at very high levels during long-term antiretroviral treatment of HIV disease. PLoS One 5:e8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Painter G. R., et al. 2007. Evaluation of hexadecyloxypropyl-9-R-[2-(phosphonomethoxy)propyl]-adenine, CMX157, as a potential treatment for human immunodeficiency virus type 1 and hepatitis B virus infections. Antimicrob. Agents Chemother. 51:3505–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pauwels R., et al. 1988. Phosphonomethoxyethyl purine derivatives, a new class of anti-human immunodeficiency virus agents. Antimicrob. Agents Chemother. 32:1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Srinivas R. V., Connely M., Fridland A. 1997. (S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine (HPMPC) inhibits HIV-1 replication in epithelial cells, but not T-lymphocytes. Antiviral Res. 35:23–27 [DOI] [PubMed] [Google Scholar]
- 30. Tchesnokov E. P., Obikhod A., Schinazi R. F., Götte M. 2008. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 283:34218–34228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Valiaeva N., et al. 2009. Antiviral evaluation of octadecyloxyethyl esters of (S)-3-hydroxy-2-(phosphonomethoxy)propyl nucleosides against herpesviruses and orthopoxviruses. Antiviral Res. 84:254–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valiaeva N., et al. Synthesis and antiviral evaluation of 9-(S)-[3-alkoxy-2-(phosphonomethoxy)propyl] nucleoside alkoxyalkyl esters: inhibitors of hepatitis C virus and HIV-1 replication. Bioorg. Med. Chem. 19:4616–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong J. L., Fuchs D. S. 1970. Competitive isomerization and dealkylation of 2,4-dialkoxypyrimidines in aqueous and nonaqueous media. J. Org. Chem. 35:3786–3791 [Google Scholar]
- 34. Xiong X., Smith J. L., Chen M. S. 1997. Effect of incorporation of cidofovir into DNA by human cytomegalovirus DNA polymerase on DNA elongation. Antimicrob. Agents Chemother. 41:594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]