Abstract
We show that 3-[4-(4-methoxyphenyl)piperazin-1-yl]piperidin-4-yl biphenyl-4-carboxylate (C10), screened out of a chemical library, selectively kills bacterial persisters that tolerate antibiotic treatment but does not affect normal antibiotic-sensitive cells. C10 led persisters to antibiotic-induced cell death by causing reversion of persisters to antibiotic-sensitive cells. This work is the first demonstration in which the eradication of bacterial persisters is based on single-chemical supplementation. The chemical should be versatile in elucidating the mechanism of persistence.
TEXT
Bacterial persistence is a phenomenon in which a subpopulation of dormant cells, or persisters, tolerates antibiotic treatment. Persistence is different from resistance in that the cells do not change or acquire inheritable genetic elements but are genetically the same as the cells affected by antibiotics. Persistence has been observed in most microbial species and in relation to different classes of antibiotics in vivo and in vitro. Bacterial persisters have been implicated in biofilms and in chronic and recurrent infections (1, 3–6, 8, 10–14). Despite this clinical relevance, there are no viable means for eradicating persisters. More importantly, low-dose antibiotics are known to enhance the probability of resistant-cell emergence (7) and the possibility of resistant-cell emergence is directly proportional to the number of persisters (9). Thus, a strategy to remove these persisters is a critical outstanding issue both for prevention of secondary infection and for prevention of resistant-cell emergence. The importance of persister eradication was highlighted in a recent study in which specific metabolic stimuli enabled the killing of persisters with aminoglycosides (2). In the present study, we show that persisters can be selectively killed by a single chemical compound without affecting the normal major bacterial population.
First, we confirmed that conventional antibiotics were effective only for the normal antibiotic-sensitive population but were not effective in killing the small fraction of persisters, which was the motivation of this study (see Fig. S1 in the supplemental material). Next, we screened for killers of persisters in a chemical library composed of 6,800 chemicals that were selected from ∼200,000 chemicals (Korea Chemical Bank, Daejeon, South Korea) based on chemical scaffolds and physicochemical properties. In the primary screening (see Fig. S2a in the supplemental material), we selected chemicals that killed all cells in combination with ampicillin (Amp). Each chemical (25 μM) was mixed together with Amp (100 μg/ml) and Escherichia coli K-12 BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) (∼108 CFU/ml) in each well of 96-well microplates. After 9 h of incubation at 37°C in Luria-Bertani (LB) medium, a 10-μl aliquot from each well was transferred onto an LB agar plate using a 48-pin replica stamp. Because the persistence frequency (number of CFU after antibiotic treatment/number of initial CFU) was ∼10−3 when cells were cultured in a 96-well microplate due to a low oxygen transfer rate (6, 10, 11), ∼1,000 transferred persisters gave rise to a large spot on the LB plate after overnight incubation at 37°C. While most chemicals did not affect spot formation, 52 chemicals did not allow formation of such large spots (see Fig. S2a in the supplemental material). In order to exclude chemicals that affected normal cell growth, we performed a secondary screening by adding only chemicals in one column of wells and both Amp and chemicals in the next column of wells (see Fig. S2b in the supplemental material). Eight chemicals did not show apparent bactericidal activity when used alone but eradicated all cells only when used together with Amp (see the coupled circles in Fig. S2b in the supplemental material). Finally, bacterial cells were cotreated with the selected 8 chemicals and 5 μg/ml norfloxacin (Nor), which has a different action mechanism from Amp. Persistence frequency was evaluated after 9 h of incubation at 37°C. Some chemicals still effectively killed the persister cells by cooperating with Nor, while other chemicals affected persistence frequency less (see Fig. S2c in the supplemental material). Three chemicals that lowered the persistence frequency by more than 10-fold compared to that in the control treated with Nor alone were finally selected.
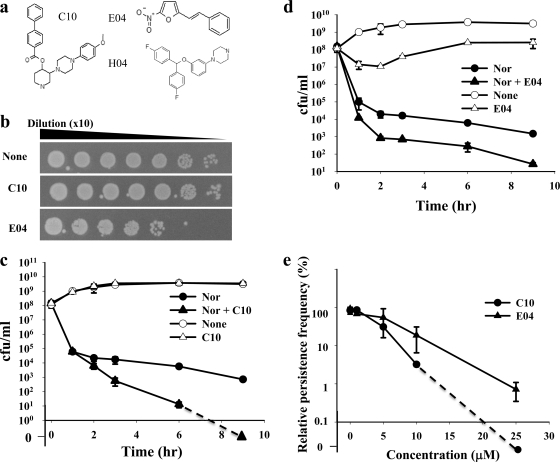
The chemical structures of the 3 selected chemicals are shown in Fig. 1a. In order to further assess the bactericidal activity of each chemical on normal cells, a serially diluted E. coli culture was treated with a 25 μM concentration of each chemical for 9 h and transferred onto an LB plate. (E)-2-Nitro-5-styrylfuran (E04) exhibited weak bactericidal effects by itself, but 3-[4-(4-methoxyphenyl)piperazin-1-yl]piperidin-4-yl biphenyl-4-carboxylate (C10) did not show any apparent inhibitory effect on normal cells (Fig. 1b). For detailed observation, we evaluated the effects of the 3 chemicals on persistence frequency as a function of time during batch culture. C10 (Fig. 1c) and 1-{3-[bis(4-fluorophenyl)methoxy]phenyl}piperazine (H04) (see Fig. S3 in the supplemental material) did not inhibit cell growth when they were used alone. C10 did not inhibit cell growth even at a 250 μM concentration (see Table S1 in the supplemental material). In contrast, E04 inhibited cell growth even without antibiotics (Fig. 1d). Cotreatment with C10 and Nor left no surviving colonies, even though C10 did not affect normal cell growth by itself (Fig. 1c). The efficacy of H04 was much lower than that of C10. Both C10 and E04 reduced persistence frequency in a concentration-dependent manner (Fig. 1e).
Fig. 1.
Effects of selected inhibitors on persister formation. (a) Chemical structures of C10, E04, and H04. (b) Effects of C10 and E04 on normal cell growth on an LB plate. (c and d) Effects of C10 (c) and E04 (d) on normal cell growth and persister killing during batch culture. (e) Concentration-dependent effects of C10 and E04 on persister formation.
Antibiotic-induced cell death of bacteria is known to exhibit two distinct phases (3). In the initial phase, normal antibiotic-sensitive cells are killed at a high rate. Then a phase of slow cell death follows, during which the death of persisters occurs. To quantitatively describe the effects of drugs on persister death, the decimal reduction time (D) was calculated. The D value is the time required to reduce CFU by a factor of 10 and is the inverse slope when the y axis is on the common logarithmic scale. In the presence of Nor alone, the D value was 0.31 h during the first phase (1 h) while the D value for persisters was 5.8 h during the second phase (2 to ∼9 h). The combination of C10 with Nor resulted in the same 0.31-h D value for the initial time period but accelerated cell death during the second phase (2 to ∼9 h), with a D value of 1.4 h (0 CFU was treated as 10−1). In contrast to C10, E04 accelerated the death rate in the first phase, with a D value of 0.26 h, but did not enhance the persister death rate. H04 showed killing profiles similar to C10, but the efficacy was lower than that of C10 (see Fig. S3 in the supplemental material). In conclusion, C10 effectively and selectively kills the persisters that tolerate antibiotic exposure but does not affect normal antibiotic-sensitive cells.
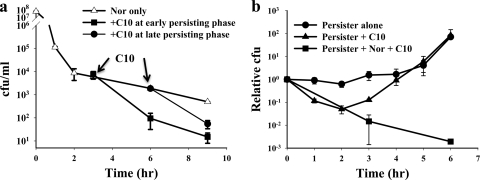
There remains the question of how C10 selectively kills only persister cells. To address this question, C10 was added at various time points during Nor-induced cell death and its killing kinetics was observed. C10 dramatically accelerated persister death regardless of the addition time (Fig. 2a), which could not be achieved by adding different classes of antibiotics (see Fig. S1 in the supplemental material). The killing of already-formed persisters by C10 suggests that C10 does not simply inhibit the conversion of antibiotic-sensitive cells to persisters but actively leads to persister cell death.
Fig. 2.
Killing persisters with C10. (a) C10 accelerated the persister cell death rate even when preformed persisters were treated. Experiments were performed in the presence of 5 μg/ml Nor and 25 μM C10. (b) C10 caused reversion of persisters to antibiotic-sensitive cells. While persisters started exponential growth after a 5-h lag period, C10 induced immediate regrowth of dormant persisters. The revertant cells were antibiotic sensitive, and hence a certain level of cell killing occurred even without the addition of external antibiotics because already-internalized antibiotics affect cells for a short period of time (see Fig. S4c in the supplemental material). Persisters were prepared by treatment with 5 μg/ml Nor for 9 h, and a trace amount of external antibiotic was thoroughly washed out before treatment with C10.
C10 may directly kill persisters by inhibiting an unknown, unique metabolic process of persisters or may accelerate reversion of persisters to antibiotic-sensitive cells. To address this question, a sample of ∼2,500 CFU/ml persisters was harvested after Nor treatment for 9 h and inoculated into fresh LB medium, and the regrowth of persisters was compared with that of normal cells by measuring optical density (see Fig. S4a in the supplemental material). Persisters exhibited a long lag period before starting exponential growth, while normal cells immediately started regrowth after inoculation (lag period, ∼0 h). This result indicated that reversion of persisters to normal antibiotic-sensitive cells is very slow, which is consistent with previous results (3). Next, we tested whether C10 alone leads persisters to death by measuring CFU. We reasoned that CFU measurement would more sensitively reflect reversion or death of persisters than optical density because there were >1,000 times more dead cells than live cells in our persister preparation. Consistent with optical density measurement, persisters started exponential growth after an ∼5-h lag period (Fig. 2b). While C10 effectively led to persister cell death in the presence of Nor, persisters started fast regrowth in the presence of C10 after a short period of cell death (Fig. 2b). This result clearly indicates that C10 caused reversion of dormant persisters to fast-replicating normal cells. These regrown cells could be eradicated when Nor was added at the end of culture (data not shown), indicating that the regrown cells were antibiotic sensitive. We exclude the possibility that C10 kills persisters by itself in spite of the apparent short cell death period. Internalized residual Nor may kill revertant cells even when C10 is applied to persisters after thorough washing. In support of this notion, the same treatment of persisters which overexpressed the multidrug efflux transporter MdtK did not cause a short cell death period because MdtK actively ejects internalized antibiotics (see Fig. S4b in the supplemental material). In conclusion, C10 leads persisters to death in combination with antibiotics by effecting reversion of persisters to normal antibiotic-sensitive cells.
In the present study, we showed that persisters can be selectively eradicated by a single compound, C10. C10 was also effective in killing persisters which were enriched during stationary growth phase (see Fig. S5a in the supplemental material). C10 dramatically reduced persistence frequency with fluoroquinolone antibiotics for both E. coli and Pseudomonas aeruginosa (see Fig. S5b in the supplemental material). While C10 did not affect normal cell growth by itself, it led persisters to death by causing reversion of persisters to antibiotic-sensitive cells. Thus, C10 seems to be a wake-up pill for persisters that ultimately returns dormant persisters to antibiotic-sensitive cells. Finding the binding target of C10 should provide pinpointing information on the mechanism of persistence. Moreover, the strategy demonstrated here, which exploits single-chemical supplementation, might be applied to prevent the advent of resistant cells as well as provide a therapy for infectious diseases.
Supplementary Material
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (through grants 2011-0006268 and 2009-0058612) and by iPET (Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries), Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Al-Dhaheri R. S., Douglas L. J. 2008. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob. Agents Chemother. 52:1884–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allison K. R., Brynildsen M. P., Collins J. J. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 4. Bigger J. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497 [Google Scholar]
- 5. Hansen S., Lewis K., Vulic M. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52:2718–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jayaraman R. 2008. Bacterial persistence: some new insights into an old phenomenon. J. Biosci. 33:795–805 [DOI] [PubMed] [Google Scholar]
- 7. Kohanski M. A., DePristo M. A., Collins J. J. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levin B. R. 2004. Noninherited resistance to antibiotics. Science 305:1578–1579 [DOI] [PubMed] [Google Scholar]
- 9. Levin B. R., Rozen D. E. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4:556–562 [DOI] [PubMed] [Google Scholar]
- 10. Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107–131 [DOI] [PubMed] [Google Scholar]
- 11. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 12. Li Y., Zhang Y. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schumacher M. A., et al. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah D., et al. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.