Abstract
Molecular markers for surveillance of Plasmodium falciparum resistance to current antimalarials are sorely needed. A 28-day efficacy study of artemether-lumefantrine in eastern Sudan identified 5 treatment failures among 100 evaluable patients; 9 further individuals were parasite positive by PCR during follow-up. Polymorphisms in pfatpase6 and pfmdr1 were evaluated by DNA sequencing. One individual carried parasites with a novel pfmdr1 polymorphism (F1044L). pfmdr1 gene amplification in parasites prior to treatment occurred in three individuals who had recurrent infection during follow-up.
TEXT
Artemisinin combination therapies (ACT) are strongly recommended for treating uncomplicated falciparum malaria (33). The most widely adopted of these combinations in sub-Saharan Africa is artemether-lumefantrine (AL) (32). The recent emergence of resistance to artemisinin on the Thai-Cambodian border (23, 5) establishes an urgent need for validated molecular markers of resistance to ACT in general and to AL in particular.
Evidence from in vitro and in vivo studies has suggested that polymorphisms in the pfatpase6 locus, encoding Plasmodium falciparum SERCA (8, 16, 17, 28, 30), and in the pfmdr1 locus, encoding the parasite multidrug resistance transporter Pgh-1 (6, 7, 24, 25), may modulate plasmodium sensitivity to artemisinins (4, 12, 14, 26). Studies in Thailand show that copy number variation (CNV) of the pfmdr1 locus is associated with reduced in vivo and in vitro sensitivity to both mefloquine and AL (24, 25, 34). CNV has not been linked to treatment outcomes with AL in Africa (4, 14), although an increased pfmdr1 copy number has been observed in isolates from Kenya and Gabon (13, 29) and confirmed in a few case studies of travelers returning from west Africa, mostly after mefloquine use (11, 35).
In Sudan, a 6-dose course of AL is currently recommended as a second-line treatment for uncomplicated falciparum malaria (18) and has reported in vivo efficacy of over 90% (9, 20, 22). In the present study, we analyzed the sequence of pfatpase6 and pfmdr1 alleles and tested for CNV at pfmdr1 in parasites before and after AL treatment for uncomplicated falciparum malaria in eastern Sudan.
Patients were recruited from among those presenting with fever or history of fever to clinics in the villages Asar, Daraweesh, and Abu Adam near Gedaref town and in a refugee camp in New Halfa, an irrigated area 150 km from Gedaref. Inclusion criteria were a positive smear for P. falciparum monoinfection, a parasite count of 1,000 to 100,000 asexual parasites/μl (Gedaref) or between 2,000 and 200,000 asexual parasites/μl (New Halfa), an axillary temperature of ≥37.5°C, and weight of >10 kg (Gedaref) or >5 kg (New Halfa). Pregnant women and patients with other underlying disease or with signs of severe malaria (36) were excluded. Clinical assessment was performed on recruitment (day 0 [D0]) and days 1, 2, 3, 7, 14, 21, and 28. Participants were treated with six doses of AL (20-mg artemether-120-mg lumefantrine tablets; Novartis). For adults, 4 tablets twice daily were administered; doses were adjusted according to weight for children under 35 kg. The first dose each day was observed; the second dose was self-administered. Patients were advised to eat fatty food or milk before each dose. Thick blood films stained with Giemsa stain were prepared and examined by microscopists on each day, and a blood spot was collected on glass fiber membranes for DNA analysis.
DNA extraction utilized a modified Chelex method (3). Amplification of both genes was attempted for all pretreatment (D0) samples. Amplification of pfmdr1 fragment 1 (6) was attempted for all posttreatment samples from day 14 and later. PCR-positive posttreatment samples were further analyzed for other pfmdr1 regions and the pfatpase6 gene, using previously described methods for nested PCR methods and DNA sequencing (14, 19). Gene copy numbers were analyzed by a duplex dual-labeled probe quantitative PCR (qPCR) assay (24); two independent experiments were performed, and in each experiment, each isolate was tested in triplicate. Control DNA from P. falciparum lines 3D7 and HB3 (1 copy of pfmdr1 each) and Dd2 (2 copies of pfmdr1) were run in parallel in each experiment. A sample was considered evaluable if it produced a duplex fluorescent signal in at least two replicates in each of the two experiments. Recrudescence was distinguished from reinfection by pfmsp1 and pfmsp2 genotyping (27).
All participants received an information sheet in English and Arabic and provided signed written informed consent. Ethical approval was obtained from the Tropical Medicine Research Institute Ethics Review Committee and the London School of Hygiene and Tropical Medicine Ethics Committee. The study was registered as a clinical trial (identification [ID] number NCT00440752).
One hundred six patients were recruited, and 100 (94.3%) completed the follow-up. Mean ages of recruits were similar in the two sites, being 16.7 years (95% confidence interval [CI], 13.0 to 20.4) in Gedaref and 20.9 years (95% CI 15.8 to 26.0) in New Halfa (P = 0.19; two-tailed Student's t test). By per-protocol analysis, 95 patients (95%) displayed adequate clinical and parasitological responses, with 5 individuals failing treatment. The PCR-corrected estimate of therapeutic efficacy was 98%, as three recurrent infections displayed different msp1 and msp2 genotypes from the original infection. Eleven individuals were positive by pfmdr1 PCR on day 14; three of these remained positive, and a further one and two individuals became positive on days 21 and 28, respectively, making a total of 14 recurrent infections identified by PCR; all five individuals identified as treatment failures by microscopy were PCR positive.
Codons 225 to 423 and 470 to 906 of the pfatpase6 gene (52% of the locus) were successfully sequenced in 78 and 87 pretreatment isolates, respectively. 569K occurred in 49.4% of isolates. The synonymous polymorphism T to A at nucleotide 2694 was detected in 59.3% of these isolates. A novel nonsynonymous change (D845N) was detected in 2 isolates. Other previously reported polymorphisms were also observed (Table 1). Three D14 isolates, one D21 isolate, and two D28 isolates were successfully sequenced, and pfatpase6 alleles were compared with those present in the same individual prior to treatment (Table 2). Considerable diversity in this locus is confirmed in this seasonal low-transmission setting, but the six evaluable recurrent infections were not associated with particular pfatpase genotypes.
Table 1.
Diversity of pfatpase6 at recruitmenta
| Study site | No. of isolates with amino acidb |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L402V |
N569K |
G639D |
E643Q |
I723V |
I735M |
D845N |
||||||||
| L | V | N | K | G | D | E | Q | I | V | I | M | D | N | |
| New Halfa | 35 | 4 | 22 | 20 | 41 | 1 | 41 | 1 | 42 | 0 | 39 | 3 | 40 | 2 |
| Gedaref | 35 | 4 | 22 | 23 | 44 | 1 | 45 | 0 | 42 | 3 | 42 | 1 | 39 | 0 |
| Total | 70 | 8 | 44 | 43 | 85 | 2 | 86 | 1 | 84 | 3 | 81 | 4 | 79 | 2 |
Amplification primers were ATP6_FOR1 (5′-AATAAAACTCCCGCTGATGC-3) and ATP6_REV1 (5′-ATCCTTCTTCTCCATCATCC-3′) (nest 1) and ATP6_FOR1 and ATP6_REV2 5′-CGTTAAAGCTTCAACATTTCC-3′ (nest 2) (800 bp).
Changed amino acids are in bold. L, leucine; V, valine; N, asparagine; K, lysine; G, glycine; D, aspartic acid; E, glutamic acid; Q, glutamine; I, isoleucine; M, methionine. The E431K polymorphism was not investigated in this study.
Table 2.
Comparison of pfatpase6 in pretreatment and posttreatment isolates
| Samplea | Sequenceb |
Treatment outcomec | |||
|---|---|---|---|---|---|
| Day 0 | Day 14 | Day 21 | Day 28 | ||
| 9 | LKGEIIDA | —NGEIIDA** | LPF day 21 | ||
| 19 | LNGEIIDA | —NGEIMDT | LCF day 28 | ||
| 56* | —KGEIIDT | —KGEIID(T/A)** | ACPR | ||
| 59 | —NGEIIDT | LNGEIIDT | LPF day 14 | ||
| 74* | VKGEIIDT | —KGEIIDT | —KGEIIDA | ACPR | |
*, PCR positive on follow-up, not a clinical failure.
Amino acid mutations seen were L402V, N569K, G639D, E643Q, I723V, I735M, and D845N, and the nucleotide mutation seen was T2694A. (Changed amino acids are in bold.) T, threonine; A, alanine; **, mixed at position T2964A; —, DNA not available.
ACPR, adequate clinical and parasitological response; LPF, late parasitological failure; LCF, late clinical failure.
A novel polymorphism, corresponding to F1044L, was observed in pfmdr1 in a single multiclonal pretreatment isolate; parasites with a wild-type sequence at this codon were also present. All isolates, pre- and posttreatment, encoded the wild-type amino acids Ser and Asn at codons 1034 and 1042, respectively, in pfmdr1. Haplotypes of pfmdr1 were constructed at codons 86, 184, and 1246 for all isolates with complete data; four isolates with unambiguous mixtures of two haplotypes were each counted as harboring both. The haplotype YFD was the most common prior to treatment, occurring in 67.4% of the 89 evaluable samples with unambiguous haplotypes, followed by YYD (15.7%), NYD (7.9%), NFD (5.6%), YYY (4.5%), YFY (2.2%), and NFY (1.1%). In contrast, NFD was relatively more abundant in posttreatment isolates (Table 3), being found alone or mixed in 5 of the 14 posttreatment isolates (odds ratio, 9.33; 95% CI, 1.72 to 48.2; Fisher's exact P, 0.004).
Table 3.
Longitudinal analysis of pfmdr1 haplotypes in 14 patients with recurrent parasitemiaa
| Site | Patient no. | Day 0 |
Day 14 haplotype | Day 28 (or day 21c) haplotype | Clinical outcome | |
|---|---|---|---|---|---|---|
| Haplotype | CNV statusb | |||||
| New Halfa | 9 | YFD | wt | NFD* | LPF | |
| 19 | YYD | wt | NFD | LCF | ||
| Gedaref | 56 | YFD | wt | NYD | ACPR | |
| 59 | YFD | 2 copies | NFD | LPF | ||
| 61 | YFD | wt | YFD | ACPR | ||
| 62 | YFD | wt | NFD | LPF | ||
| 63 | YFY | 2 copies | NFD | ACPR | ||
| 64 | NYD | wt | NYY | LCF | ||
| 65 | YFD | wt | NYD | ACPR | ||
| 68 | YYD | wt | YFD | ACPR | ||
| 73 | YFD | wt | YFD | ACPR | ||
| 74 | YFD | wt | YFD | ACPR | ||
| 75 | YFD | 2 copies | NYD | ACPR | ||
| 82 | YFD | — | NYD | ACPR | ||
Individuals shown include those with submicroscopic PCR-positive parasitemia.
wt, wild-type with respect to pfmdr1 CNV; —, amplification assay unsuccessful.
*, day 21 haplotype.
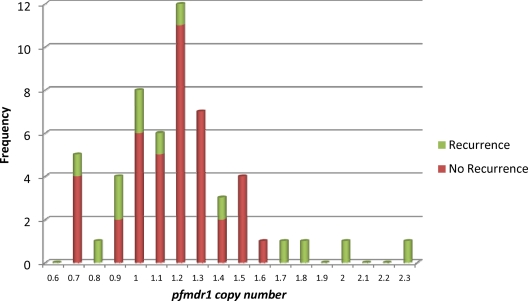
Seventy-four pretreatment isolates and 14 posttreatment isolates were investigated for evidence of pfmdr1 amplification. Copy number estimates of 1.03, 0.81, and 2.19 were obtained for 3D7, HB3, and Dd2, respectively. Reliable estimates of pfmdr1 copy numbers from at least two independent experiments were obtained for 57 pretreatment isolates and 2 posttreatment isolates. The average copy number in pretreatment isolates was 1.24 (range, 0.73 to 2.33; 95% CI, 1.16 to 1.32); pfmdr1 copy number estimates above 1.8 in at least two independent experiments were obtained for three pretreatment isolates (Fig. 1). Each of these individuals also had recurrent parasitemia during follow-up; one case was microscopy positive on day 14 and typed by pfmsp2 genotyping as a recrudescence, whereas the remaining two cases were classified as having an adequate clinical and parasitological response (ACPR) but positive by PCR on day 14. There was thus an association between carriage of parasites with amplified pfmdr1 copy numbers, prior to treatment, and recurrent parasitemia after AL (Fisher's exact test, P = 0.011). Two posttreatment isolates were successfully tested, and both of these carried one copy of the pfmdr1 gene. Interestingly, pfmdr1-amplified isolates in this study carried the 86Y allele instead of the N86 seen in Southeast Asia (Table 3). This observation is consistent with other African reports (13, 29; Maja Malmberg [Malaria Research Lab, Department of Medicine, Karolinska Institutet, Stockholm, Sweden], personal communication).
Fig. 1.
Frequency distribution of pfmdr1 copy number estimates. Estimates of pfmdr1 locus copy numbers obtained from 55 pretreatment isolates with complete follow-up data are grouped in bins of 0.1 copy units. The values shown represent the means of at least two independent experiments; each DNA sample in each experiment was run in duplicate. Red data represent pretreatment parasite isolates from patients without subsequent recurrent parasitemia. Green data represent pretreatment parasite isolates from patients with later recurrent parasitemia by microscopy and/or PCR. Isolates with copy number estimates of 1.8 and above were considered true duplications.
The NFD allele is suggested to appear in reinfecting rather than recrudescing parasites following AL treatment (4, 12, 14, 26). Consistent with this view, two of the clinical failures in this study were classified as new infections by msp1 and msp2 genotyping. These were late failures from New Halfa, where the irrigation canals provide a stable mosquito habitat. In contrast, some earlier cases of parasite recurrence were identified by PCR in Gedaref, where the probability of reinfection is low. The observation of a new substitution at the Phe1044Leu codon, as well as the amplification of pfmdr1, suggests that diversification of this locus may be occurring. However, as the number of observations in our study was small, these preliminary findings need to be confirmed in larger studies.
AL proved efficacious for treating uncomplicated malaria caused by P. falciparum in this study across two sites in eastern Sudan. We identified subpatent parasitemia in 10 patients with ACPR but cannot rule out the possibility that gametocytes of P. falciparum were the origin of this DNA in at least some of these individuals. Selection by AL for genotypes at the pfatpase6 locus was not observed, but we found evidence of selection by AL for the pfmdr1 haplotype NFD in recurrent parasitemia as early as D14 after treatment. We present the first evidence from an African efficacy study that amplification of the pfmdr1 locus may contribute to recurrent P. falciparum parasitemia following AL therapy. This association needs to be confirmed in larger studies, particularly as we lack locus amplification data for posttreatment isolates. There is published evidence that injectable artemether monotherapy has been in use by medical practitioners in northern Sudan (1, 10, 21), and this may have led to selection of parasites carrying this gene amplification (15, 31); no such amplification was observed among 24 isolates collected in 1989 (2). Continual surveillance of pfmdr1 and other loci implicated in antimalarial treatment response is justified as large-scale use of ACT continues in sub-Saharan Africa.
Acknowledgments
N.G. was supported by an IAEA training fellowship (reference SUD/6/025) and a WHO/TDR research training grant. C.J.S. is supported by the United Kingdom Health Protection Agency. Resistance marker studies were funded by the MALACTRES consortium (EC FP7). I.A. acknowledges the financial support of the University of Khartoum.
We appreciate the provision of drugs by the National Malaria Control Program in Sudan and the continuous cooperation of the study participants. We are also thankful to the field teams in both Gedaref and New Halfa. We thank Rachel Hallett for helpful discussions and assistance with sequencing protocols.
Footnotes
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Adam I., Elwasila E., Mohammed Ali A. D., Elansari E., Elbashir M. I. 2004. Artemether in the treatment of falciparum malaria during pregnancy in eastern Sudan. Trans. R. Soc. Trop. Med. Hyg. 98:509–513 [DOI] [PubMed] [Google Scholar]
- 2. Babiker H. A., Creasey A. M., Bayoumi R. A., Walliker D., Arnot D. E. 1991. Genetic diversity of Plasmodium falciparum in a village in eastern Sudan. 2. Drug resistance, molecular karyotypes and the mdr1 genotype of recent isolates. Trans. R. Soc. Trop. Med. Hyg. 85:578–583 [DOI] [PubMed] [Google Scholar]
- 3. Dlamini S. V., Beshir K., Sutherland C. J. 2010. Markers of anti-malarial drug resistance in Plasmodium falciparum isolates from Swaziland: identification of pfmdr1-86F in natural parasite isolates. Malar. J. 9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dokomajilar C., Nsobya S. L., Greenhouse B., Rosenthal P. J., Dorsey G. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 50:1893–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dondorp A. M., et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duraisingh M. T., et al. 2000. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 108:13–23 [DOI] [PubMed] [Google Scholar]
- 7. Duraisingh M. T., Roper C., Walliker D., Warhurst D. C. 2000. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 36:955–961 [DOI] [PubMed] [Google Scholar]
- 8. Eckstein-Ludwig U., et al. 2003. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424:957–961 [DOI] [PubMed] [Google Scholar]
- 9. Elamin S. B., et al. 2010. Descriptive study on the efficacy of artemether-lumefantrine in the treatment of uncomplicated Plasmodium falciparum malaria in Sudan. Eur. J. Clin. Pharmacol. 66:231–237 [DOI] [PubMed] [Google Scholar]
- 10. Elmardi K. A., et al. 2011. Self-reported fever, treatment actions and malaria infection prevalence in the northern states of Sudan. Malar. J. 10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gay F., et al. 1990. Mefloquine failure in child contracting falciparum malaria in West Africa. Lancet 335:120–121 [DOI] [PubMed] [Google Scholar]
- 12. Hastings I. M., Ward S. A. 2005. Coartem (artemether-lumefantrine) in Africa: the beginning of the end? J. Infect. Dis. 192:1303–1305 [DOI] [PubMed] [Google Scholar]
- 13. Holmgren G., Bjorkman A., Gil J. P. 2006. Amodiaquine resistance is not related to rare findings of pfmdr1 gene amplifications in Kenya. Trop. Med. Int. Health 11:1808–1812 [DOI] [PubMed] [Google Scholar]
- 14. Humphreys G. S., et al. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 51:991–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imwong M., et al. 2010. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob. Agents Chemother. 54:2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jambou R., et al. 2005. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet 366:1960–1963 [DOI] [PubMed] [Google Scholar]
- 17. Jambou R., et al. 2010. Geographic structuring of the Plasmodium falciparum sarco(endo)plasmic reticulum Ca2+ ATPase (PfSERCA) gene diversity. PLoS One 5:e9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malik E. M., Khalafalla O. M. 2004. Malaria in Sudan: past, present and the future. Gezira J. Health Sci. 1:47–53 [Google Scholar]
- 19. Menegon M., Sannella A. R., Majori G., Severini C. 2008. Detection of novel point mutations in the Plasmodium falciparum ATPase6 candidate gene for resistance to artemisinins. Parasitol. Int. 57:233–235 [DOI] [PubMed] [Google Scholar]
- 20. Mohamed A. O., Eltaib E. H., Ahmed O. A., Elamin S. B., Malik E. M. 2006. The efficacies of artesunate-sulfadoxine-pyrimethamine and artemether-lumefantrine in the treatment of uncomplicated, Plasmodium falciparum malaria, in an area of low transmission in central Sudan. Ann. Trop. Med. Parasitol. 100:5–10 [DOI] [PubMed] [Google Scholar]
- 21. Mohammed-Ali A. R., Kheir M. M., Adam I. 2006. Intramuscular artemether in the treatment of uncomplicated Plasmodium falciparum malaria in Sudanese patients. Saudi Med. J. 27:122–123 [PubMed] [Google Scholar]
- 22. Mukhtar E. A., et al. 2007. A comparative study on the efficacy of artesunate plus sulphadoxine/pyrimethamine versus artemether-lumefantrine in eastern Sudan. Malar. J. 6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noedl H., et al. 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359:2619–2620 [DOI] [PubMed] [Google Scholar]
- 24. Price R. N., et al. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Price R. N., et al. 2006. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria Clin. Infect. Dis. 42:1570–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sisowath C., et al. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J. Infect. Dis. 191:1014–1017 [DOI] [PubMed] [Google Scholar]
- 27. Snounou G., et al. 1999. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R. Soc. Trop. Med. Hyg. 93:369–374 [DOI] [PubMed] [Google Scholar]
- 28. Uhlemann A. C., et al. 2005. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat. Struct. Mol. Biol. 12:628–629 [DOI] [PubMed] [Google Scholar]
- 29. Uhlemann A. C., Ramharter M., Lell B., Kremsner P. G., Krishna S. 2005. Amplification of Plasmodium falciparum multidrug resistance gene 1 in isolates from Gabon. J. Infect. Dis. 192:1830–1835 [DOI] [PubMed] [Google Scholar]
- 30. Valderramos S. G., Scanfeld D., Uhlemann A. C., Fidock D. A., Krishna S. 2010. Investigations into the role of the Plasmodium falciparum SERCA (PfATP6) L263E mutation in artemisinin action and resistance. Antimicrob. Agents Chemother. 54:3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vinayak S., et al. 2010. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J. Infect. Dis. 201:1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. von Seidlein L., et al. 1998. A randomized controlled trial of artemether/benflumetol, a new antimalarial and pyrimethamine/sulfadoxine in the treatment of uncomplicated falciparum malaria in African children. Am. J. Trop. Med. Hyg. 58:638–644 [DOI] [PubMed] [Google Scholar]
- 33. White N. J. 1999. Delaying antimalarial drug resistance with combination chemotherapy. Parassitologia 41:301–308 [PubMed] [Google Scholar]
- 34. Wilson C. M., et al. 1993. Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol. Biochem. Parasitol. 57:151–160 [DOI] [PubMed] [Google Scholar]
- 35. Witkowski B., et al. 2010. pfmdr1 amplification associated with clinical resistance to mefloquine in West Africa: implications for efficacy of artemisinin combination therapies. J. Clin. Microbiol. 48:3797–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. World Health Organization 2000. Severe falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 94(Suppl. 1):S1–S90 [PubMed] [Google Scholar]