Abstract
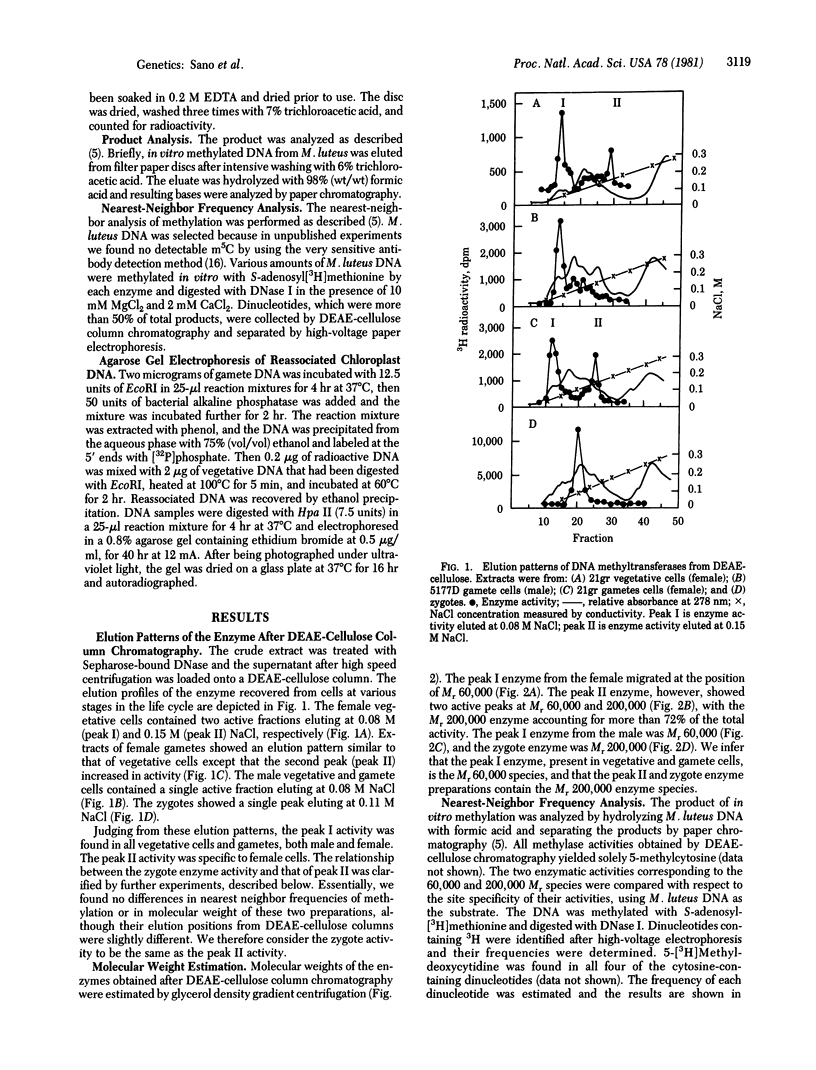
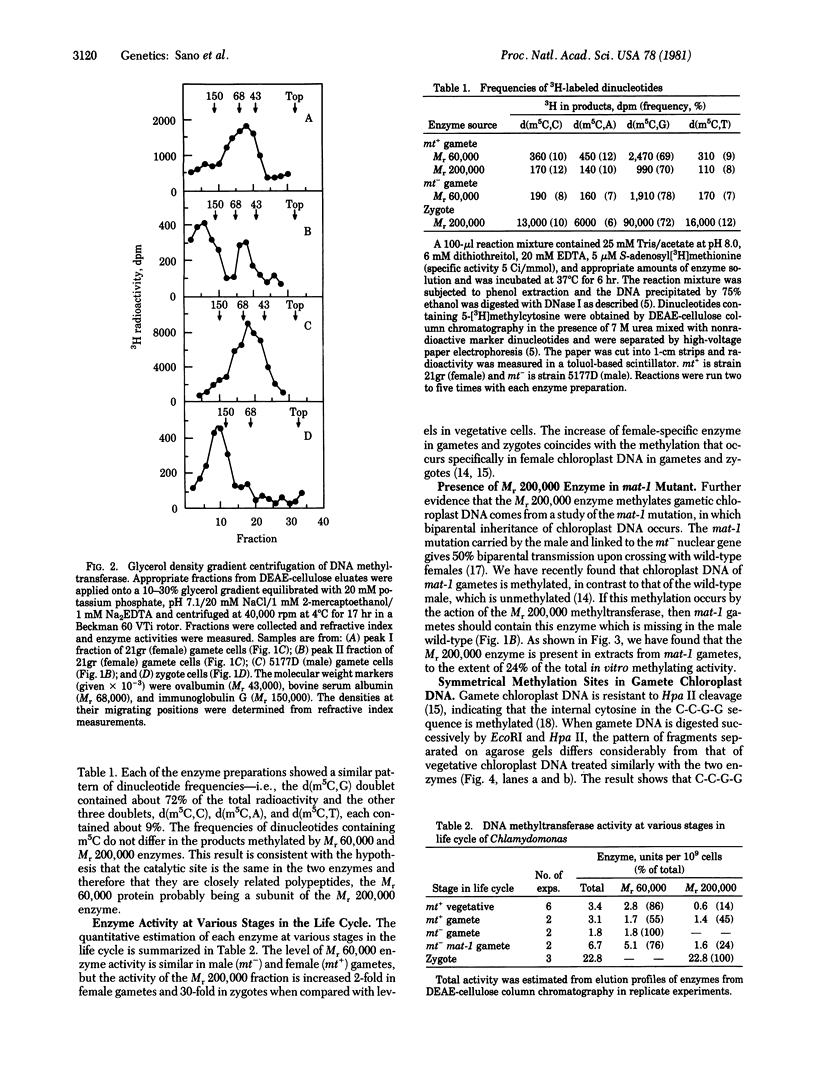
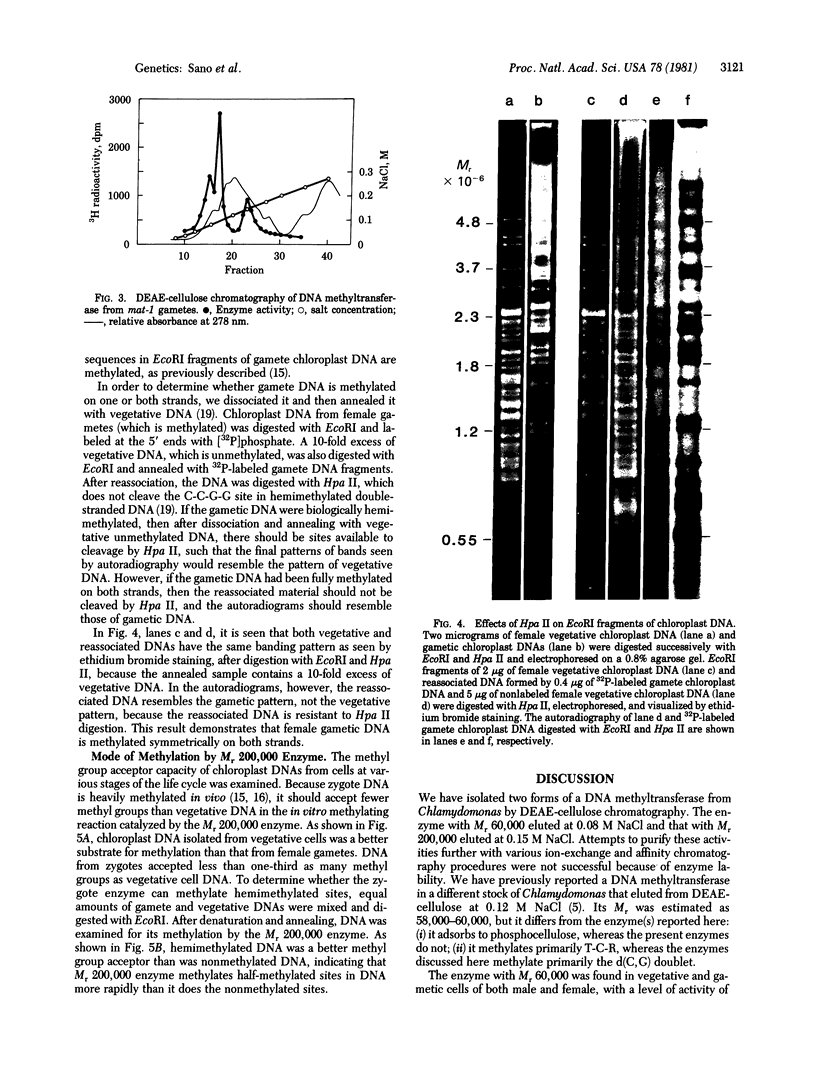
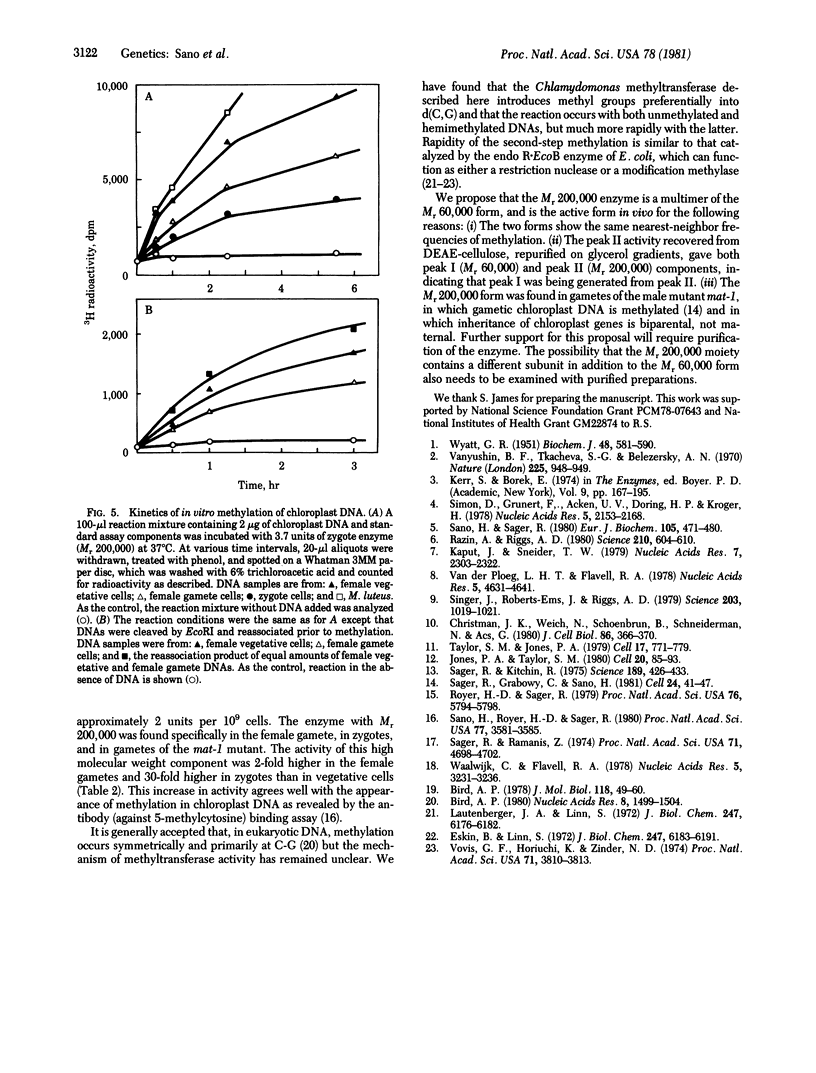
Two molecular weight forms of DNA (cytosine-5-)-methyltransferase [S-adenosyl-L-methionine:DNA (cytosine-5-)- methyltransferase, EC 2.1.1.37], both active in assays in vitro, were isolated from the green alga Chlamydomonas reinhardi at various stages of the life cycle. The enzyme with Mr 60,000 was found in vegetative cells and gametes of both male (mt-) and female (mt+) mating types. The enzyme with Mr 200,000 was specific to gametic cells and zygotes, which are the only stages at which methylation of chloroplast DNA occurs in vivo. Chloroplast DNA from gametes was shown to be methylated on both strands at most if not all methylation sites and the Mr 200,000 enzyme was shown to methylate both unmethylated and hemimethylated sites, the latter at an elevated rate. Micrococcus luteus DNA showed the same nearest-neighbor frequencies of methylation after methylation by each molecular weight component. The data suggest strongly that the Mr 200,000 enzyme is the active multimeric form of the Mr 60,000 enzyme and that it acts as both initiation and maintenance methylase. It is proposed that methylation of chloroplast DNA in female gametes and zygotes is regulated by assembly of the multimeric Mr 200,000 active enzyme, which in turm determines the maternal inheritance of chloroplast DNA.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Christman J. K., Weich N., Schoenbrun B., Schneiderman N., Acs G. Hypomethylation of DNA during differentiation of Friend erythroleukemia cells. J Cell Biol. 1980 Aug;86(2):366–370. doi: 10.1083/jcb.86.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin B., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. II. Purification, subunit structure, and catalytic properties of the restriction endonuclease. J Biol Chem. 1972 Oct 10;247(19):6183–6191. [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980 May;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. I. Purification, subunit structure, and catalytic properties of the modification methylase. J Biol Chem. 1972 Oct 10;247(19):6176–6182. [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Royer H. D., Sager R. Methylation of chloroplast DNAs in the life cycle of Chlamydomonas. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5794–5798. doi: 10.1073/pnas.76.11.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Kitchin R. Selective silencing of eukaryotic DNA. Science. 1975 Aug 8;189(4201):426–433. [PubMed] [Google Scholar]
- Sager R., Ramanis Z. Mutations that alter the transmission of chloroplast genes in Chlamydomonas. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4698–4702. doi: 10.1073/pnas.71.12.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Royer H. D., Sager R. Identification of 5-methylcytosine in DNA fragments immobilized on nitrocellulose paper. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3581–3585. doi: 10.1073/pnas.77.6.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Sager R. Deoxyribonucleic acid methyltransferase from the eukaryote, Chlamydomonas reinhardi. Eur J Biochem. 1980 Apr;105(3):471–480. doi: 10.1111/j.1432-1033.1980.tb04522.x. [DOI] [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Riggs A. D. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979 Mar 9;203(4384):1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979 Aug;17(4):771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Vovis G. F., Horiuchi K., Zinder N. D. Kinetics of methylation of DNA by a restriction endonuclease from Escherichia coli B. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3810–3813. doi: 10.1073/pnas.71.10.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WYATT G. R. The purine and pyrimidine composition of deoxypentose nucleic acids. Biochem J. 1951 May;48(5):584–590. doi: 10.1042/bj0480584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]



