Abstract
Indoleamine 2,3-dioxygenase (IDO) catalyzes the initial and rate-limiting step of tryptophan catabolism in a specific pathway, resulting in a series of extracellular messengers collectively known as kynurenines. IDO has been recognized as an authentic regulator of immunity not only in mammalian pregnancy, but also in infection, autoimmunity, inflammation, allergy, transplantation, and neoplasia. Its suppressive effects are mostly mediated by dendritic cells (DCs) and involve tryptophan deprivation and/or production of kynurenines, which act on IDO-negative DCs as well as CD4+ and CD8+ T cells. We have found that mouse IDO contains two tyrosine residues within two distinct putative immunoreceptor tyrosine-based inhibitory motifs, VPY115CEL and LLY253EGV. We have also found that Suppressor of Cytokine Signaling 3 (SOCS3)—known to interact with phosphotyrosine-containing peptides and be selectively induced by interleukin 6 (IL-6)—binds mouse IDO, recruits the ECS (Elongin-Cullin-SOCS) E3 ligase, and targets the IDO/SOCS3 complex for proteasomal degradation. This event underlies the ability of IL-6 to convert otherwise tolerogenic, IDO-competent DCs into immunogenic cells. Thus onset of immunity in response to antigen within an early inflammatory context demands that IDO be degraded in tolerogenic DCs. These studies support the finding that IDO is regulated by proteasomal degradation in response to immunogenic and inflammatory stimuli.
Keywords: IL-6, SOCS proteins, tryptophan catabolism, dendritic cells
Introduction
Suppressor of cytokine signaling (SOCS) proteins have emerged as critical modulators of cytokine-mediated processes.1 A series of studies with various experimental approaches have demonstrated that SOCS3 modulates interferon (IFN)-γ-inducible genes and is involved in the prevention of IFN-γ-like responses in hepatocytes and macrophages stimulated with interleukin (IL)-6.2–4 It has been suggested that, in the absence of SOCS3, IL-6 may become immunosuppressive, activating genes typically induced by IFNs.5 Not only does the feedback inhibitor SOCS3 attenuate IL-6 signaling,6 but also upregulation of SOCS3 by IL-6 is responsible for inhibiting the IFN-γ–driven transcriptional expression of IDO.7 In fact, although SOCS3 may be an important regulator of IDO in response to nitric oxide,8 an inducer of SOCS3,9 the underlying mechanisms could be broader than simply opposing IFN-γ signaling and the IFN-γ–like actions of IL-6.10 SOCS proteins are, in general, critical modulators of immune responses,11 and they possess an Src homology 2 (SH2) domain, which binds phosphotyrosine-containing peptides, and a SOCS box. The latter domain participates in the formation of an E3 ubiquitin ligase complex and targets several signaling proteins for proteasomal degradation.12–15 In murine dendritic cells (DCs), treatment with IL-6 up-regulates SOCS3 and prevents IDO activation by IFN-γ.7,16 We found an inverse correlation between SOCS3 and IDO expression.10,17,18 Not only does SOCS3 influence IL-6 transcriptional programs in DCs, but it can also contribute to posttranscriptional events that directly shape the presentation pattern of DC subsets programmed to direct tolerance. In this study we demonstrate that IL-6 exerts immunogenic effects on otherwise tolerogenic CD8+ DCs that are contingent on functional SOCS3 in vivo and impaired tryptophan catabolism in vitro.
Materials and Methods
Mice and reagents
Eight- to ten-week-old female DBA/2J (H-2d) mice were obtained from Charles River Breeding Laboratories. All in vivo studies were conducted in compliance with National (Italian Parliament DL 116/92) and Perugia University Animal Care and Use Committee guidelines. Murine rIL-6 was from Peprotech. Rabbit monoclonal anti-mouse IDO antibody (cv152) was obtained as described.19 Anti-SOCS3 and anti-Flag M2 antibodies were purchased from Abcam and Sigma-Aldrich, respectively. Anti-ubiquitin was from Cell Signaling. The P815AB tumor peptide (amino acid sequence LPYLGWLVF) was synthesized and purified as described.20,21 Biotinylated phosphorylated and unphosphorylated peptides for pull-down experiments spanning the ITIM-contained tyrosines of mouse IDO (ITIM1: SGSGNIAVPY115CELSE; pITIM1: SGSGNIAVP-pY115CELSE; ITIM2: SGSGPEGLLY253EGVWD; pITIM2: SGSGPEGLLpY253 EGVWD) were obtained from Sigma-Genosys. The IDO inhibitor 1-MT (d,l-isomers; Sigma-Aldrich) and the proteasome inhibitor MG132 (Calbiochem) were also used.
DC purification and treatments
Splenic DCs were purified using CD11c MicroBeads (Miltenyi Biotec) in the presence of EDTA to disrupt DC-T cell complexes.22 Cells were >99% CD11c+, >99% MHC I-A+, >98% B7-2+, <0.1% CD3+, and appeared to consist of 90%–95% CD8−, 5%–10% CD8+, and 1%–5% B220+ cells. DC populations were further fractionated according to CD8 expression in order to obtain purified CD8+ DCs by means of CD8α MicroBeads (Miltenyi Biotec). After cell fractionation, the CD8+ fraction was made up of >95% CD8+ DCs. Less than 1% CD8+ and <5% CD8− DCs expressed the B220 marker, respectively23 DCs were exposed to 20 ng/ml rIL-6 for 24 h at 37 °C. Also used, in specific experiments, were 1-MT (4 μM) and MG132 (10 μM).
RT-PCR analysis
Expression of SOCS3 (sense, 5′-CAGCCTGCGCCT CAAGACCTT-3′; antisense, 5′-GCACCAGCTTGA GTACACAGTCG-3′) transcripts were evaluated by RT-PCR analysis using specific primers. RT-PCR products were also normalized to murine Gapdh.
siRNA synthesis and transfection
These procedures have been described previously.10,24 Briefly, the siRNA sequences specific for murine SOCS3 (sense, 5′-GGAGCAAAAGGGUCAGAGGtt-3′; antisense, 5′-CCUCUGACCCUUUUGCUCCtt-3′) were selected, synthesized and annealed by the manufacturer (Ambion). For transfection, siRNAs (5 μg) in 30 μl of transfection buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) were pipetted into a sterile Eppendorf tube. In a separate polystyrene tube, 6.7 μg of 1,2 dioleoyl-3-trimethylammonium-propane (DOTAP) was mixed with 30 μl of transfection buffer and then both solutions were mixed gently by pipetting several times. After incubation at room temperature for 20 min, the mixture was added to 1 ml of complete medium containing 106 DCs and incubated for 24 h at 37 °C in the presence of rIL-6. Cells were then recovered, washed and immediately used for in vivo experiments. siRNA treatment resulted in the complete disappearance of SOCS3 transcripts at 24–48 h, as described.10,24 Control treatments consisted of cells treated with negative control siRNA (Ambion). After siRNA transfection, DC viability always exceeded 70%.
Construction and expression of mouse IDO-flag
Constructs expressing mouse IDO were generated amplifying the cDNA from purified DCs (Indo gene) with primers (Table 1) containing SpeI (sense) and NotI (antisense) restriction enzyme site sequences. The antisense primer also contained an N-terminal Flag-encoding sequence and a linker sequence coding for Gly3 to ensure flexibility of the resulting Flag-tagged protein. PCR products were digested with the appropriate restriction enzymes and cloned into a pEF-BOS plasmid. Since immunostimulatory sequences present in plasmid DNA (i.e. hypomethylated CpG motifs) may produce nonspecific effects, we transfected DCs by means of mRNAs. Briefly, plasmids obtained as above were linearized, purified using a Geneprep kit (Ambion), and used as templates for the in vitro transcription reaction using the mMES-SAGE mMACHINE T7 Ultra Kit (Ambion). Concentration and quality of in vitro-transcribed mRNAs were assessed by spectrophotometry and agarose gel electrophoresis. Control mRNA was obtained from the pTRI-Xef plasmid (supplied by manufacturer) containing the Xenopus elongation factor 1α gene, which codes for a 50.2 kDa protein. One million DCs were transfected with 2 μg mRNA, using DOTAP as described above for siRNA transfection.7
Table 1.
Primer sequences.
| Construct | Sense and antisense oligonucleotides |
|---|---|
| IDO | S, 5′-GGCCAAACTAGTGGTCAGTGGAGTAGACAGCA-3′ |
| AS, 5′-TCTAGAGTCGCGGCCGCTTACTTATCGTCATCGTCTTTGTAATCTCCTCCTCCAGGCCAACTCAGAAGAGCTT-3′ |
Immunization and skin test assay
For immunization in vivo, fractionated DCs, either as such or treated and/or transfected as described above, were loaded with the P815AB peptide in vitro (5 μM, 2 h at 37 °C), before intravenous injection into DBA/2J recipient hosts. Three × 105 peptide-loaded CD8+ DCs were injected. A skin test assay was used for measuring class I-restricted, delayed-type hypersensitivity responses to synthetic peptides as previously described.7,25 The response to intrafootpad challenge with the eliciting peptide was measured at 2 weeks, and results were expressed as the increase in footpad weight of peptide-injected footpads over that of respective, vehicle-injected counterparts.7 Data are the mean ± SD for at least six mice per group.
Kynurenine assay
IDO functional activity was measured in vitro in terms of the ability of DCs to metabolize tryptophan to kynurenine, the concentrations of which were measured by HPLC as described previously.19,26
Peptide pull-down experiments, Immunoprecipitation, and immunoblot analyses
Cells [2–5 × 105/sample for immunoblots of whole cell lysates; 3 × 106 DCs/sample for immunoprecipitation] were lysed on ice in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% Na-deoxycholate, 1 mM EDTA, 1.4 mM Na3VO4, and protease inhibitors). Lysates were either immunoprecipitated with the anti-Flag antibody or directly run on SDS-PAGE. For pull-down experiments, cell lysates were incubated sequentially with 10 μg biotinylated peptide (2 h) and streptavidin agarose beads (2 h). Immunoblots involved the use of specific antibodies in combination with the appropriate horseradish peroxidase conjugates, followed by ECL.
Statistical analysis
Student’s t-test was used to analyze the results of in vitro studies in which data are mean values (± SD). In the in vivo skin test assay, statistical analysis was performed using two-tailed paired t-test by comparing the mean weight of experimental footpads with that of control, saline injected counterparts.21 Data are mean values (±SD) of three experiments with at least six mice per group per experiment, as computed by power analysis to yield a power of at least 80% with an α level of 0.05.27
Results and Discussion
SOCS3 is required for the effect of IL-6 on CD8+ DCs
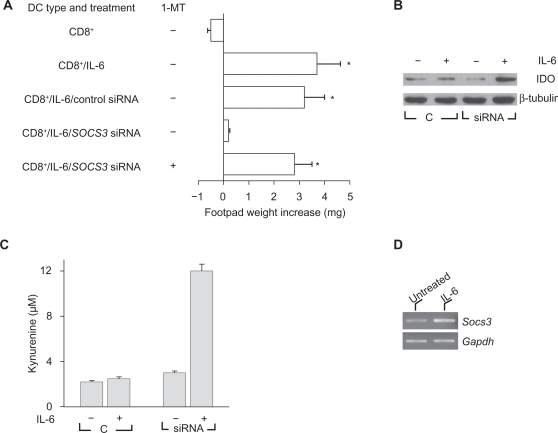
Mouse splenic DCs can present peptide antigens in an immunogenic or tolerogenic way, with the distinction depending on either the occurrence of specialized DC subsets or the maturation or activation state of the DC.28 Environmental factors are crucial in conditioning the outcome of DC presentation of the synthetic tumor/self nonapeptide P815AB,21,29 a poorly immunogenic antigen of mouse mastocytoma P1.HTR.30 DC populations in the spleens of DBA/2 mice consist of CD8− (∼90%) and CD8+ (∼10%) fractions that mediate the respective immunogenic and tolerogenic presentation of the synthetic nonapeptide P815AB.31 Upon transfer into recipient hosts, peptide-loaded CD8− DCs initiate immunity, and CD8+ DCs initiate anergy when Ag-specific skin test reactivity is measured at 2 wk after cell transfer.20,29 Consistent with previous results of adjuvant activity by IL-6,7 otherwise tolerogenic CD8+ DCs were made capable of immunogenic presentation of P815AB when exposed to IL-6 before peptide loading and transfer into recipient hosts (Fig. 1A). IL-6 is a multifunctional cytokine that regulates inflammatory responses, and overproduction of IL-6 is associated with autoimmunity and chronic inflammatory diseases.5 It has been shown that IL-6 strongly activates STAT3 and induces the expression of IFN-responsive genes in SOCS3-deficient macrophages, indicating that IL-6 might mimic the actions of IFNs.2,3 We examined whether SOCS3 expression is required for IL-6 effects on CD8+ DCs. Splenic DCs were fractionated, and specific SOCS3 gene silencing was achieved in the CD8+ fraction with siRNA technology. P815AB-pulsed CD8+ DCs were injected either untreated or treated with rIL-6, with or without concomitant siRNA-SOCS3 treatment (Fig. 1A). The results showed that SOCS3 was an absolute requirement for the occurrence of regulatory effects by rIL-6 on CD8+ DCs. Thus, in the absence of SOCS3, CD8+ DCs are refractory to immunomodulation by externally added IL-6, leaving their default tolerogenic program apparently unaffected. Also, the effect of IL-6 on SOCS3-deficient CD8+ cells was due to active suppression involving IDO because it could be reversed by the addition of the enzyme inhibitor 1-MT during cell exposure to the cytokine (Fig. 1A). On analyzing IDO expression by Western blot using rabbit monoclonal IDO-specific Ab, we found increased detection of the enzyme protein after IL-6 treatment in cells lacking SOCS3 (Fig. 1B). This was reflected by enhanced conversion of tryptophan to kynurenine (Fig. 1C). Therefore, IL-6 strongly activates tryptophan catabolism in DCs with silenced expression of SOCS3. It is also of interest that IL-6 may increase SOCS3 transcriptional expression (Fig. 1D).
Figure 1.
IL-6 becomes immunosuppressive in DCs lacking SOCS3, and modulation of SOCS3 by gene silencing enhances IDO expression, and tryptophan catabolism in response to IL-6. A) The effects of IL-6 on CD8+ DCs require SOCS3. P815AB-pulsed CD8+ DCs were transferred into recipient mice to be assayed for skin test reactivity at 2 wk. The CD8+ DC fraction was used as such or after treatment with rIL-6, with or without concomitant SOCS3 gene silencing by siRNA. Control cells consisted of CD8+ treated with DOTAP alone. The effect of IL-6 on SOCS3-deficient cells was also studied in the presence of 1-MT treatment. *, P < 0.001, experimental vs control footpads. B) CD8+ DCs, either with control treatment C) or subjected to siRNA treatment, were exposed to IL-6 and assayed for expression of IDO protein by Western blot analyses. C) IL-6 was studied for the ability to induce tryptophan conversion to kynurenine in CD8+ DCs with or without concomitant Socs3 gene silencing by siRNA. Kynurenine levels in supernatants were measured by HPLC, and the results are the mean ± SD of triplicate samples in one of three experiments. Control treatment (c) consisted of DOTAP alone. D) RT-PCR analysis of SOCS3 expression in CD8+ DCs treated with IL-6 for 2 h. Results are from one experiment representative of four. PCR bands were quantified by scanning densitometry and represented as fold change in IL-6 treated (2.5 ± 0.7, P = 0.045) relative to untreated control (in which fold change is 1).
Proteasome–mediated degradation of IDO
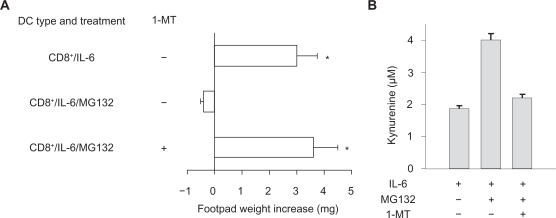
The mechanisms of action of SOCS proteins include SOCS box targeting of bound proteins to ubiquitin-proteasome–mediated degradation. The proteasome is a major protein-degrading enzyme, which catalyzes degradation of oxidized and aged proteins, signal transduction factors and cleaves peptides for antigen presentation. As mentioned above, 1-MT is a specific and widely used inhibitor of IDO activity,32 and MG132 is a specific proteasome inhibitor. We examined the inverse relationship between SOCS3 and IDO functions by using the two inhibitors in combination. In a skin test assay with P815AB, CD8+ DCs rendered immunogenic by IL-6 (Fig. 2A) would revert their phenotype when co-treated with MG132, yet the addition of 1-MT restored immunogenicity. Studies of IDO function in vitro with CD8+ DCs treated with IL-6 confirmed that MG132 activated the metabolic conversion of tryptophan to kynurenine—the initial IDO-dependent catabolite—and it did so in a 1-MT–sensitive manner (Fig. 2B). Therefore, an inverse relationship appeared to occur in DCs between functional IDO and SOCS3-proteasome–mediated effects.
Figure 2.
The proteasome inhibitor MG132 confers IDO-dependent, immunosuppressive properties on IL-6 in CD8+ DCs. CD8+ DCs were conditioned by overnight incubation with IL-6. The proteasome inhibitor, MG132, was added at 10 μM for 1 h before addition of the stimuli. The IDO inhibitor, 1-MT, was added to selective cultures at 4 μM. A) Conditioned CD8+ DCs were pulsed with the P815AB peptide and injected into recipients hosts that were assayed for the development of P815AB-specific skin test reactivity at 2 weeks after cell transfer. Data are mean (± SD). *, P < 0.001, experimental vs control footpads. B) IDO activity was evaluated in terms of kynurenine production in culture supernatants. *, P < 0.005. In both A and B, results are mean values (± SD) of three experiments.
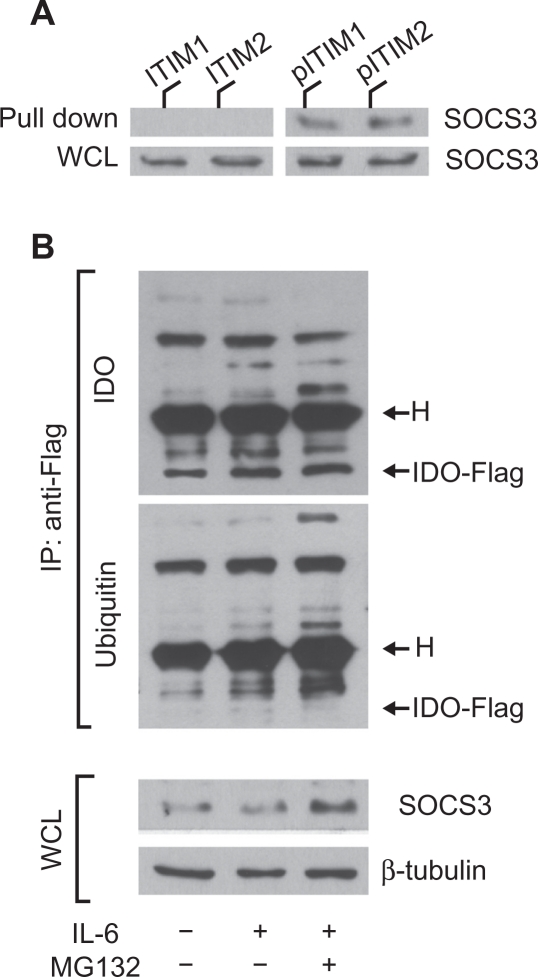
SOCS3 binds IDO via specific phosphotyrosine binding and mediates ubiquitination and proteasomal degradation of IDO
SOCS3-associated SH2 domains bind protein sequences shared by inhibitory receptors,12,33 i.e. ITIMs. A prototypic ITIM is the I/V/L/SxYxxL/V sequence,34 where x denotes any amino acid. As previously recognized by our studies, IDO contains two tyrosines within two distinct canonical ITIMs (ITIM1, VPY115CEL; ITIM2, LLY253EGV).18 The occurrence of ITIM domains in mouse IDO raised the possibility that the enzyme undergoes ubiquitin-proteasome–mediated degradation after tyrosine phosphorylation and SOCS3 binding via SH2 domains with high affinity for ITIM phosphotyrosine. To verify whether the putative ITIMs in IDO could represent docking sites for SOCS3, biotinylated peptides with phosphorylated or unphosphorylated mouse IDO ITIM1 or ITIM2 sequences were used in a pull-down assay of SOCS3. Lysates from IL-6–treated DCs were treated with unphosphorylated or phosphorylated IDO peptides and immunoblotted with anti-SOCS3 (Fig. 3A). We found that SOCS3 is associated with the phosphorylated forms of IDO peptides. We investigated whether the SOCS3-dependent immunoadjuvant activity of IL-6 in DCs is associated with posttranslational modification and proteasomal degradation of IDO. Unfractionated DCs were transfected with IDO-Flag mRNA and treated with IL-6 in the presence or absence of MG132. Cells were lysed and IDO-Flag was immunoprecipitated with anti-Flag. Sequential immunoblotting was conducted using anti-ubiquitin and an IDO-specific monoclonal antibody. Aliquots of whole cell lysates from parallel samples were blotted with SOCS3- and β-tubulin-specific antibodies (Fig. 3B). Treatment of IDO-Flag–transfected in DCs with IL-6 caused the appearance of several ubiquitinated proteins (as detected by specific anti-ubiquitin antibody), immunoprecipitated by anti-Flag antibody, which were greater in size than the IDO-Flag protein (>42 kDa). Interestingly, the bands corresponding to ubiquitinated proteins became more intense in DCs stimulated with IL-6 in the presence of MG132. Moreover, SOCS3 expression also increased in cells treated with the combination of IL-6 and MG132, thus suggesting that, in accordance with previous results,12,33 the SOCS3 protein itself is degraded by the proteasome concomitantly with its target protein.
Figure 3.
IDO contains ITIM sequences that are necessary for SOCS3-mediated ubiquitination and proteasomal degradation. A) IL-6 treated DCs were pulled down with unphosphorylated (ITIM1 and ITIM2) or phosphorylated (pITIM1 and pITIM2) IDO peptides and immunoblotted with anti–SOCS3 antibodies, which were also used in parallel Western blot analyses of WCL (whole cell lysates). B) Unfractionated DCs were transfected with IDO-Flag mRNA and treated or not with IL-6, in the presence or absence of MG132. Cells were lysed and IDO-Flag was immunoprecipitated (IP) with anti–Flag. Sequential immunoblotting was conducted using anti–IDO and –ubiquitin antibodies. One-tenth aliquots of WCL from parallel samples were used in parallel Western blot analyses. β-tubulin expression was evaluated as a loading control. H, heavy chain of the anti–Flag antibody (55 kDa). One experiment of three.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors report no conflicts of interest.
References
- 1.O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–87. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croker BA, Krebs DL, Zhang JG, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4:540–45. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 3.Lang R, Pauleau AL, Parganas E, et al. SOCS3 regulates the plasticity of gp130 signaling. Nat Immunol. 2003;4:546–50. doi: 10.1038/ni932. [DOI] [PubMed] [Google Scholar]
- 4.Yasukawa H, Ohishi M, Mori H, et al. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol. 2003;4:551–6. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 5.Johnston JA, O’Shea JJ. Matching SOCS with function. Nat Immunol. 2003;4:507–9. doi: 10.1038/ni0603-507. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orabona C, Grohmann U, Belladonna ML, et al. CD28 induces immunostimulatory signals in dendritic cells via CD80 and CD86. Nat Immunol. 2004;5:1134–42. doi: 10.1038/ni1124. [DOI] [PubMed] [Google Scholar]
- 8.Hucke C, MacKenzie CR, Adjogble KD, Takikawa O, Daubener W. Nitric oxide-mediated regulation of gamma interferon-induced bacteriostasis: inhibition and degradation of human indoleamine 2,3-dioxygenase. Infect Immun. 2004;72:2723–30. doi: 10.1128/IAI.72.5.2723-2730.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goren I, Linke A, Muller E, Pfeilschifter J, Frank S. The suppressor of cytokine signaling-3 is upregulated in impaired skin repair: implications for keratinocyte proliferation. J Invest Dermatol. 2006;126:477–85. doi: 10.1038/sj.jid.5700063. [DOI] [PubMed] [Google Scholar]
- 10.Orabona C, Belladonna ML, Vacca C, et al. Cutting edge: silencing suppressor of cytokine signaling 3 expression in dendritic cells turns CD28-Ig from immune adjuvant to suppressant. J Immunol. 2005;174:6582–6. doi: 10.4049/jimmunol.174.11.6582. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–65. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 12.Orr SJ, Morgan NM, Elliott J, et al. CD33 responses are blocked by SOCS3 through accelerated proteasomal-mediated turnover. Blood. 2007;109:1061–8. doi: 10.1182/blood-2006-05-023556. [DOI] [PubMed] [Google Scholar]
- 13.Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol Cell Biol. 2002;22:3316–26. doi: 10.1128/MCB.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong PK, Egan PJ, Croker BA, et al. SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. J Clin Invest. 2006;116:1571–81. doi: 10.1172/JCI25660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machado FS, Esper L, Dias A, et al. Native and aspirin-triggered lipoxins control innate immunity by inducing proteasomal degradation of TRAF6. J Exp Med. 2008;205:1077–86. doi: 10.1084/jem.20072416. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Logue EC, Sha WC. CD28–B7 bidirectional signaling: a two-way street to activation. Nat Immunol. 2004;5:1103–5. doi: 10.1038/ni1104-1103. [DOI] [PubMed] [Google Scholar]
- 17.Fallarino F, Orabona C, Vacca C, et al. Ligand and cytokine dependence of the immunosuppressive pathway of tryptophan catabolism in plasmacytoid dendritic cells. Int Immunol. 2005;17:1429–38. doi: 10.1093/intimm/dxh321. [DOI] [PubMed] [Google Scholar]
- 18.Orabona C, Pallotta MT, Volpi C, et al. SOCS3 drives proteasomal degradation of indoleamine 2,3-dioxygenase (IDO) and antagonizes IDO-dependent tolerogenesis. Proc Natl Acad Sci U S A. 2008;105(52):20828–33. doi: 10.1073/pnas.0810278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romani L, Fallarino F, De Luca A, et al. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451:211–5. doi: 10.1038/nature06471. [DOI] [PubMed] [Google Scholar]
- 20.Grohmann U, Fallarino F, Bianchi R, et al. IL-6 inhibits the tolerogenic function of CD8α+ dendritic cells expressing indoleamine 2,3-dioxygenase. J Immunol. 2001;167:708–14. doi: 10.4049/jimmunol.167.2.708. [DOI] [PubMed] [Google Scholar]
- 21.Grohmann U, Bianchi R, Orabona C, et al. Functional plasticity of dendritic cell subsets as mediated by CD40 versus B7 activation. J Immunol. 2003;171:2581–7. doi: 10.4049/jimmunol.171.5.2581. [DOI] [PubMed] [Google Scholar]
- 22.Grohmann U, Fallarino F, Bianchi R, et al. A defect in tryptophan catabolism impairs tolerance in nonobese diabetic mice. J Exp Med. 2003;198:153–60. doi: 10.1084/jem.20030633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fallarino F, Asselin-Paturel C, Vacca C, et al. Murine plasmacytoid dendritic cells initiate the immunosuppressive pathway of tryptophan catabolism in response to CD200 receptor engagement. J Immunol. 2004;173:3748–54. doi: 10.4049/jimmunol.173.6.3748. [DOI] [PubMed] [Google Scholar]
- 24.Fallarino F, Grohmann U, You S, et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–61. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 25.Grohmann U, Bianchi R, Fioretti MC, et al. CD8+ cell activation to a major mastocytoma rejection antigen, P815AB: requirement for tumor helper peptides in priming for skin test reactivity to a P815AB-related peptide. Eur J Immunol. 1995;25:2797–802. doi: 10.1002/eji.1830251013. [DOI] [PubMed] [Google Scholar]
- 26.Grohmann U, Orabona C, Fallarino F, et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat Immunol. 2002;3:1097–101. doi: 10.1038/ni846. [DOI] [PubMed] [Google Scholar]
- 27.Grohmann U, Volpi C, Fallarino F, et al. Reverse signaling through GITR ligand enables dexamethasone to activate IDO in allergy. Nat Med. 2007;13:579–86. doi: 10.1038/nm1563. [DOI] [PubMed] [Google Scholar]
- 28.Shortman K, Heath WR. Immunity or tolerance? That is the question for dendritic cells. Nat Immunol. 2001;2:988–9. doi: 10.1038/ni1101-988. [DOI] [PubMed] [Google Scholar]
- 29.Grohmann U, Fallarino F, Silla S, et al. CD40 ligation ablates the tolerogenic potential of lymphoid dendritic cells. J Immunol. 2001;166:277–83. doi: 10.4049/jimmunol.166.1.277. [DOI] [PubMed] [Google Scholar]
- 30.Uyttenhove C, Pilotte L, Thèate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–74. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 31.Fallarino F, Grohmann U, Bianchi R, Vacca C, Fioretti MC, Puccetti P. Th1 and Th2 cell clones to a poorly immunogenic tumor antigen initiate CD8+ T cell-dependent tumor eradication in vivo. J Immunol. 2000;165:5495–501. doi: 10.4049/jimmunol.165.10.5495. [DOI] [PubMed] [Google Scholar]
- 32.Munn DH, Zhou M, Attwood JT, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–3. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 33.Orr SJ, Morgan NM, Buick RJ, et al. SOCS3 targets Siglec 7 for proteasomal degradation and blocks Siglec 7-mediated responses. J Biol Chem. 2007;282:3418–22. doi: 10.1074/jbc.C600216200. [DOI] [PubMed] [Google Scholar]
- 34.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84–9. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]