Abstract
Sporadic tumours, which account for the majority of all human cancers, arise from the acquisition of somatic, genetic and epigenetic alterations leading to changes in gene sequence, structure, copy number and expression. Within the last decade, the availability of a complete sequence-based map of the human genome, coupled with significant technological advances, has revolutionized the search for somatic alterations in tumour genomes. Recent landmark studies, which resequenced all coding exons within breast, colorectal, brain and pancreatic cancers, have shed new light on the genomic landscape of cancer. Within a given tumour type there are many infrequently mutated genes and a few frequently mutated genes, resulting in incredible genetic heterogeneity. However, when the altered genes are placed into biological processes and biochemical pathways, this complexity is significantly reduced and shared pathways that are affected in significant numbers of tumours can be discerned. The advent of next-generation sequencing technologies has opened up the potential to resequence entire tumour genomes to interrogate protein-encoding genes, non-coding RNA genes, non-genic regions and the mitochondrial genome. During the next decade it is anticipated that the most common forms of human cancer will be systematically surveyed to identify the underlying somatic changes in gene copy number, sequence and expression. The resulting catalogues of somatic alterations will point to candidate cancer genes requiring further validation to determine whether they have a causal role in tumourigenesis. The hope is that this knowledge will fuel improvements in cancer diagnosis, prognosis and therapy, based on the specific molecular alterations that drive individual tumours. In this review, I will provide a historical perspective on the identification of somatic alterations in the pre- and post-genomic eras, with a particular emphasis on recent pioneering studies that have provided unprecedented insights into the genomic landscape of human cancer.
Keywords: neoplasia, gene, sequence, mutation, history, cancer, somatic, genomic, personalized medicine
Introduction
In 1914, Theodor Boveri hypothesized that numerical alterations in the chromosome content of a cell might contribute to tumourigenesis [1]. However, it would take almost another 50 years before the first specific chromosomal abnormality, a marker chromosome termed the Philadelphia chromosome, was identified within cancer genomes of patients with chronic myelogenous leukaemia (CML) [2]. We now know that sporadic human tumours, which account for the majority of all human cancers, result from the accumulation of numerous genetic and epigenetic alterations, leading to the dysregulation of protein-encoding genes as well as non-coding RNAs (ncRNAs) [3–6]. Such alterations underlie the acquired attributes shared by most cancer cells, namely their capacity to generate their own mitogenic signals, evade apoptosis, resist exogenous growth-inhibitory signals, proliferate without limits, and acquire angiogenic, invasive and metastatic properties [7].
Early attempts to identify protein-encoding cancer genes were hampered by both the limited resolution of technologies available to detect genomic alterations and the enormous complexity of tumour genomes. This complexity was first recognized from cytogenetic studies cataloguing numerical and structural abnormalities within solid tumours [8]. Unlike haematological malignancies and certain sarcomas, which were often characterized by a signature chromosome abnormality, most solid tumours of epithelial origin exhibited numerous chromosomal aberrations, both clonal and nonclonal [8–10]. The karyotypic heterogeneity of tumour genomes led to the proposal by Peter Nowell, in the mid-1970s, that tumourigenesis occurs by a stepwise evolutionary process [11]. Nowell suggested that, following an initiating event that converts a normal cell to a neoplastic cell, cancer progression results from the acquisition of genetic instability, leading to the accumulation of genetic alterations and the continual selective outgrowth of variant subpopulations of tumour cells with a proliferative advantage. This model not only accounted for the genetic heterogeneity of solid tumours, but also led Nowell to speculate that each cancer patient might require individual specific therapy [11].
In the ensuing years, cancer gene identification was a laborious task, significantly impeded by the lack of a sequence-based map of the normal human genome. This changed in 2003 with the completion of the Human Genome Project [12]. In the so-called post-genomic era, the availability of a complete map of the human genome and significant technological advances together powered systematic interrogations of cancer genomes, at unprecedented resolution and throughput, to catalogue somatic alterations and thus pinpoint candidate cancer genes. Within the last decade the concept of individualized therapy, based on the presence of specific molecular alterations within a patient's tumour, has become a reality, spurring the research community to systematically catalogue the somatic alterations that underlie all human cancers [13–26]. Here I review progress that has been made in the search for the genetic basis of sporadic cancers, with a particular emphasis on recent efforts to comprehensively and systematically catalogue somatic alterations using integrated genomic approaches. For comprehensive reviews of other factors that shape the cancer landscape, including the role of ncRNAs, dysregulation of protein synthesis, altered splicing, changes in chromatin structure, epi-genetic alterations and germline cancer susceptibility, the reader is referred to additional articles within this issue and elsewhere [27–38].
Searching for cancer genes in the pre-genomics era
To appreciate the significance of recent developments in the search for cancer genes, it is useful to understand the pace of cancer gene discovery between 1960 and 2003, the years marking the identification of the first cancer-specific chromosomal abnormality and the completion of the Human Genome Project (Figure 1) [2,12]. In the late 1970s it was discovered that the genes responsible for the oncogenicity of certain retroviruses were actually altered versions (oncogenes) of normal cellular genes (proto-oncogenes). The first proto-oncogene, termed c-src, was identified in 1976, based on homology to the transforming gene (v-src) of the Rous sarcoma virus, a retrovirus that induces sarcomas in chickens [39,40].
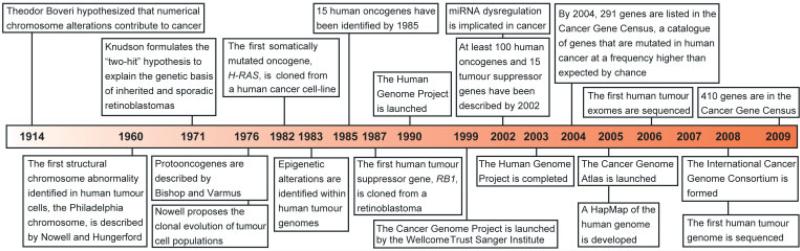
Figure 1.
Timeline of seminal hypotheses, research discoveries and research initiatives that have led to an improved understanding of the genetic aetiology of human tumourigenesis within the last century [1,2,4,5,11,12,39,41–44,58,61,65,66,135,148,240]. The consensus cancer gene data were obtained from the Wellcome Trust Sanger Institute Cancer Genome Project website (http://www.sanger.ac.uk/genetics/CGP)
The significance of proto-oncogenes in relation to human cancer causation became clear in 1982, when a transforming gene isolated from a human bladder carcinoma cell line was discovered to be a mutated version of a normal cellular proto-oncogene [41–44]. This marked the first description of a sequence change, a point mutation resulting in the oncogenic activation of H-RAS, in a human cancer gene.
The development of high-resolution chromosome banding techniques in the early 1970s allowed for detailed karyotypic analysis of cancer genomes and the identification of recurrent chromosomal abnormalities in haematological malignancies, cancer-derived cell lines and, to a lesser extent, solid tumours (reviewed in [8]). The molecular characterization of genes positioned at the breakpoints of chromosomal translocations, or those contained within homogeneously staining regions and double minutes, revealed that gene rearrangement, amplification and increased expression also resulted in oncogene activation in human cancers [45–54] (reviewed in [55]). In the pre-genomics era, oncogene identification largely relied on cloning genes at the site of proviral integrations, functional assays to isolate cellular genes capable of inducing foci formation in NIH3T3 cells, and positional cloning following the identification of structural and numerical chromosomal abnormalities, using both conventional and, later, molecular cytogenetic approaches (reviewed in [56,57]). By 2002, more than 100 oncogenes had been described [58]. Many of these encoded protein tyrosine kinases that modulate mitogenic signal transduction pathways [59]. Their oncogenic activation was associated with increased kinase activity and cellular transformation, properties that would have important therapeutic implications in later years (reviewed in [60]).
In 1971, by studying the inheritance pattern of retinoblastoma, a childhood cancer of the eye with a familial component, Alfred Knudson hypothesized the existence of a distinct class of cancer gene, acting recessively and requiring ‘two hits’ to contribute to tumourigenesis [61]. The idea that neoplasia was sometimes a genetically recessive phenotype was supported by the cell fusion experiments of Henry Harris. The retinoblastoma gene (RB1), the first of the co-called tumour suppressor genes, was cloned 16 years later [62–68]. As Knudson had predicted, familial forms of retinoblastoma are caused by the inheritance of a germline alteration within one allele of RB1 and the acquisition of a somatic alteration in the other allele (the second hit) during the patient's lifetime. In contrast, sporadic forms of retinoblastoma arise later in life after a single cell acquires two independent somatic alterations, leading to a complete loss of RB1 function. A variety of genetic alterations lead to tumour suppressor gene inactivation, including point mutations, gene deletions and epigenetic alterations leading to gene silencing [64,69]. The earliest tumour suppressor genes identified were isolated by positional cloning, following the delineation of regions of linkage to disease susceptibility in cancer kindreds [62–68,70–88]. By 1993, 13 tumour suppressor genes had been positionally cloned or mapped to specific chromosomal locations [89].
Later, candidate gene approaches were also successful in uncovering cancer susceptibility genes [90–92]. With the development of molecular methods to search for somatic loss-of-heterozygosity and homozygous deletions in sporadic tumours, additional recessively acting cancer genes were described [70,71,93–95]. In 1986, Renato Dulbecco contended that the research community was faced with a choice — either to continue searching for cancer genes by a piecemeal approach or to decode the human genome and lay a foundation for future systematic searches for cancer genes [96]. Four years later saw the launch of the Human Genome Project, a publicly funded, international effort to decode the human genome, led by the International Human Genome Sequencing Consortium (IHGSC).
Searching for cancer genes in the post-genomics era
In 2001, the IHGSC and Celera Genomics reported draft versions of the human genome sequence [97,98]. Shortly thereafter, the complete human genome sequence was reported by the IHGSC [12]. The availability of a sequence-based map of the human genome and the development of high-throughput Sanger sequencing, a by-product of the human genome project, provided new opportunities to systematically catalogue somatic mutations within both causal and candidate cancer genes on a larger scale, and at a faster pace, than previously possible. Given the clinical successes of genotype-directed therapies in treating HER2-positive breast cancer patients with trastuzumab, and treating BCR-ABL-positive CML patients with imatinib, many of the earliest high-throughput resequencing efforts centred on protein kinase-encoding genes [13,99,100]. As a result, a number of novel therapeutic targets were revealed for genotypically-defined subgroups of cancer patients [101–118] (reviewed in [119]). This paved the way for other high-throughput resequencing efforts targeting specific biochemical pathways, large gene families and large numbers of candidate cancer genes [120–130]. Not all cancer genes encode proteins that are amenable to direct therapeutic intervention. However, identifying synthetic lethal partners of these so-called ‘undruggable targets’ holds promise in leveraging such alterations for targeted cancer therapy [23–26,131] (reviewed in [132]).
In parallel with the human genome project, the development of array-based comparative genomic hybridization and progressive refinements of this technology significantly increased the resolution and throughput at which genome-wide somatic alterations in DNA copy number could be ascertained (reviewed in [133]). The completion of the human genome project also facilitated the construction of the HapMap, a map of naturally occurring human genomic variation, in the form of single nucleotide polymorphisms (SNPs) and its underlying genomic structure [134–136]. This allowed for the development of high-density SNP genotyping arrays that could be used to screen not only for copy number alterations but also for copy-neutral loss-of-heterozygosity throughout the genome, at unparalleled resolution [137–143] (reviewed in [133]). The implementation of genome-wide expression profiling revealed novel transcriptional signatures that can be utilized to molecularly classify cancer subtypes or predict clinical phenotypes, and was also instrumental in facilitating the identification of previously unrecognized gene fusions in common epithelial tumours (reviewed in [144,145]). Furthermore, integrated studies that combined genome-wide searches for copy number alterations with global gene expression profiling significantly increased the power to hone in on candidate cancer genes [146,147]. Nonetheless, the capacity to understand the full compendium of genomic alterations that drive human tumourigenesis was limited by the inability to rapidly and systematically sequence entire tumour genomes.
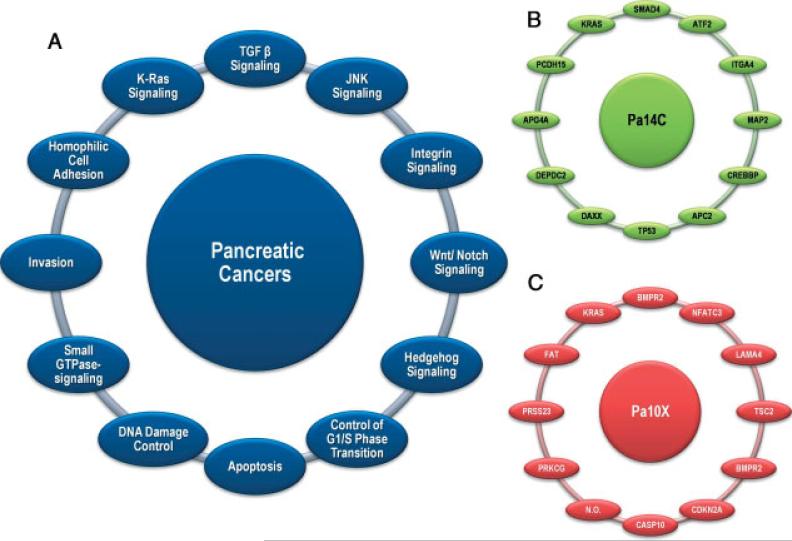
Our first glimpse into the true genetic complexity of human cancers has come from recent pioneering studies that sequenced the exomes, the coding exons of more than 18 000 protein-encoding genes, from a series of breast, colorectal, pancreatic and brain cancers [148–151]. Remarkably, these explorations were achieved using high-throughput Sanger sequencing, and were integrated with genomic analyses to interrogate gene expression and copy number [149,150,152]. These studies revealed that cancer genomes are highly complex, with an average of 48–101 somatic alterations in each tumour, depending on the cancer type [149,150]. Within a given cancer type there is considerable inter-tumour heterogeneity, resulting in large numbers of altered genes. However, this complexity is reduced significantly by considering the biological pathways and processes on which altered genes converge, rather than the altered genes themselves [151]. For example, 12 core biological processes or pathways appear to be deregulated in the majority of pancreatic tumours, although precisely how this is achieved varies from tumour to tumour (Figure 2) [149]. As has been noted, this may have practical implications for the development of targeted therapeutics, in that it may be more prudent to consider targeting functional pathways or processes rather than individual proteins encoded by mutated genes [151]. Prior to these investigations, most cancer genes had been identified because they were frequently altered in tumourigenesis. However, the resequencing of tumour exomes revealed that, for a given type of cancer, the majority of somatically mutated genes are altered in just a fraction of tumours. This new view of the genomic landscape of human cancer suggested that the acquisition of numerous somatic mutations, each with a small fitness advantage, may also drive tumourigenesis [151,153].
Figure 2.
Signalling pathways and processes. (A) The 12 pathways and processes whose component genes were genetically altered in most pancreatic cancers. (B, C) Two pancreatic cancers (Pa14C and Pa10X) and the specific genes that are mutated in them. The positions around the circles in (B, C) correspond to the pathways and processes in (A). Several pathway components overlapped, as illustrated by the BMPR2 mutation that presumably disrupted both the SMAD4 and Hedgehog signalling pathways in Pa10X. Additionally, not all 12 processes and pathways were altered in every pancreatic cancer, as exemplified by the fact that no mutations known to affect DNA damage control were observed in Pa10X. N.O., not observed. From Jones et al. Science 2008; 321: 1801–1806 [149]. Reprinted with permission from AAAS
Somatic alterations can be so-called driver mutations that confer a selective growth advantage to the tumour cell, or passenger mutations that have no effect on tumourigenesis [101,154]. Thus, the identification of a somatically altered gene indicates a candidate cancer gene rather than a causal cancer gene. What necessarily follows are detailed biochemical and cellular studies comparing the functional properties of the wild-type and mutant proteins. To guide such studies, statistical calculations, based on the frequency and nature of the observed somatic mutations, can be applied to prioritize or rank candidate cancer genes based on the likelihood that they represent driver genes [155]. The statistical assumptions that are most appropriate to use in this type of predictive modelling have been the subject of some debate, because of the inherent difficulty in setting a background mutation rate for each tumour type [155–158]. Other computational approaches predict driver mutations rather than driver genes [159–161]. One such method has estimated that approximately 8% of missense mutations identified by exomic sequencing of glioblastomas are likely to be functionally significant, with the majority of these affecting infrequently mutated genes [161]. Although synonymous somatic mutations are generally not considered in statistical predictions because they do not result in amino acid changes, it is worth noting that they can, on occasion, encode proteins with altered functional activity [162]. It is important to note that, in addition to statistical predictions, functional genetic screens in mice and large-scale RNA interference screens can also guide the identification of causal cancer genes [131,163–172] (reviewed in [173–175]).
Although exomic resequencing of cancer genomes captures the spectrum of mutations within protein-encoding genes, it does not assess the sequence integrity of non-coding regions of the genome. These regions contain functionally relevant elements, including ultra-conserved elements and ncRNAs, which are being systematically mapped by the Encyclopedia of DNA Elements (ENCODE) project [176–180]. Non-coding RNAs have been implicated in a variety of processes, including the regulation of transcription and chromosome structure, RNA processing and modification, mRNA stability and translation and protein stability and transport (reviewed in [181]). Within the past few years, our vision of the cancer landscape has been reshaped with the realization that the dysregulation of micro-RNAs (miRNAs), a subset of ncRNAs, contributes to tumourigenesis (reviewed in [32,35]). MiRNAs are small ncRNAs that negatively regulate gene expression, including that of protein-encoding cancer genes. Dysregulated miRNAs have been described in human cancers and in some instances are associated with oncogenic properties, tumour-suppressive properties or both, depending on the cellular context (reviewed in [32]). Furthermore, miRNA expression profiling of a mouse model of pancreatic cancer revealed distinct miRNA expression signatures at each step in the progression of tumourigenesis, correlating with the acquired attributes shared by most cancer cells [6,7]. The full extent to which ncRNAs contribute to cancer has yet to be revealed. In addition to miRNAs, another class of ncRNAs, represented by transcribed ultraconserved regions of the genome, has also been implicated in tumourigenesis [179,180]. Moreover, inherited mutations within the gene encoding DICER1, an endonuclease that regulates the processing of ncRNAs, have been linked to familial pleuropulmonary blastoma [182].
Next-generation sequencing
The impetus for the human genome project was to generate a sequence-based map that would allow researchers to identify the germline and somatic variation that underlies human disease, including cancer [96]. One implication is that of personalized medicine, the ability to predict individual disease risk and drug response based on personal genomics. In the context of sporadic tumourigenesis, decoding individual tumour genomes holds the promise of more accurate diagnosis and prognosis and of guiding personalized treatment strategies for cancer patients.
Sanger sequencing has been the gold standard of DNA sequencing since it was first described over 30 years ago [183]. However, the expense and throughput of this approach prohibits its application in sequencing large numbers of genomes. Therefore, in order to make personalized medicine incorporating whole-genome sequencing a reality, several new and revolutionary sequencing technologies have been developed in the past few years. These so-called next-generation sequencing approaches have been reviewed in detail elsewhere [184–188]. Suffice it to say that, by sequencing DNA in a massively parallel fashion, next-generation sequencing methodologies have significantly lowered the cost and time required to decode an entire human genome [189–193]. Nonetheless, the short reads obtained, coupled with the relatively high error rate, require deep coverage of each genome, keeping current costs relatively high. In addition, data analysis and confirmation of sequence variants are non-trivial. To bring whole-genome sequencing in line with other clinical diagnostic tests, the goal of a $1000 genome has been set (reviewed in [194,195]). Although this has not yet been attained, it is believed to be within reach with further improvements in technology. In addition to their higher throughput and lowered costs, next-generation sequencing methodologies have several other advantages compared with Sanger sequencing that are key to comprehensively deciphering tumour genomes. They are quantitative in nature and can be used to simultaneously determine both nucleotide sequence and copy number. They are more sensitive than Sanger sequencing, and therefore can detect somatic mutations present in just a subset of tumour cells [196,197]. In addition, these methods can be coupled with a so-called paired-end read strategy, in which both ends of individual clones are sequenced and mapped back to the genome [198]. This strategy can be used to identify structural alterations, including insertions, deletions, duplications and rearrangements [199–201]. It is important to note that sequencing entire genomes encompasses not only the cellular genome but also the mitochondrial genome and, potentially, any virally-associated genomes. Prior to the development of next-generation sequencing, searching for changes in sequence, copy number and structure required the integration of data from multiple platforms. As next-generation sequencing eventually becomes routine, it will transform the analyses of cancer genomes, using a single platform.
Next-generation sequencing of whole tumour genomes and transcriptomes
Decoding entire genomes by next-generation sequencing is feasible, but is not yet commonplace [189–193]. One consideration that pertains to whole-genome sequencing of cancer genomes is the current need to survey both the tumour and the constitutional genomes from the same individual, to accurately discriminate polymorphic variants from potential somatic mutations. At the time of this writing, the genomes of two cytogenetically normal cases of acute myeloid leukaemia (AML), as well as DNA from the normal skin cells of these individuals, have been resequenced and partially analysed. The extraordinary number of potential somatic mutations identified (20 000–30 000 in each tumour), together with the high error rate, necessitated the prioritization of mutations for confirmatory sequencing by other methods. To date, only those mutations localizing within protein-encoding genes of the first AML genome have been validated [202]. For the second AML genome, mutations within the coding regions of annotated genes and RNA genes, as well as those within highly conserved non-genic regions and regulatory regions, have been validated [203]. Eight to twelve true somatic alterations were identified within the coding regions of each tumour genome [202,203]. Most of the altered genes were not previously implicated in the pathogenesis of AML and would not have been obvious choices for resequencing using a candidate gene approach. The coding regions of the genome comprise only 1–2% of the entire human genome. It has been estimated that 500–1000 additional somatic mutations exist within the non-coding regions of each AML genome. Most are anticipated to be passenger mutations with no contribution to tumourigenesis, but this awaits experimental confirmation [203].
Other studies have applied next-generation sequencing to search for somatic alterations within only the transcribed genes, or transcriptome, of tumours and tumour-derived cell lines, by a process known as RNA sequencing [204–213]. Because transcriptomes are significantly smaller than whole genomes, RNA sequencing represents a more cost-effective alternative than whole-genome sequencing to search for mutations within coding genes. In addition, transcriptome analysis can provide insights into the nature and abundance of alternative splice forms. However, one disadvantage of transcriptome sequencing versus exome or whole-genome sequencing is that unstable or low-abundance transcripts might escape detection [214]. Notably, this approach has thus far led to the identification of novel candidate cancer genes within malignant mesotheliomas, as well as a novel genetic signature within granulosa-cell tumours of the ovary [205,206]. A recurrent somatic mutation within the FOXL2 gene was identified within almost all adult cases of ovarian granulosa cell tumours, opening up the possibility for improved diagnostic classification of this rare malignancy [205]. The relevance of massively parallel transcriptome analysis to histopathology is reviewed elsewhere in this issue [215]. Paired end-read transcriptome sequencing has also been applied to search for gene fusions resulting from chromosomal rearrangements within tumours [216,217]. Likewise, massively parallel sequencing, using a paired end-read strategy, has been used to catalogue structural rearrangements and copy number alterations present within two lung cancer cell lines [199]. Approximately one-third of somatic rearrangements identified among these cell lines had escaped detection by other methods, testifying to the sensitivity of this approach to catalogue genomic rearrangements.
Large-scale initiatives to search comprehensively for somatic alterations in human cancers
The past few years have seen the formation of increasingly more organized efforts to comprehensively and systematically search for the somatic alterations that underlie human cancer. The Cancer Genome Project of the Wellcome Trust Sanger Institute (UK) spearheaded the systematic search for somatic alterations in human tumours and tumour-derived cell lines, and continues this mission. Ongoing projects include the resequencing of 4000 candidate cancer genes from a series of cancer cell lines derived from a variety of solid tumours and from a collection of clear cell renal carcinomas (http://www.sanger.ac.uk/genetics/CGP/Studies/). Additionally, 800 tumour-derived cell lines are being interrogated for copy number alterations.
In 2005, the National Human Genome Research Institute and the National Cancer Institute (USA) launched The Cancer Genome Atlas (TCGA), a publicly-funded initiative to systematically catalogue genomic alterations present within the major forms of human cancer (http://cancergenome.nih.gov/about/mission.asp). This began with a 3 year pilot study interrogating glioblastoma multiforme, ovarian cancer and lung cancer, using integrated genomic approaches to search for changes in gene copy number, sequence and expression. In the pilot phase, ~600 candidate cancer genes were selected for Sanger sequencing [218]. The analysis of glioblastomas by the TCGA network revealed novel somatic alterations in this disease, including mutations within the PIK3R1 gene, the regulatory subunit of PI3K, and indicated that disruption of the TP53, RB1 and receptor tyrosine kinase-mediated signalling pathways is a core element of gliomagenesis. It also pointed to a possible mechanistic link between the methylation of the MGMT gene, which identifies subgroups of glioblastoma patients sensitive to temozolomide treatment, and the appearance of MSH6 mutations, which are associated with temozolomide resistance [218, 219]. TCGA is moving out of its pilot phase with the objective of comprehensively identifying somatic alterations in 20–25 major tumour types by 2014 (http://www.cancer.gov/recovery). The data are publicly available through both open and controlled access via the Cancer Genome Workbench (CGWB), a feature of the Cancer Biomedical Informatics Grid (caBIG) [220,221].
One limitation experienced in the pilot phase of TCGA was the difficulty in obtaining sufficient amounts of tumour tissue for integrated genomic analyses using multiple platforms [218]. This is likely to become less of a limitation as next-generation sequencing, a single method that requires significantly less starting material for analysis, is incorporated into this endeavour. Building on the experience gained in the pilot phase of TCGA, the NCI and the Foundation for the National Institutes of Health launched the Childhood Cancer Therapeutically Applicable Research to Generate Effective Treatments (TARGET) Initiative (http://target.cancer.gov/). TARGET aims to identify therapeutic targets for childhood cancers, including acute lymphoblastic leukaemia (ALL) and neuroblastoma, by characterizing cancer genomes and transcriptomes. Already studies within this initiative have revealed that somatic alterations within the IKZF1 and janus kinase (JAK ) genes occur in a subset of paediatric patients with B cell progenitor ALL and are predictive of poor outcome in such cases [222]. This is also significant because it suggests the potential for therapeutic intervention of molecularly defined subsets of paediatric ALL patients with JAK inhibitors [223].
In 2008, the International Cancer Genome Consortium (ICGC) was formed in an effort to standardize the approaches by which genomic alterations are identified in human cancers (http://www.icgc.org/). The consortium aims to produce comprehensive catalogues of the genomic alterations of up to 50 clinically and societally significant cancer types and subtypes over the course of a decade. Tumours of the pancreas, ovary, stomach, liver, breast and oral cavity, as well as chronic lymphocytic leukaemia, have thus far been prioritized for analysis.
Catalogues of genetic alterations in cancer
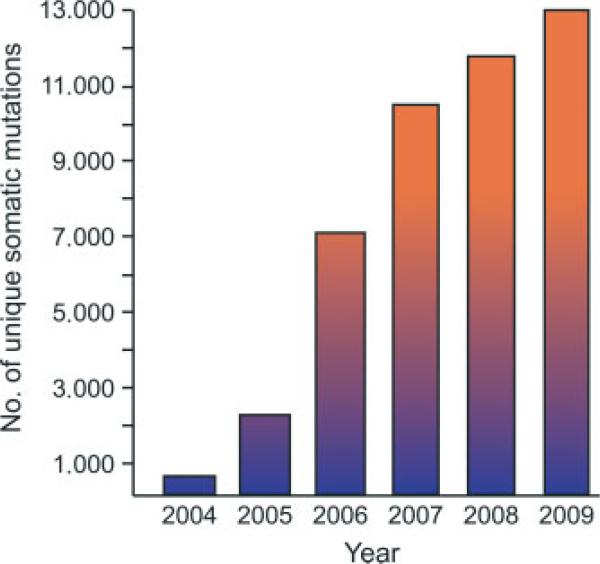
With the emergence of the post-genomic era, and the potential to interrogate human tumour genomes for somatic alterations at unprecedented resolution, came the need to organize and integrate mutation data into searchable catalogues. In 2004, the Wellcome Trust Sanger Institute established the Cancer Gene Census, a catalogue of genes mutated in human cancer at a higher frequency than expected by chance alone [224]. This catalogue includes genes that have undergone somatic mutations in sporadic tumours as well as those that are mutated in the germline of cancer families. From 2004 to 2009 the number of consensus cancer genes increased by 40%, from 291 to 410 genes, coinciding with concerted efforts to systematically interrogate cancer genomes for somatic alterations in gene sequence and copy number. A complementary resource, the Catalogue of Somatic Mutations in Cancer (COSMIC), catalogues somatic mutation frequencies in benign and malignant tumours as well as tumour-derived cell lines [225,226]. The genotypic information available through COSMIC is not limited to just the consensus cancer genes but also includes mutated genes that do not meet the requirement for a consensus cancer gene, as well as genes that have been found not to harbour cancer-associated mutations. Currently, almost 5000 genes and ~340 000 tumours have been curated, resulting in a catalogue of approximately 13 000 unique mutations (Figure 3). Yet other catalogues of mutations serve more specific purposes. The Mitelman Database of Chromo-some Aberrations in Cancer systematically catalogues structural and numerical chromosomal alterations and their clinical associations, as reported in the literature [227,228]. The Genetic Alterations in Cancer database curates mutations associated with exposures to specific chemical, physical and biological agents implicated in tumourigenesis [229]. Numerous other databases catalogue the frequency and spectrum of mutations within individual cancer genes with the goal of facilitating the clinical interpretation of variants (reviewed in [230]).
Figure 3.
Number of unique somatic mutations catalogued in human cancers. The mutation data were obtained from the Wellcome Trust Sanger Institute Catalogue Of Somatic Mutations In Cancer (COSMIC) website (http://www.sanger.ac.uk/cosmic) [225,226,241]. The mutation data displayed for 2004–2008 are derived from the year-end COSMIC data. The mutation data for 2009 is derived from the COSMIC v42 release. The COSMIC v42 release does not incorporate data from large-scale resequencing projects [102,148,150,151]
As the number of somatic mutations identified in cancer genomes continues to grow, it seems likely that additional, gene-specific catalogues will emerge. In this regard, guidelines to standardize the reporting of both somatically mutated and non-mutated tumour samples have recently been proposed, in order to establish a foundation for the accurate interpretation of their clinical and biological significance [230]. Setting and adhering to such guidelines will be critical as we enter a new era in cancer genomics, in which next-generation sequencing of tumour genomes will undoubtedly lead to an explosion in the wealth of mutation data.
Concluding remarks
Over the course of the next decade we will gain an unparalleled appreciation of the somatic alterations present within human cancers, within protein-coding genes, non-coding RNA genes and non-coding regions of the genome, as well as the mitochondrial genome. However, this is just a first step towards identifying bone fide cancer genes. What necessarily follows will be the detailed functional characterization of individual candidate cancer genes, to determine whether and how they contribute to a tumourigenic phenotype. In parallel, functional genetic screens in mice and large-scale RNA interference screens may also guide the confirmation of causal cancer genes [131,163–172] (reviewed in [173–175]).
Once a causal cancer gene has been identified, it is important to recognize that not all somatic mutations within the gene will be functionally equivalent; some mutations might be consequential and others functionally insignificant [154]. For example, certain EGFR mutations predict clinical responses to targeted therapies, whereas other mutations within this gene predict clinical resistance [115,231–236]. Likewise, the larger genomic context of a mutated gene can also affect its interpretation. For example, in glioblastomas it is the co-expression of a mutated form of EGFR (EGFRvIII) and PTEN that correlates with clinical sensitivity to small molecule inhibitors of EGFR [237]. Conversely, the absence of a somatic mutation within tumour cells can also be used to guide molecularly directed therapies. For example, in both the USA and Europe the drugs cetuximab and panitumumab (monoclonal antibodies against EGFR) are recommended only for advanced colorectal patients whose tumours contain a non-mutated form of the KRAS gene (reviewed in [238]). Therefore, before new tests are introduced into the clinical setting it will be imperative to establish the clinicopathological associations of individual mutations, or lack thereof.
How the systematic cataloguing of somatic alterations in cancer genomes will transform diagnostic and clinical practices remains to be seen. One could envision a new battery of specific tests for each tumour type and subtype, based on altered genes or altered pathways, or eventually routine sequencing of individual tumour genomes. Such approaches might ultimately be coupled with methods that noninvasively isolate circulating tumour cells from peripheral blood samples, permitting the serial analyses of cancer genomes to dynamically monitor tumour evolution and adjust clinical management accordingly (reviewed in [239]). Whatever the final outcome, the systematic analyses of cancer genomes using new technologies [242] that will take place over the next decade has the potential to create a paradigm shift in the diagnosis, prognosis and treatment of individual cancer patients, as Nowell speculated over 30 years ago [11].
Supplementary Material
Acknowledgements
I would like to extend my sincere thanks to Dr Cariappa Annaiah and members of my laboratory for critical reading of the manuscript. I thank Julia Fekecs and Darryl Leja for graphical expertise. Funded by the Intramural Program of the National Human Genome Research Institute at NIH (to DWB).
Footnotes
Conflict of interest statement: I am employed by the National Human Genome Research Institute (NHGRI)/NIH, but I am not a member of The Cancer Genome Atlas research network, which is partially funded by NHGRI. I am an inventor on a patent describing EGFR mutations, which is licensed to Genzyme Corporation.
Teaching materials
PowerPoint slides of the figures from this review are supplied as supporting information in the online version of this article.
References
- 1.Boveri T. Zur Frage der Entstehung Maligner Tumouren. Gustav Fischer; Jena: 1914. [Google Scholar]
- 2.Nowell P, Hungerford D. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497. doi: 10.1126/science.144.3623.1229. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal tumour development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, et al. MicroRNA dynamics in the stages of tumourigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 8.Wolman Karyotypic progression in human tumours. Cancer Metast Rev. 1983;2:257–293. doi: 10.1007/BF00048481. [DOI] [PubMed] [Google Scholar]
- 9.Sandberg AA. Cytogenetics and molecular genetics of bone and soft-tissue tumours. Am J Med Genet. 2002;115:189–193. doi: 10.1002/ajmg.10691. [DOI] [PubMed] [Google Scholar]
- 10.Atkin NB. Solid tumour cytogenetics. Progress since 1979. Cancer Genet Cytogenet. 1989;40:3–12. doi: 10.1016/0165-4608(89)90140-4. [DOI] [PubMed] [Google Scholar]
- 11.Nowell PC. The clonal evolution of tumour cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 12.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 13.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 14.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 15.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 16.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 17.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 18.Guilhot F, Apperley J, Kim DW, Bullorsky EO, Baccarani M, Roboz GJ, et al. Dasatinib induces significant hematologic and cytogenetic responses in patients with imatinib-resistant or - intolerant chronic myeloid leukemia in accelerated phase. Blood. 2007;109:4143–4150. doi: 10.1182/blood-2006-09-046839. [DOI] [PubMed] [Google Scholar]
- 19.Kantarjian H, Pasquini R, Hamerschlak N, Rousselot P, Holowiecki J, Jootar S, et al. Dasatinib or high-dose imatinib for chronic-phase chronic myeloid leukemia after failure of first-line imatinib: a randomized phase 2 trial. Blood. 2007;109:5143–5150. doi: 10.1182/blood-2006-11-056028. [DOI] [PubMed] [Google Scholar]
- 20.Kantarjian HM, Giles F, Gattermann N, Bhalla K, Alimena G, Palandri F, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110:3540–3546. doi: 10.1182/blood-2007-03-080689. [DOI] [PubMed] [Google Scholar]
- 21.Kantarjian H, Giles F, Wunderle L, Bhalla K, O'Brien S, Wassmann B, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 22.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 23.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 24.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 25.Jones C, Plummer ER. PARP inhibitors and cancer therapy — early results and potential applications. Br J Radiol. 2008;81(spec no. 1):S2–5. doi: 10.1259/bjr/30872348. [DOI] [PubMed] [Google Scholar]
- 26.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 27.Davidson E. The Regulatory Genome. Gene Regulatory Networks in Development and Evolution. Academic Press; Amsterdam: 2006. [Google Scholar]
- 28.Lever E, Sheer D. The role of nuclear organization in cancer. J Pathol. 2009 doi: 10.1002/path.2651. DOI:10.1002/path.2651. [DOI] [PubMed] [Google Scholar]
- 29.Cazier J- B, Tomlinson I. General lessons from large-scale studies to identify human cancer predisposition genes. J Pathol. 2009 doi: 10.1002/path.2650. DOI:10.1002/path.2650. [DOI] [PubMed] [Google Scholar]
- 30.Le Quesne JP, Spriggs KA, Bushell M, Willis AE. Dysregulation of protein synthesis and disease. J Pathol. 2009 doi: 10.1002/path.2627. DOI:10.1002/path.2627. [DOI] [PubMed] [Google Scholar]
- 31.Hall PA, Russell SH. New perspectives on neoplasia and the RNA world. Hematol Oncol. 2005;23:49–53. doi: 10.1002/hon.748. [DOI] [PubMed] [Google Scholar]
- 32.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kampranis SC, Tsichlis PN. Histone demethylases and cancer. Adv Cancer Res. 2009;102:103–169. doi: 10.1016/S0065-230X(09)02004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- 35.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 38.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 40.Rous P. A sarcoma of the fowl transmissible by an agent seperable from the tumour cells. J Exp Med. 1911;13:397–410. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy EP, Reynolds RK, Santos E, Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature. 1982;300:149–152. doi: 10.1038/300149a0. [DOI] [PubMed] [Google Scholar]
- 42.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 43.Tabin CJ, Bradley SM, Bargmann CI, Weinberg RA, Papageorge AG, Scolnick EM, et al. Mechanism of activation of a human oncogene. Nature. 1982;300:143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- 44.Taparowsky E, Suard Y, Fasano O, Shimizu K, Goldfarb M, Wigler M. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 1982;300:762–765. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- 45.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 46.de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 47.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 48.Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G. Structural organization of the bcr gene and its role in the Ph′ translocation. Nature. 1985;315:758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- 49.Konopka JB, Watanabe SM, Singer JW, Collins SJ, Witte ON. Cell lines and clinical isolates derived from Ph1-positive chronic myelogenous leukemia patients express c-abl proteins with a common structural alteration. Proc Natl Acad Sci USA. 1985;82:1810–1814. doi: 10.1073/pnas.82.6.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 51.Alitalo K, Schwab M, Lin CC, Varmus HE, Bishop JM. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci USA. 1983;80:1707–1711. doi: 10.1073/pnas.80.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human cmyc oncogene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erikson J, Nishikura K, ar-Rushdi A, Finan J, Emanuel B, Lenoir G, et al. Translocation of an immunoglobulin κ locus to a region 3′ of an unrearranged cmyc oncogene enhances c-myc transcription. Proc Natl Acad Sci USA. 1983;80:7581–7585. doi: 10.1073/pnas.80.24.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishikura K, ar-Rushdi A, Erikson J, Watt R, Rovera G, Croce CM. Differential expression of the normal and of the translocated human c-myc oncogenes in B cells. Proc Natl Acad Sci USA. 1983;80:4822–4826. doi: 10.1073/pnas.80.15.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2009 doi: 10.1002/path.2632. DOI:10.1002/ path.2632. [DOI] [PubMed] [Google Scholar]
- 56.Weinberg RA. Fewer and fewer oncogenes. Cell. 1982;30:3–4. doi: 10.1016/0092-8674(82)90003-4. [DOI] [PubMed] [Google Scholar]
- 57.Sozzi G, Testi MA, Croce CM. Advances in cancer cytogenetics. J Cell Biochem. 1999;32–33(suppl):173–182. doi: 10.1002/(sici)1097-4644(1999)75:32+<173::aid-jcb21>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 58.Weinstein IB. Cancer. Addiction to oncogenes — the Achilles heel of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 59.Hunter T, Cooper JA. Protein–tyrosine kinases. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 60.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 61.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knudson AG, Jr, Meadows AT, Nichols WW, Hill R. Chromosomal deletion and retinoblastoma. N Engl J Med. 1976;295:1120–1123. doi: 10.1056/NEJM197611112952007. [DOI] [PubMed] [Google Scholar]
- 63.Francke U, Kung F. Sporadic bilateral retinoblastoma and 13q-chromosomal deletion. Med Pediatr Oncol. 1976;2:379–385. doi: 10.1002/mpo.2950020404. [DOI] [PubMed] [Google Scholar]
- 64.Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 65.Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 66.Lee WH, Shew JY, Hong FD, Sery TW, Donoso LA, Young LJ, et al. The retinoblastoma susceptibility gene encodes a nuclear phosphoprotein associated with DNA binding activity. Nature. 1987;329:642–645. doi: 10.1038/329642a0. [DOI] [PubMed] [Google Scholar]
- 67.Fung YK, Murphree AL, T'Ang A, Qian J, Hinrichs SH, Benedict WF. Structural evidence for the authenticity of the human retinoblastoma gene. Science. 1987;236:1657–1661. doi: 10.1126/science.2885916. [DOI] [PubMed] [Google Scholar]
- 68.Huang HJ, Yee JK, Shew JY, Chen PL, Bookstein R, Friedmann T, et al. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science. 1988;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- 69.Ferguson-Smith AC, Reik W, Surani MA. Genomic imprinting and cancer. Cancer Surv. 1990;9:487–503. [PubMed] [Google Scholar]
- 70.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, et al. Allelotype of colorectal carcinomas. Science. 1989;244:207–211. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 71.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 72.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 73.Chen PL, Chen YM, Bookstein R, Lee WH. Genetic mechanisms of tumour suppression by the human p53 gene. Science. 1990;250:1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]
- 74.Goldgar DE, Green P, Parry DM, Mulvihill JJ. Multipoint linkage analysis in neurofibromatosis type I: an international collaboration. Am J Hum Genet. 1989;44:6–12. [PMC free article] [PubMed] [Google Scholar]
- 75.Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 76.Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 77.European Chromosome 16 Tuberous Sclerosis Consortium Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 78.van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 79.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harsh-man K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 80.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 81.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, et al. Identification of the von Hippel–Lindau disease tumour suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 82.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 83.Haber DA, Buckler AJ, Glaser T, Call KM, Pelletier J, Sohn RL, et al. An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms’ tumour. Cell. 1990;61:1257–1269. doi: 10.1016/0092-8674(90)90690-g. [DOI] [PubMed] [Google Scholar]
- 84.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 85.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 86.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 87.Joslyn G, Carlson M, Thliveris A, Albertsen H, Gelbert L, Samowitz W, et al. Identification of deletion mutations and three new genes at the familial polyposis locus. Cell. 1991;66:601–613. doi: 10.1016/0092-8674(81)90022-2. [DOI] [PubMed] [Google Scholar]
- 88.Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumour suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- 89.Knudson AG. Anti-oncogenes and human cancer. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, et al. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 91.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 92.Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 93.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, et al. A cell cycle regulator potentially involved in genesis of many tumour types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 94.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 95.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, et al. KLF6, a candidate tumour suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 96.Dulbecco R. A turning point in cancer research: sequencing the human genome. Science. 1986;231:1055–1056. doi: 10.1126/science.3945817. [DOI] [PubMed] [Google Scholar]
- 97.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 98.McPherson JD, Marra M, Hillier L, Waterston RH, Chinwalla A, Wallis J, et al. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 99.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 100.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 101.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 102.Parsons DW, Wang TL, Samuels Y, Bardelli A, Cummins JM, DeLong L, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 103.Loriaux MM, Levine RL, Tyner JW, Frohling S, Scholl C, Stoffregen EP, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–4796. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 106.Rand V, Huang J, Stockwell T, Ferriera S, Buzko O, Levy S, et al. Sequence survey of receptor tyrosine kinases reveals mutations in glioblastomas. Proc Natl Acad Sci USA. 2005;102:14344–14349. doi: 10.1073/pnas.0507200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci USA. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brown JR, Levine RL, Thompson C, Basile G, Gilliland DG, Freedman AS. Systematic genomic screen for tyrosine kinase mutations in CLL. Leukemia. 2008;22:1966–1969. doi: 10.1038/leu.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 110.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 111.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bignell G, Smith R, Hunter C, Stephens P, Davies H, Greenman C, et al. Sequence analysis of the protein kinase gene family in human testicular germ-cell tumours of adolescents and adults. Genes Chromosomes Cancer. 2006;45:42–46. doi: 10.1002/gcc.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 114.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 115.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 116.Jiang J, Paez JG, Lee JC, Bo R, Stone RM, DeAngelo DJ, et al. Identifying and characterizing a novel activating mutation of the FLT3 tyrosine kinase in AML. Blood. 2004;104:1855–1858. doi: 10.1182/blood-2004-02-0712. [DOI] [PubMed] [Google Scholar]
- 117.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 118.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 120.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 121.Tyner JW, Erickson H, Deininger MW, Willis SG, Eide CA, Levine RL, et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood. 2009;113:1749–1755. doi: 10.1182/blood-2008-04-152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Teng DH, Chen Y, Lian L, Ha PC, Tavtigian SV, Wong AK. Mutation analyses of 268 candidate genes in human tumour cell lines. Genomics. 2001;74:352–364. doi: 10.1006/geno.2001.6551. [DOI] [PubMed] [Google Scholar]
- 123.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Link DC, Kunter G, Kasai Y, Zhao Y, Miner T, McLellan MD, et al. Distinct patterns of mutations occurring in de novo AML versus AML arising in the setting of severe congenital neutropenia. Blood. 2007;110:1648–1655. doi: 10.1182/blood-2007-03-081216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang TL, Rago C, Silliman N, Ptak J, Markowitz S, Will-son JK, et al. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Z, Cummins JM, Shen D, Cahill DP, Jallepalli PV, Wang TL, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64:2998–3001. doi: 10.1158/0008-5472.can-04-0587. [DOI] [PubMed] [Google Scholar]
- 128.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Palavalli LH, Prickett TD, Wunderlich JR, Wei X, Burrell AS, Porter-Gill P, et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat Genet. 2009;41:518–520. doi: 10.1038/ng.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 132.Martin SA, Hewish M, Lord CJ, Ashworth A. Genomic instability and the selection of treatments for cancer. J Pathol. 2009 doi: 10.1002/path.2631. DOI:10.1002/path.2631. [DOI] [PubMed] [Google Scholar]
- 133.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39:S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 135.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Greshock J, Feng B, Nogueira C, Ivanova E, Perna I, Nathanson K, et al. A comparison of DNA copy number profiling platforms. Cancer Res. 2007;67:10173–10180. doi: 10.1158/0008-5472.CAN-07-2102. [DOI] [PubMed] [Google Scholar]
- 138.Cox C, Bignell G, Greenman C, Stabenau A, Warren W, Stephens P, et al. A survey of homozygous deletions in human cancer genomes. Proc Natl Acad Sci USA. 2005;102:4542–4547. doi: 10.1073/pnas.0408593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 140.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS One. 2007;2:e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, et al. Highly sensitive method for genome-wide detection of allelic composition in nonpaired, primary tumour specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 145.Prensner JR, Chinnaiyan AM. Oncogenic gene fusions in epithelial carcinomas. Curr Opin Genet Dev. 2009;19:82–91. doi: 10.1016/j.gde.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 147.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 148.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 149.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 152.Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Frohling S, Scholl C, Levine RL, Loriaux M, Boggon TJ, Bernard OA, et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell. 2007;12:501–513. doi: 10.1016/j.ccr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 155.Parmigiani G, Lin J, Boca S. Response to comments on ‘The consensus coding sequences of human breast and colorectal cancers’. Science. 2007;317:1500d. [Google Scholar]
- 156.Forrest WF, Cavet G. Comment on ‘The consensus coding sequences of human breast and colorectal cancers’. Science. 2007;317:1500. doi: 10.1126/science.1138179. [DOI] [PubMed] [Google Scholar]
- 157.Getz G, Hofling H, Mesirov JP, Golub TR, Meyerson M, Tibshirani R, et al. Comment on ‘The consensus coding sequences of human breast and colorectal cancers’. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 158.Rubin AF, Green P. Comment on ‘The consensus coding sequences of human breast and colorectal cancers’. Science. 2007;317:1500. doi: 10.1126/science.1138956. [DOI] [PubMed] [Google Scholar]
- 159.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 163.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 167.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 168.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, et al. A genetic screen for candidate tumour suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 169.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hij-mans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 170.Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumourigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 171.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 172.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer. 2009;9:389–399. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- 174.Quigley D, Balmain A. Systems genetics analysis of cancer susceptibility: from mouse models to humans. Nat Rev Genet. 2009;10:651–657. doi: 10.1038/nrg2617. [DOI] [PubMed] [Google Scholar]
- 175.Zender L, Lowe SW. Integrative oncogenomic approaches for accelerated cancer-gene discovery. Curr Opin Oncol. 2008;20:72–76. doi: 10.1097/CCO.0b013e3282f31d5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 178.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 180.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 181.Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 182.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shendure JA, Porreca GJ, Church GM. Overview of DNA sequencing strategies. Curr Protoc Mol Biol. 2008;7 doi: 10.1002/0471142727.mb0701s81. Unit 7.1. [DOI] [PubMed] [Google Scholar]
- 185.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genom Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 186.Ansorge WJ. Next-generation DNA sequencing techniques. Nat Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 187.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 188.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 189.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 193.Pushkarev D, Neff NF, Quake Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27:847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mardis ER. Anticipating the 1000 dollar genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Service RF. Gene sequencing. The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 196.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 197.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ng P, Tan JJ, Ooi HS, Lee YL, Chiu KP, Fullwood MJ, et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, et al. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci USA. 2009;106:1886–1891. doi: 10.1073/pnas.0812945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shah SP, Kobel M, Senz J, Morin RD, Clarke BA, Wiegand KC, et al. Mutation of FOXL2 in granulosa-cell tumours of the ovary. N Engl J Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 206.Sugarbaker DJ, Richards WG, Gordon GJ, Dong L, De Rienzo A, Maulik G, et al. Transcriptome sequencing of malignant pleural mesothelioma tumours. Proc Natl Acad Sci USA. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 208.Bainbridge MN, Warren RL, Hirst M, Romanuik T, Zeng T, Go A, et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genom. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tsuchihara K, Suzuki Y, Wakaguri H, Irie T, Tanimoto K, Hashimoto S, et al. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37:2249–2263. doi: 10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hashimoto S, Qu W, Ahsan B, Ogoshi K, Sasaki A, Nakatani Y, et al. High-resolution analysis of the 5′ -end transcriptome using a next generation DNA sequencer. PLoS One. 2009;4:e4108. doi: 10.1371/journal.pone.0004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li H, Lovci MT, Kwon YS, Rosenfeld MG, Fu XD, Yeo GW. Determination of tag density required for digital transcriptome analysis: application to an androgen-sensitive prostate cancer model. Proc Natl Acad Sci USA. 2008;105:20179–20184. doi: 10.1073/pnas.0807121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 215.Aparicio SA, Huntsman DG. Does massively parallel DNA resequencing signify the end of histopathology as we know it? J Pathol. 2009 doi: 10.1002/path.2636. DOI:10.1002/path.2636. [DOI] [PubMed] [Google Scholar]
- 216.Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci USA. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumour progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang J, Finney RP, Rowe W, Edmonson M, Yang SH, Dracheva T, et al. Systematic analysis of genetic alterations in tumours using Cancer GenomeWorkBench (CGWB). Genome Res. 2007;17:1111–1117. doi: 10.1101/gr.5963407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.von Eschenbach AC, Buetow K. Cancer Informatics Vision: caBIG. Cancer Inform. 2007;2:22–24. [PMC free article] [PubMed] [Google Scholar]
- 222.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2009;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet. 2008;10 doi: 10.1002/0471142905.hg1011s57. Unit 10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations in Cancer. 2009 http://cgapncinihgov/Chromosomes/Mitelman.
- 228.Mitelman F. Catalogue of chromosome aberrations in cancer. Cytogenet Cell Genet. 1983;36:1–515. doi: 10.1159/000131930. [DOI] [PubMed] [Google Scholar]
- 229.Jackson MA, Lea I, Rashid A, Peddada SD, Dunnick JK. Genetic alterations in cancer knowledge system: analysis of gene mutations in mouse and human liver and lung tumours. Toxicol Sci. 2006;90:400–418. doi: 10.1093/toxsci/kfj101. [DOI] [PubMed] [Google Scholar]
- 230.Olivier M, Petitjean A, Teague J, Forbes S, Dunnick JK, den Dunnen JT, et al. Somatic mutation databases as tools for molecular epidemiology and molecular pathology of cancer: proposed guidelines for improving data collection, distribution, and integration. Hum Mutat. 2009;30:275–282. doi: 10.1002/humu.20832. [DOI] [PubMed] [Google Scholar]
- 231.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 232.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 233.Bean J, Riely GJ, Balak M, Marks JL, Ladanyi M, Miller VA, et al. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin Cancer Res. 2008;14:7519–7525. doi: 10.1158/1078-0432.CCR-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 235.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 238.Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6:519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 239.Alix-Panabieres C, Riethdorf S, Pantel K. Circulating tumour cells and bone marrow micrometastasis. Clin Cancer Res. 2008;14:5013–5021. doi: 10.1158/1078-0432.CCR-07-5125. [DOI] [PubMed] [Google Scholar]
- 240.Sandberg AA. Application of cytogenetics in neoplastic diseases. Crit Rev Clin Lab Sci. 1985;22:219–274. doi: 10.3109/10408368509165844. [DOI] [PubMed] [Google Scholar]
- 241.Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, et al. Cosmic 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.McCaughan F, Dear PH. Single-molecule genomics. J Pathol. 2009 doi: 10.1002/path.2647. DOI:10.1002/path.2647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.