Abstract
The integrin αvβ6 is an emergent biomarker for non-small cell lung cancer (NSCLC) as well as other carcinomas. We previously developed a tetrameric peptide, referred to as H2009.1, which binds αvβ6 and displays minimal affinity for other RGD-binding integrins. Here we report the use of this peptide to actively deliver paclitaxel to αvβ6–positive cells. We synthesized a water soluble paclitaxel-H2009.1 peptide conjugate in which the 2′-position of paclitaxel is attached to the tetrameric peptide via an ester linkage. The conjugate maintains its specificity for αvβ6–expressing NSCLC cells, resulting in selective cytotoxicity. Treatment of αvβ6–positive cells with the conjugate results in cell cycle arrest followed by induction of apoptosis in the same manner as free paclitaxel. However, initiation of apoptosis and the resultant cell death is delayed compared to free drug. The conjugate demonstrates anti-tumor activity in a H2009 xenograft model of NSCLC with efficacy comparable to treatment with free paclitaxel.
1. Introduction
Non-small cell lung cancer (NSCLC) is a notoriously deadly cancer with a 5-year survival rate of less than 15%.1 Paclitaxel (TAXOL®) is a widely used cytotoxic agent for the treatment of NSCLC as well as various other carcinomas. However, its clinical utility is hampered by poor aqueous solubility and non-selective toxicity. Delivery systems that specifically target paclitaxel to the tumor site are anticipated to (a) reduce systemic toxicity, (b) improve efficiency of paclitaxel delivery to tumors and increase its dwell time within the tumor, (c) lower the paclitaxel dosage required to achieve effective tumor growth reduction and (d) improve paclitaxel’s solubility. The nanoparticle formulation of paclitaxel, Abraxane® (Nab-paclitaxel), improves solubility and negates the need for formulation in polyethoxylated castor oil (Cremophor EL).2,3 Additionally, increased accumulation of paclitaxel is observed in tumors due to passive targeting of the nanoparticle that occurs as a result of the enhanced permeability and retention effect (EPR). However, patients treated with Nab-paclitaxel still suffer from neutropenia and neuropathy due to non-specific uptake in non-diseased tissues. As such, there is a continuing need to develop active targeting drug delivery systems that utilize homing ligands which transport therapeutics specifically to tumors that express a specific cell surface biomarker. Active targeting is anticipated to further improve drug accumulation and/or increase cellular uptake of a therapeutic while decreasing non-specific uptake in other organs.
Most efforts in active tumor targeting have focused on conjugating paclitaxel to monoclonal antibodies (MAbs) specific for cell surface cancer biomarkers in order to deliver paclitaxel preferentially to the tumor.4-9 Yet, chemically modifying antibodies is challenging, production costs are substantial, and, post-translational modifications on MAbs can trigger severe hypersensitivity reactions.10 Synthetic peptides have attracted attention as targeting molecules.11,12 Peptides are smaller than antibody-based targeting agents, can be synthesized in large quantities and are amenable to regiospecific derivatization13. The toxicity profile of peptides is low. Peptides can be chemically modified to alter affinity, charge, hydrophobicity, stability, and solubility.
Several peptide-paclitaxel conjugates have been reported. Cell penetrating peptides have been employed as carriers of paclitaxel.14,15 In addition, peptides that bind cell surface biomarkers have served as delivery vehicles for paclitaxel. Most of these are based on naturally occurring peptides such as bombesin and somatostatin or on the well-characterized αvβ3 binding peptide RGD.4,16-20 More recently, paclitaxel peptide conjugates that target HER-221 and GRP7815,22 have met with success in vitro. However, the number of targeting ligands needs to expand in order to address the diversity of cancer phenotypes found in the patient population. Moreover, most of these ligands are unsuitable for the treatment of NSCLC.
We previously identified a peptide from a phage displayed peptide library via biopanning on the NSCLC cell line H2009.23 This peptide, referred to as H2009.1, binds the integrin αvβ6 and displays minimal affinity for other RGD-binding integrins24. Expression of αvβ6 is widespread in early stage human NSCLC, and it is associated with poor patient survival.24,25 Additionally, this integrin is over-expressed in many epithelial derived carcinomas but is not found in normal primate tissues.26-36 In sum, the integrin αvβ6 is emerging as an important target for anticancer therapies. As such, our lab is focusing on developing the H2009.1 peptide for clinical use.
Tetramerization of the H2009.1 peptide dramatically increases its affinity for its cellular target resulting in a ligand with a half-maximal binding affinity of 50 pM.37 In addition, the H2009.1 tetrameric peptide mediates cellular uptake allowing for the chemotherapeutics to be delivered intracellularly. Finally, we have demonstrated that the peptide accumulates specifically in αvβ6–positive tumors but not in αvβ6–negative tumors nor in normal healthy tissues. Thus, this peptide has the properties necessary to be an effective homing ligand for drug delivery. In the present study, we report the synthesis and characterization of a water soluble, cancer cell specific delivery drug system, in which H2009.1 tetrameric peptide is conjugated to paclitaxel (Schemes I and II).
Scheme I.
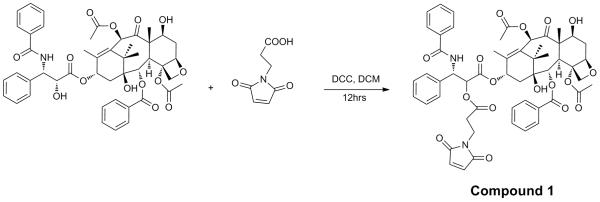
Synthesis of 2maleimido-paclitaxel was prepared via coupling paclitaxel with 3-maleimidopropionic acid (compound 1).
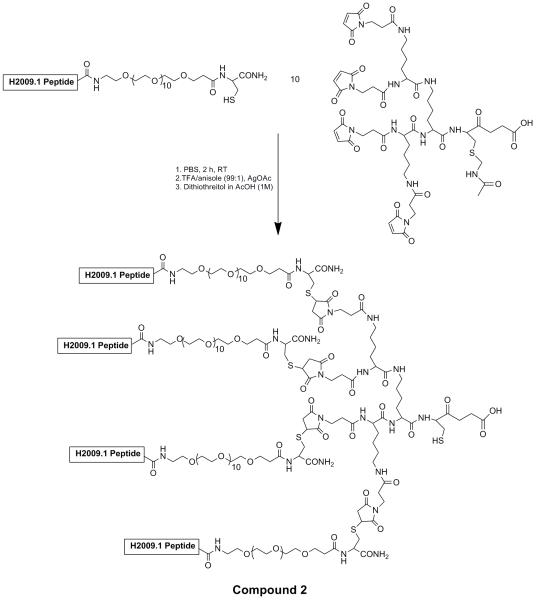
Scheme II. Synthesis of tetrameric H2009 peptide by a convergent coupling reaction (compound 2).
Michael addition of cysteine containing monomeric peptides with the maleimide trilysine core results in formation of the tetrameric peptide. A PEG11 linker is placed between the peptide and the core to improve solubility and prevent peptide aggregation. Removal of the acetamidomethyl protein group from the trilysine core by treatment with the Ag(OAc) reveals a unique thiol for coupling to compound 1.
2. Results and discussion
2.1. Synthesis and characterization of H2009.1 peptide-paclitaxel conjugate
The 2′-hydroxyl and 7′-hydroxyl groups of paclitaxel are suitable sites for conjugation to a targeting ligand, with preferential modification occurring at the 2′-hydroxyl group due to steric hindrance at the 7′-hydroxyl group. 2′-Maleimido-paclitaxel was prepared in 68% yield through coupling paclitaxel with 3-maleimidopropionic acid (Scheme I, compound 1). 1NMR analysis revealed a shift of the C-2′ proton to 5.47 ppm compared to 4.71 ppm in free paclitaxel, and the resonance of the 2′-OH proton at 3.55 ppm in paclitaxel disappeared. The resonance of the C-7′ proton was unaffected. These data confirm the formation of an ester bond at the 2′ hydroxyl group of paclitaxel.
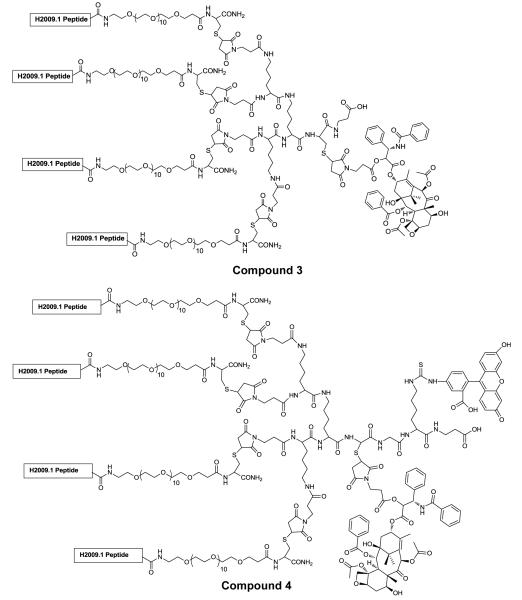
The H2009.1 10mer tetrameric peptide (Scheme II, compound 2) was synthesized separately by a convergent strategy published previously by our lab.37 Chemoselective reaction between the thiol of the trilysine peptide core with the 2′-maleimido-paclitaxel results in formation of the peptide-drug conjugate in ~90% yield (Figure 1, compound 3). The same reaction conditions were employed using a scrambled sequence tetrameric peptide (noted as scH2009.1) in order to provide a control peptide-paclitaxel conjugate. These two constructs have the same chemical composition but a different linear sequence. The H2009.1 peptide-paclitaxel conjugate and scrambled peptide-paclitaxel conjugate were obtained as white powder, which were soluble in cold water as well as in most polar organic solvents such as chloroform and methyl alcohol. The increased solubility of paclitaxel allows for use of the drug in buffered saline solutions and avoids the need for the use of Cremophor® EL.
Figure 1. Structure of H2009.1 peptide-paclitaxel conjugate (compound 3) and FITC-labeled H2009.1 peptide-paclitaxel conjugate (compound 4).
The H2009.1 peptide displays the sequence RGDLATLRQL and the scrambled H2009.1 peptide is DALRLQGTLR.
2.2. Cell-specific binding of H2009.1 tetrameric peptide-paclitaxel conjugate
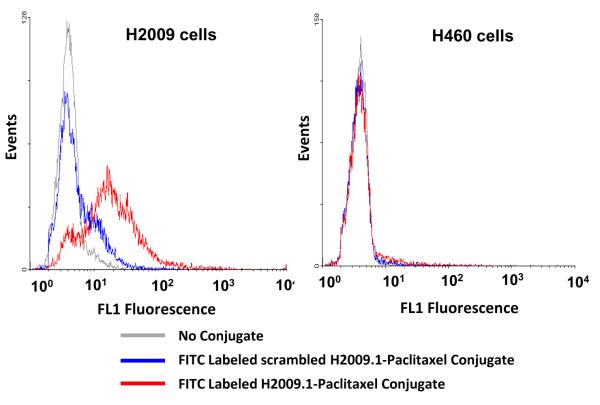
Although paclitaxel is incorporated in the trilysine core away from the peptide binding domain, it is important to verify that the addition of this hydrophobic drug does not destroy H2009.1’s binding nor affect its unique cell specificity. To address this issue, the peptide-paclitaxel conjugates were labeled with fluorescein (Figure 1, compound 4) and binding was quantified by flow cytometric analysis. When H2009 cells were treated with scrambled peptide-paclitaxel conjugate, no binding was observed (Figure 2). However, H2009 cells treated with labeled H2009.1 peptide-paclitaxel had a mean fluorescence intensity (MFI) 3.3-fold greater than that observed for the scrambled peptide conjugate, corroborating that binding is peptide-mediated and not affected by attachment of paclitaxel. H460 cells express the integrins αvβ3, α5β1, and αvβ5 but do not express αvβ6.24 As such, these cells are a good control with which to address specificity of the conjugate for αvβ6. H460 cells exhibit minimal binding to the H2009.1 peptide-paclitaxel conjugate, indicating that H2009.1 tetrameric peptide-paclitaxel selectively targets αvβ6 positive cells and addition of the hydrophobic drug does not increase binding to non-αvβ6 expressing cells.
Figure 2. The paclitaxel-H2009.1 conjugate binds specifically to αvβ6–expressing cells.
H2009 cells (αvβ6-positive) and H460 cells (αvβ6- negative) were incubated for 1 hour with 1 M paclitaxel-H2009.1 conjugate labeled with FITC (compound 4) or scrambled peptide-paclitaxel-dye conjugate. Binding of the conjugate was measured by flow cytometry. A total of 10,000 events were evaluated for fluorescence in channel 1 (excitation at 488 nm, emission at 500-550 nm).
2.2. Drug release study
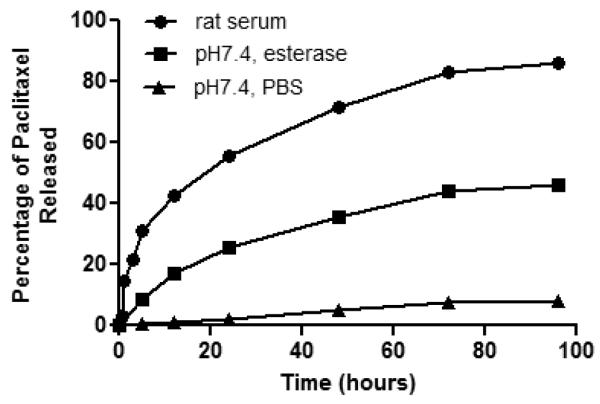
Paclitaxel must be released from the peptide carrier to be active; yet, the conjugate must remain stable until it reaches its tumor target. Our linker design is based on the hypothesis that the ester linkage will be stable until internalized into cells where increased concentration of esterases and proteases will lead to hydrolysis thus liberating paclitaxel. As the 2′-hydroxyl group is required for tubulin binding, the peptide-paclitaxel conjugate is anticipated to be inactive unless internalized38-40. To assure that the drug-conjugate is stable at neutral pH but hydrolyzed by esterases, release of paclitaxel from the H2009.1 peptide-paclitaxel conjugate was determined in both PBS buffer (pH 7.4) and esterase containing PBS buffer. In PBS, minimal paclitaxel was released even after 96 hours (Figure 3). Similar results were observed under acidic condition (pH 5.6 buffer, data not shown). The hydrolysis rate was increased in the presence of porcine esterase; 34.1% of paclitaxel was released by 48 h. This 7-fold increase of paclitaxel release indicates that hydrolysis is enhanced in the presence of esterase; however, drug release remains inefficient. By comparison, 70% of paclitaxel is released from the peptide-paclitaxel conjugate after 48 h incubation in rat serum, with a half-life of approximately 20 h. The half-life of the conjugate in serum is similar to that observed for other paclitaxel ester linkers.41 The increased release is likely due to participation of other proteases or esterases present in rat serum. This also raises some concern about the stability of the conjugate while in circulation; however, 86% of the conjugate remains intact in rat serum at 1 h. As the overall size of the complex is below the renal filtration cutoff, we believe most of the conjugate that does not reach the tumor will be cleared by the kidney within this time frame. Of note, the tetrameric peptide itself has a half-life of 12 h indicating that peptide stability will not be an issue within this context.
Figure 3. Paclitaxel is released from the conjugate in the presence of esterase and serum.
Compound 3 was dissolved in phosphate-buffered solutions (PBS, 0.01M) at pH 7.4 (▴), or esterase (18u/mL) in PBS (∎), or rat serum (●), individually. The solutions were incubated at 37° C. At time points 3, 8, 24, 48, 72, 96 h during incubation, the release of paclitaxel was analyzed by HPLC.
2.3. Evaluation of in vitro cytotoxicity
To evaluate the ability of the peptide conjugates to deliver active paclitaxel in a peptide-specific manner, the cytotoxicity of free paclitaxel, H2009.1 tetrameric peptide-paclitaxel conjugate and scrambled peptide-paclitaxel conjugate was determined (Table I). The IC50 value for H2009.1 peptide-paclitaxel conjugate is 460 nM on H2009 cells whereas the scrambled peptide conjugate does not reach a 50% reduction in cell viability, even at 1 μM. Thus, H2009.1 peptide is mediating cellular delivery of paclitaxel in a sequence dependent manner. There is a reduction in the potency of the paclitaxel when it is conjugated to the peptide (IC50 of free paclitaxel: 15 nM). This could be due to reduced uptake of the drug, slow release of paclitaxel from its carrier, and/or drug delivery to and retention in an inappropriate cellular compartment. Drug conjugates often have reduced effectiveness compared to their non-conjugated partners. Other peptide-paclitaxel conjugates have shown reduced efficacy under short incubation times but the reason for the observation has not been determined.16,41 However, improved biodistribution of the targeted drug conjugates in vivo can often overcome the loss of activity.
Table I.
IC50 Values of Paclitaxel Conjugatesa
| H2009 Cells | H460.1 | |
|---|---|---|
| Paclitaxel | 14.8 ± 3.3 nM | 23.6 ± 7.4 nM |
| H2009.1-Paclitaxel | 460 ± 75 nM | Not Reachedb |
| Scramble H2009.1-Paclitaxel | Not Reachedb | Not Reachedb |
Cells were exposed to drug for 10 min followed by a 120 h recovery in media. The short incubation time assures that cytotoxicity is due to the peptide conjugate and not free pacliatxel released prematuraly from the conjugate.
Not reached at highest concentration used, 1 μM.
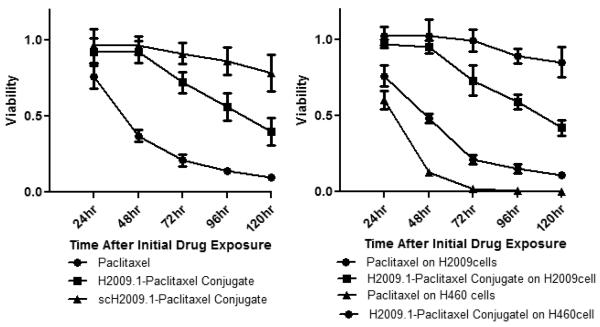
To understand the time frame in which cell death is initiated, H2009 cells were incubated with 1 μM of each compound for 10 min followed by different time points of recovery. In the presence of free paclitaxel, a rapid reduction of cell viability was observed within the first 48 h. Neither H2009.1 10mer tetrameric peptide-paclitaxel conjugate nor scrambled peptide-paclitaxel conjugate exhibited any effect within the first 48 h. However, H2009.1-paclitaxel conjugate began to reduce cell viability at 72h and inhibition of 40% is observed at 120 h. By contrast, the scrambled peptide-paclitaxel conjugate exhibited less than 15% cell growth inhibition at 120 h. As the cells are only exposed to the drug conjugates for 10 minutes, the delayed response is not due to a slow accumulation of conjugate within the cells. Instead, this data suggests that drug release is a limiting factor and that sufficient exposure time is essential for the drug to be released from the conjugate. This is consistent with the in vitro drug release assays which show release of paclitaxel from the peptide to be inefficient. However, we cannot rule out the possibility that drug release is rapid yet transportation to the cytoplasm is rate limiting.
To determine whether the H2009.1 conjugate is specifically cytotoxic toward αvβ6 positive cells, H2009 and αvβ6-negative H460 cells were incubated in the presence of 1 μM of unconjugated paclitaxel or of H2009.1 peptide-paclitaxel conjugate (Figure 4). Both cell types are sensitive to free paclitaxel; the IC50 of paclitaxel on H460 cells is 24 nM, similar to that observed on H2009. However, after treatment with H2009.1 peptide-paclitaxel conjugate, the αvβ6 negative H460 cells remained almost unaffected while the H2009 cells were sensitive to the conjugate. At 120 h, the difference in viability between the two cell types is 3.9-fold. In sum, conjugation of paclitaxel to H2009.1 converts paclitaxel from a general cytotoxic agent to one that only affects αvβ6–positive cells. Thus, while decreased in vitro efficacy is observed with the H2009.1-paclitaxel conjugate compared to free paclitaxel, the conjugate has a wider therapeutic window.
Figure 4. The H2009.1-paclitaxel conjugate displays αvβ6–selective cytotoxicity that is time dependent.
Left Panel: H2009 cells were exposed to 1 μM Taxol, H2009.1 peptide-paclitaxel (compound 3) and scH2009.1 peptide-paclitaxel for 10min, washed, and incubated further in fresh medium for different time point. The cell viability is normalized to untreated control cells. Right Panel: H2009 cells and H460 cells were exposed to 1 μM Taxol and compound 3 for 10min, washed, and incubated further in fresh medium for indicated times. The short exposure time to the conjugates is necessary to assure that the affects observed are due to the conjugate and not paclitaxel released from the peptide before internalization.
2.4. Competition assay
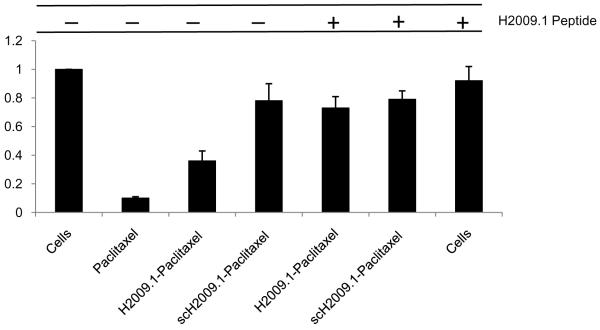
To further assure that the H2009 peptide delivers paclitaxel in a receptor dependent fashion, we sought to determine if free H2009.1 peptide could block the toxicity of the H2009.1-paclitaxel conjugate. H2009 cells were treated with a 10-fold excess of free H2009.1 peptide, followed by treatment with the H2009.1 peptide-paclitaxel conjugate. At 120 h after exposure to the conjugate, cells treated first with free peptide were significantly protected from the cytotoxic effects of the H2009.1-paclitaxel conjugate; cell viability was 73 ± 0.08% for samples pre-treated with free H2009.1 vs. 36 ± 0.07% for cells that were incubated with the drug conjugate alone (Figure 5), restoring viability to that observed with the scrambled peptide-paclitaxel conjugate (79 ± 0.06%). Additionally, free H2009.1 tetrameric peptide showed only slight inhibition of cell proliferation on H2009 cells (0.92 ± 0.10 % cell viability). These data support that conjugate internalization is a receptor-mediated process. The need for a 10-fold excess of free peptide to block the peptide-paclitaxel conjugate’s cytotoxicity can be attributed to the high avidity of the conjugate and/or a rapid recycling of αvβ6.
Figure 5. The affect of H2009.1-paclitaxel conjugate on cell viability is blocked by pretreatment with H2009.1 peptide.
H2009 cells were treated with or without 10 μM H2009.1 10-mer tetrameric peptide for 1 hour, washed, and then treated with 1 μM H2009.1-paclitaxel conjugate (compound 3) or scH2009.1 peptide-paclitaxel. Viability was determined at 96 h.
2.5. Cell cycle analysis
Paclitaxel induces G2/M cell cycle arrest resulting in apoptosis. Flow cytometry analysis was performed to analyze the cell cycle perturbations of H2009 cells treated with the H2009.1 peptide-paclitaxel conjugate to assure that the conjugate is functioning in a similar manner as free paclitaxel (Table II). At 24h, G2/M cell cycle arrest was observed for H2009 cells treated with 1 μM free paclitaxel. When H2009 cells were treated with 1 μM H2009.1 peptide-paclitaxel conjugate, the cell cycle remains unperturbed at 24 h. However, at 48 h, the number of cells in G2/M increases by ~2-fold and a 3-fold increase in polyploid cells is observed. Increasing the peptide-paclitaxel conjugate concentration also increases the percentage of cells in cycle arrest. Approximately 50% more G2/M arrest occurred with a decrease of cells in G1 phase at 24 h. The effect of paclitaxel on the cell-cycle progression of H2009 cells thus occurs in a dose-dependent manner. These data support the conclusion that the H2009.1-paclitaxel conjugate induces cell cycle arrest as observed for the free drug. However, the conjugate shows a delayed effect as time is required to accumulate sufficient concentrations of liberated paclitaxel.
Table II.
Cell Cycle Analysis of H2009 Cells Treated with Paclitaxel-Peptide Conjugatesa
| 0 μM | 1 μM | 5 μM | ||||
|---|---|---|---|---|---|---|
| control cells | paclitaxel | conjugate 24 h |
conjugate 48 h |
conjugate 24 h |
conjugate 48 h |
|
| G1 | 50.9 | 8.5 | 44.9 | 11.4 | 35.8 | 23.9 |
| S | 11.2 | 4.1 | 8.1 | 4.5 | 6.7 | 8.5 |
| G2/M | 31.1 | 73.5 | 39.0 | 57.1 | 45.8 | 53.9 |
| Polyploid | 6.8 | 14 | 8.0 | 26.9 | 11.7 | 23.6 |
Represents the average of two experiments
2.6. Characterization of drug-induced apoptosis
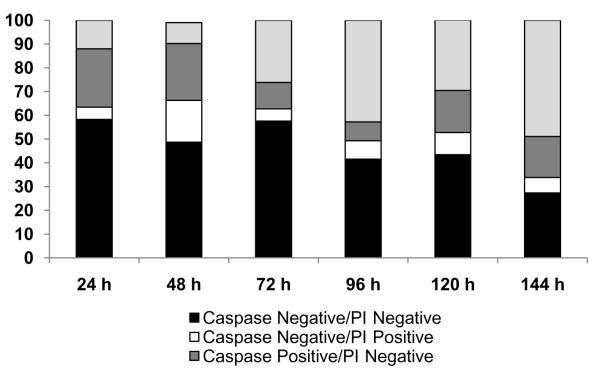
Initiation of apoptosis is characterized by activation of a system of caspases. To determine the time frame in which this process begins upon treatment with H2009.1 peptide-paclitaxel conjugate, caspase activation was followed by flow cytometry using Vybrant ® FAM Poly Caspases Assay. Propidium iodine was employed to detect non-viable cells. At 24h, 37% of cells treated with H2009.1 peptide-paclitaxel conjugate were positive for caspase activation. Of this subpopulation, 70% are PI negative, suggestive of cells in early apoptosis. As time progressed, the number of caspase and PI double positive cells increased (Figure 6). At day 4, 43% of the cell population is positive for caspase activation and PI staining, indicating the cells are in late apoptosis and have lost integrity of the plasma membrane. By comparison, cells treated with the control peptide conjugate exhibit minimal caspase activation with a maximum of 14% caspase positive cells at 96h. These data again confirm that the H2009.1 peptide specifically delivers paclitaxel. The time course of H2009.1 peptide-conjugate induced apoptosis is similar to that observed for both the cell cycle arrest and cytotoxicity studies, again indicating that the H2009.1-paclitaxel conjugate shows delayed action. However, it should be noted that at 96-h, 40% of the cells treated with the H2009.1-paclitaxel conjugate still remain caspase negative and PI negative. At 144 h, this population drops to 27%. Thus there is a significant subpopulation of viable cells that are not affected. It is possible that there is a subpopulation of cells that do not internalize the peptide or do not accumulate enough paclitaxel during the 10 min exposure to be effective. This is consistent with the flow cytometry data which reveals a small subpopulation of cells that do not bind significant amounts of the conjugate. This highlights a concern in using targeted therapies; due to heterogeneity of the tumor, small subsets of cells may remain refractory to treatment. Expanding the suite of tumor targeting ligands is anticipated to minimize this problem by allowing a greater percentage of the tumor to be targeted.
Figure 6. H2009.1-pacliatxel activates caspases and initiates apoptosis.
H2009 cells were exposed to 1 μM H2009.1 H2009.1-paclitaxel conjugate for 10 min, washed, and incubated in fresh media for indicated times. Floating and attached cells were collected and subjected to caspases/PI staining using Vybrant® FAM Poly Caspases Assay and analyzed by flow cytometry. 10,000 events were measured using channel 1 (excitation 488 nm, emission 500 nm) and channel 3 (excitation 488 nm, emission 650 nm). The data represents the average of two experiments.
2.7. Tumor growth studies
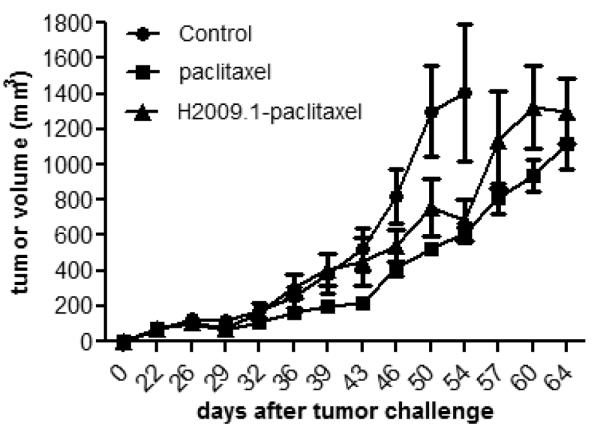
While drug conjugates often have reduced efficacy in vitro compared to their non-conjugated partners, the opposite is often true in vivo as improved biodistribution of the targeted drug conjugates can overcome the loss of activity. We determined the efficacy of the H2009.1-paclitaxel conjugate in a H2009 xenograph model. H2009 cells were implanted subcutaneously on the flank of immunocompromised mice. At day 18 when palpable tumors had formed, the animals were treated with saline, 5 mg/kg paclitaxel or 5 mg/kg H2009.1-paclitaxel (based on the total amount of paclitaxel). A total of 5 injections per mouse were given every three days. While the H2009.1-paclitaxel conjugate reduced the rate of tumor growth compared to saline, there is no statistical difference between free paclitaxel and the conjugate in terms of tumor growth rate or survival (Figure 7). Similarly, the animal weights remained steady for both groups suggesting that no gross toxicity was observed (data not shown). Thus, conjugation to the targeting peptide did not provide any obvious benefit over the free drug in vivo. Interestingly, the effect of the H2009.1-paclitaxel is delayed; the growth rates for the control animals and those treated with H2009.1-paclitaxel conjugate do not begin to diverge until day 46. This is 7 days after the free drug begins to show an effect on tumor growth. Also of note, the conjugate does not decrease the performance of paclitaxel despite its higher IC50 found in vitro. Complete biodistribution and pharmacokinetic experiments are needed to determine if the observed results are due to a lack of drug targeting, inefficient drug release, premature drug release, or poor distribution throughout the tumor. As the conjugate is below the renal filtration limit, rapid clearance by the kidneys may contribute to the lack of efficacy; however free paclitaxel is also cleared by the kidneys and previous studies have demonstrated accumulation of the peptide in positive tumors despite the rapid renal filtration. Additionally, maximally tolerated dosage needs to be examined; it is possible that higher doses of paclitaxel can be administered as a conjugate because of reduced off target effects.
Figure 7. Tumor growth rate is slowed by free paclitaxel and H2009.1-paclitaxel conjugate by similar amounts.
Subcutaneous tumors were established on the flank of NOD/SCID mice (n=5). At day 18, saline, free paclitaxel and compound 3 were injected intravenously via the tail vein with 5mg/kg, based on paclitaxel weight. Animals were treated at days 18, 21, 24, 27, and 30. Measurements were made with calipers by an independent scientist. Tumor volumes are calculated as V = l × 2w/2. Saline (●), Paclitaxel (∎), H2009.1-paclitaxel (▴).
3. Conclusions
In sum, the H2009.1 peptide is able to selectively deliver a chemotherapeutic agent to αvβ6 positive cells, thus opening the therapeutic window. Our data indicate that this conjugate effects cell death via cell cycle arrest followed by the induction of apoptosis in the same manner as paclitaxel. However, the time frame of activity is delayed compared to free drug in vitro. We believe this is due to inefficient release of paclitaxel or poor intracellular trafficking to the cytoplasm where paclitaxel exerts its effects. Ester based conjugates to paclitaxel have met with mixed results in vitro and in vivo. Future optimization of drug linkers will be needed for peptide-paclitaxel conjugates to be viable. Recently, self-immolative linkers based on para-aminobenzylalcohol and ethylenediamine have shown promising results in vitro and in vivo.14,15,42,43 These linkers undergo spontaneous drug release may be more appropriate for paclitaxel delivery. However, drug release will depend on the subcellular localization of the conjugate. As such, effective linkers for one targeting ligand may not be suitable for another. In addition, further studies need to be performed to determine absolute uptake of paclitaxel in order to assure that this is not limiting the conjugate’s effectiveness.
4. Experimental
4.1. Materials
All reagents, unless specified, were of analytical grade and purchased commercially. Paclitaxel was obtained from LC Laboratories (Woburn, MA). N,N’-Dicyclohexyl-carbodiimide (DCC) and N-Methylmorpholine (NMM) were purchased from ACROS Organics (Geel, Belgium). All Fmoc-protected amino acids and 2-(1H-benzotriazole-l-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were purchased from EMD Biosciences Inc (San Diego, CA). Anhydrous Hydroxybenzotriazole (HOBt) was purchased from SynBioSci (Livermore, CA). 3-Maleimidopropionic, Piperidine, Porcine liver esterase, and rat serum were purchased from Sigma-Aldrich Inc (St. Louis, MO).
4.2. Cell lines
Human lung cancer cell lines were obtained from the Hamon Center for Therapeutic Oncology Research (UTSW) and were maintained according to standard protocols.44 RPMI1640 was purchased from Mediatech (Herndon, VA). Fetal Bovine Serum (FBS) was purchased from Gemini Bio-products (Woodland, CA). Enzyme-free Cell Dissociation buffer was from Invitrogen (Grand Island, NY). R5 media consists of RPMI supplemented with 5% FBS.
4.3. Synthesis of 2′-maleimido-paclitaxel (compound 1)
Paclitaxel (8.6 mg, 10 μmol) was incubated with 3-Maleimidopropionic acid (1.0 mg, 6 μmol) and dicyclohexylcarbodiimide (4.1 mg, 20 μmol ) in 500 μL anhydrous dichloromethane for 12 hrs at RT (Scheme 1). Dicyclohexylurea was removed by filtration. The organic solvent was removed under vacuum and the solid was resuspended in 1 mL CH3OH:H2O (50:50 V/V) containing 0.1% trifluoroacetic acid. The reaction mixture was purified by reverse phase HPLC with eluents of H2O/0.1% TFA (eluent A) and acetonitrile/0.1%TFA (eluent B). The following elution profile (referred to as Method A) was utilized: 0-1 minute, 70%A, 30%B; 1-71 minutes, eluent B was increased from 30-100% at a flow rate of 10 mL /minute. Elution of the conjugate was monitored by UV absorbance at 220 nM. The purified conjugate was characterized by MALDI Mass Spectra and 1H NMR. Compound 2 (C54H56N2O17 ): MALDI MNa+ (monoisotopic mass calculated/found: 1027.36/1027.19). 1H NMR reported in PPM (CDCl3): 1.82 (s; 1H; 1-OH); 5.68 (d; 1H; H-2); 3.80 (d; 1H; H-3); 2.45 (s; 3H; 4-OAc); 4.97 (d; 1H; H-5); 2.54 (m; 1H; Ha-6); 1.98 (m; 1H; Hb-6); 4.43( m; 1H; H-7); 2.40 (d; 1H; 7-OH); 6.28 (s; 1H; H-10); 2.23 (s; 3H; 10-OAc); 6.19 (t; 1H; H13); 2.20-2.30 (band; 2H; H14); 1.24 (s; 3H; Me-16); 1.14 (s; 3H; Me-17); 1.68 (s; 3H; Me-18); 1.93 (s; 3H; Me-19); 4.32 (d; 1H; Ha-20); 4.21 (d; 1H; Hb-20); 5.47 (d; 1H; H-2′); 6.06 (dd; 1H; H-3′); 6.98 (d; 1H; 3′-NH); 7.62-8.13 (m; 5H; C2-OBz); 7.32-7.52 (m; 5H; C3′-Ph); 7.49-7.77 (m; 5H; C3′-NBz) and 2.75 (t; 2H); 3.84 (t; 2H); 6.51 (s; 2H) contributed to 3-maleimidopropionic acid.
4.4. Synthesis of H2009.1 10-mer tetrameric peptide (compound 2)
H2009.1 10-mer tetrameric peptide with a free thiol group was obtained by convergent synthesis followed by deprotection of the acetamidomethyl-cysteine to reveal a free thiol as previously described.37 The peptide was purified by reverse phase HPLC using a Vydac PR-C18 column (250mm ×22mm, 10 μm) on a Breeze™ HPLC (Water Inc.) with eluents of H2O/0.1% TFA (eluent A) and acetonitrile/0.1%TFA (eluent B). The following elution profile (referred to as Method B) was utilized: 0-1 minute, 90%A,10%B; 1-61 minutes, eluent B was increased from 10-40% at a flow rate of 10 mL /minute. Elution of the peptides was monitored by UV absorbance at 220 nM. The peptide mass was confirmed by matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI MS) in linear mode using sinapinic acid as matrix on a Voyager DE™ Pro instrument (Applied Biosystems Inc., Foster City, CA). Compound 1 (C364H652N92O130S5 ): MALDI MH+ (average mass calculated/found: 8557.94/8558.38).
4.5. Conjugation of H2009.1 10mer tetrameric peptide to 2′-maleimido-paclitaxel (compound 3)
A solution of compound 1 (17.1 mg, 2 μmol) was prepared in 0.8 mL Ar-purged 1×PBS/0.01M EDTA. A solution of compound 2 (3.0 mg, 3 μmol) in 0.2 mL DMF was added to the peptide solution. The reaction mixture was stirred at RT for 30 min. The product, H2009.1 10-mer tetrameric peptide-paclitaxel conjugate (compound 3) was then purified with RP-HPLC using the Method A and characterized by MALDI Mass Spectra. Compound 3 (C418H708N94O147S5): MALDI MH+ (average mass calculated/found: 9562.96/9561.75).
4.6. Synthesis of H2009.1 peptide-Paclitaxel-FITC (compound 4)
The maleimido tetrameric core for fluorescein isothiocyanate (FITC) labeling was synthesized on Fmoc-β-Ala-CLEAR™ Acid Resin, (substitution level 0.52 mmol/g) using standard protocols. Fmoc-Lys(Boc)-OH and Fmoc-Cys(Acm)-OH was coupled at a 5-fold excess using HBTU, HOBt and NMM coupling (45 min). Piperidine in DMF (20%) was employed to remove N-terminal Fmoc protecting groups. Fmoc-Lys(Fmoc)-OH and 3-maleimidopropionic acid was coupled in the same fashion. Upon completion of the synthesis, the maleimido tetrameric core was cleaved from the resin using a TFA: triisopropylsilane: H2O cocktail (95%:2.5%:2.5%) and precipitated in cold diethyl ether. The crude tetrameric core was purified by reverse phase HPLC using method B.
The free ε-amino group of Lys on the maleimido tetrameric core was labeled with FITC. A solution of maleimido tetrameric core with Lys (ε-NH2) (2.9 mg, 2 μmol) was prepared in 0.5 mL 0.1M borate buffer ( pH 9.3). A solution of FITC (1.2 mg, 3 μmol) in 0.5 mL DMF was added to the tetrameric core solution. The reaction mixture was stirred at RT in the dark for 3 h and purified by reverse phase HPLC using method B. The FITC labeled maleimido tetrameric core was applied for convergent synthesis of H2009.1 10-mer tetrameric peptide and Paclitaxel conjugation as described above.
4.7. In vitro release of Paclitaxel from H2009.1 10-mer tetrameric peptide-Paclitaxel conjugate
Compound 3 was dissolved in phosphate-buffered solutions (PBS, 0.01M) at pH 7.4, or esterase (18u/mL) in PBS, or rat serum, individually. The solutions were incubated at 37° C. At time points 3, 8, 24, 48, 72, 96 h during incubation, aliquots were removed and mixed with 100 μL of ethanol to precipitate the serum proteins. The mixture was centrifuged at 3,000 rpm for 5 min. The pellets were resuspended with 80% ethanol, followed by centrifugation at 3,000 rpm again for 5 min. The pellets were rinsed and washed one more time. Then the pooled supernatants were analyzed by HPLC to determine the amounts of paclitaxel liberated from the conjugate. Paclitaxel was quantified by HPLC using a standard curve prepared with authentic paclitaxel. UV-Visible detection at 220 nm was used for data collection and analysis.
4.8. Cell viability assay
H2009 and H460 cells were seeded in 96 well culture plates at 1000 cells/well in a volume of 50 μL R5 medium and allowed to adhere overnight at 37° C in humidified atmosphere of 5% CO2. Paclitaxel or H2009.1 peptide-paclitaxel conjugate or scrambled peptide-paclitaxel conjugate were added to quadruplicate wells in 50 μL R5 media to produce the final concentrations indicated (ranging from 1 nM to 1 μM, all based on total paclitaxel concentration). Untreated control cells received 50 μL R5 only and were cultured as a set of eight replicate wells per plate. After 10 min of exposure to the drug, media was aspirated from all wells, and the wells were washed four times with 200 μL R5 before continuing the culture in 100 μL R5 for varying periods of time (from 24 h to 120 h). At the end of the post-treatment incubation cell viability was assessed using CellTiter Glo™ reagent (Promega, Madison, WI). Luminescence was detected using a plate reader 30-60 min after the addition of the CellTiter Glo™ reagent.
4.9. Binding of H2009.1 tetrameric peptide-paclitaxel-FITC by flow cytometry
Approximately 100,000 cells were seeded per well in 12 well culture plates and allowed to adhere overnight. Cells were incubated for 1h with 1 μM H2009.1 10mer tetrameric peptide-paclitaxel-FITC or scrambled peptide-paclitaxel-FITC. Cells were washed four times with PBS+ containing 0.1% BSA, followed by 2 brief rinses with 20 mM HCl-glycine, pH 2.2, 150 mM NaCl, and a final one with PBS. Cells were removed from the wells by incubation on ice for 30 mins in 1 mL/well of Enzyme-free Cell Dissociation Buffer. Cells were scraped from the plate and prepared as a single cell suspension by passage through a 27 gauge needle. FITC-peptide-paclitaxel binding was assessed for 10,000 cells per treatment group by flow cytometry using a CellQuanta™ flow cytometer. Cells were gated by size and side scatter properties, and FITC assayed by fluorescence in channel 1 (excitation 395 nm, emission 488 nm).
4.10 Drug competition assay
H2009 cells were plated at 1000 cell/well in 96-well plates one day prior to the treatment to allow the cells to adhere to the plate. The next day, the cells were pre-incubated with 10 μM H2009.1 10-mer tetrameric peptide for 1h then 1 μM H2009.1 10mer tetrameric peptide-paclitaxel conjugate was added. After 10 min, cells were washed three times with 200 μL R5 before continuing culture in 100 μL R5 for 120 H. Cell viability was analyzed as described in section 4.8.
4.11. Cell cycle analysis
Cell cycle perturbations induced by H2009.1 10mer tetrameric peptide-paclitaxel conjugate were analyzed by propidium iodide (PI) DNA staining with flow cytometric analysis. H2009 cells were seeded in 12 well culture plates at approximately 100,000 cells per well and allowed to adhere overnight. Cells were treated with paclitaxel or H2009.1 10mer tetrameric peptide-paclitaxel conjugate at different concentrations for 10 min. Drugs were removed and the cells washed with R5 three times followed by recovering for 24 or 48 h. At the end of each treatment, cells were collected, resuspended in 0.5 mL PBS and fixed in 70% ethanol for 12 h at 4° C. Ethanol-suspended cells were centrifuged at 3000 rpm for 5 min and washed twice in PBS to remove residual ethanol. Cell pellets were suspended in 1 mL of PBS containing 0.02mg/mL of PI, 0.5mg/mL of DNase-free RNase A and incubated at 37°C for 1h. Cell cycle profiles were studied using CellQuanta™ flow cytometer and data were analyzed by WinMDI 2.9 software.
4.12. Apoptosis assay
Drug induced apoptosis was analyzed by Vybrant® FAM Poly Caspases Assay which detects activated caspases 1, 3, 4, 5, 6, 7, 8, and 9 (Invitrogen, Carlsbad, CA). Briefly, H2009 cells were plated and exposed to 1 μM drug for 10 min followed by different recovery times. Both suspended and adherent cells were collected and subjected to caspases/PI staining using Vybrant® FAM Poly Caspases Assay Kit according to the protocol provided by the manufacturer. Stained cells were analyzed on a CellQuanta™ flow cytometer. Data were analyzed by WinMDI 2.9 software.
4.13 In vivo tumor growth
Female NOD/SCID mice were injected with 1 million H2009 cells in the right flank. At day 18, after palpable tumors had formed, mice (5 per group) were injected via tail vein with PBS, 5mg/kg of free paclitaxel, or 5mg/kg of H2009.1 tetrameric peptide-paclitaxel. The mice were treated every 3 days for a total of 5 injections. Tumor growth was determined by measuring tumor length and width with calipers, and tumor volume was calculated using the formula v = (l × w2)/2, where l is length and w is width.
Supplementary Material
Acknowledgments
This work was supported by the Welch Foundation (I1622 to KCB) and a Career Development Award to KCB from the Specialized Program of Research Excellence in Lung Cancer (grant P50CA70907). BPG is supported by a fellowship from the Cancer Research and Prevention Institute of Texas (RP 101496).
Abbreviations
- NSCLC
Non-small cell lung cancer
- MAb
Monoclonal antibody
- H2009.1
Peptide sequence: RGDLATLRQL
- scH2009.1
Peptide sequence: DALRLQGTLR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary materials The 1HNMR of compound 1 and analytical HPLC traces for compounds 3 and 4 are included.
References and Notes
- 1.Jemal A, Siegel R, Xu J, Ward E. CA Cancer J Clin. 2010;60:277. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Ibrahim NK, Desai N, Legha S, Soon-Shiong P, Theriault RL, Rivera E, Esmaeli B, Ring SE, Bedikian A, Hortobagyi GN, Ellerhorst JA. Clin Cancer Res. 2002;8:1038. [PubMed] [Google Scholar]
- 3.Henderson IC, Bhatia V. Expert Rev Anticancer Ther. 2007;7:919. doi: 10.1586/14737140.7.7.919. (25) [DOI] [PubMed] [Google Scholar]
- 4.Safavy A. Curr Drug Del. 2008;5:42. doi: 10.2174/156720108783331005. [DOI] [PubMed] [Google Scholar]
- 5.Quiles S, Raisch KP, Sanford LL, Bonner JA, Safavy A. J Med Chem. 2009;53:586. doi: 10.1021/jm900899g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safavy A, Bonner JA, Waksal HW, Buchsbaum DJ, Gillespie GY, Khazaeli MB, Arani R, Chen D, Carpenter M, Raisch KP. Bioconj. Chem. 2003;14:302. doi: 10.1021/bc020033z. [DOI] [PubMed] [Google Scholar]
- 7.Safavy A, Georg GI, Velde DV, Raisch KP, Safavy K, Carpenter M, Wang W, Bonner JA, Khazaeli MB, Buchsbaum DJ. Bioconj Chem. 2004;15:1264. doi: 10.1021/bc049868v. [DOI] [PubMed] [Google Scholar]
- 8.Wu AM, Senter PD. Nat. Biotech. 2005;23:1137. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert CW, McGowan EB, Seery GB, Black KS, Pegram MD. J Exp Ther Oncol. 2003;3:27. doi: 10.1046/j.1359-4117.2003.01064.x. [DOI] [PubMed] [Google Scholar]
- 10.Chung CH, Mirakhur B, Chan E, Le Q-T, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, Slebos RJ, Zhou Q, Gold D, Hatley T, Hicklin DJ, Platts-Mills TAE. N Engl J Med. 2008;358:1109. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reubi JC, Maecke HR. J Nucl Med. 2008;49:1735. doi: 10.2967/jnumed.108.053041. [DOI] [PubMed] [Google Scholar]
- 12.Chen K, Conti PS. Adv Drug Deliv Rev. 2010;62:1005. doi: 10.1016/j.addr.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Bray BL. Nat. Rev. Drug Discov. 2003;2:587. doi: 10.1038/nrd1133. [DOI] [PubMed] [Google Scholar]
- 14.Dubikovskaya E, Thorne S, Pillow T, Contag C, Wender P. Proc Natl Acad Sci USA. 2008;105:12128. doi: 10.1073/pnas.0805374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoneda Y, Steiniger SCJ, Capková K, Mee JM, Liu Y, Kaufmann GF, Janda KD. Bioorg Med Chem Lett. 2008;18:1632. doi: 10.1016/j.bmcl.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryppa C, Mann-Steinberg H, Biniossek ML, Satchi-Fainaro R, Kratz F. Int J Pharm. 2009;368:89. doi: 10.1016/j.ijpharm.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 17.Papas S, Akoumianaki T, Kalogiros C, Hadjiarapoglou L, Theodoropoulos PA, Tsikaris V. J Pept Sci. 2007;13:662. doi: 10.1002/psc.899. [DOI] [PubMed] [Google Scholar]
- 18.Huang C-M, Wu Y-T, Chen S-T. Chem Biol. 2000;7:453. doi: 10.1016/s1074-5521(00)00131-9. [DOI] [PubMed] [Google Scholar]
- 19.Safavy A, Raisch KP, Khazaeli MB, Buchsbaum DJ, Bonner JA. J Med Chem. 1999;42:4919. doi: 10.1021/jm990355x. [DOI] [PubMed] [Google Scholar]
- 20.Safavy A, Raisch K, Matusiak D, Bhatnagar S, Helson L. Bioconjug Chem. 2006;17:565. doi: 10.1021/bc050224c. [DOI] [PubMed] [Google Scholar]
- 21.Guillemard V, Nedev HN, Berezov A, Murali R, Saragovi HU. DNA Cell Biol. 2005;24:351. doi: 10.1089/dna.2005.24.351. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y, Lillo AM, Steiniger SCJ, Liu Y, Ballatore C, Anichini A, Mortarini R, Kaufmann GF, Zhou B, Felding-Habermann B, Janda KD. Biochemistry. 2006;45:9434. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- 23.Oyama T, Sykes KF, Samli KN, Minna JD, Johnston SA, Brown KC. Cancer Lett. 2003;202:219. doi: 10.1016/j.canlet.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Elayadi AN, Samli KN, Prudkin L, Liu YH, Bian A, Xie XJ, Roth JA, Wistuba II, McGuire MJ, Brown KC. Cancer Res. 2007;67:5889. doi: 10.1158/0008-5472.CAN-07-0245. [DOI] [PubMed] [Google Scholar]
- 25.Prudkin L, Liu D, Ozburn N, Sun M, Behrens C, Tang X, Brown K, Bekele B, Moran C, Wistuba I. Mod Pathol. 2009;22:668. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed N, Riley C, Rice GE, Quinn MA, Baker MS. J Histochem Cytochem. 2002;50:1371. doi: 10.1177/002215540205001010. [DOI] [PubMed] [Google Scholar]
- 27.Arihiro K, Kaneko M, Fujii S, Inai K, Yokosaki Y. Breast Cancer. 2000;7:19. doi: 10.1007/BF02967183. [DOI] [PubMed] [Google Scholar]
- 28.Bates RC, Bellovin DI, Brown C, Maynard E, Wu B, Kawakatsu H, Sheppard D, Oettgen P, Mercurio AM. J. Clin. Invest. 2005;115:339. doi: 10.1172/JCI23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breuss JM, Gallo J, DeLisser HM, Klimanskaya IV, Folkesson HG, Pittet JF, Nishimura SL, Aldape K, Landers DV, Carpenter W, Gillette M, Sheppard D, Matthay MA, Albelda SM, Kramer RH, Pytela R. J. Cell Sci. 1995;108:2241. doi: 10.1242/jcs.108.6.2241. [DOI] [PubMed] [Google Scholar]
- 30.Kawashima A, Tsugawa S, Boku A, Kobayashi M, Minamoto T, Nakanishi I, Oda Y. Pathol. Res. Pract. 2003;199:57. doi: 10.1078/0344-0338-00355. [DOI] [PubMed] [Google Scholar]
- 31.Hamidi S, Salo T, Kainulainen T, Epstein J, Lerner K, Larjava H. Br J Cancer. 2000;82:1433. doi: 10.1054/bjoc.1999.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazelbag S, Kenter G, Gorter A, Dreef E, Koopman L, Violette S, Weinreb P, Fleuren G. J Pathol. 2007;212:316. doi: 10.1002/path.2168. [DOI] [PubMed] [Google Scholar]
- 33.Hecht J, Dolinski B, Gardner H, Violette S, Weinreb P. Appl Immunohistochem Mol Morphol. 2008;16:543. doi: 10.1097/PAI.0b013e31816bc5ee. [DOI] [PubMed] [Google Scholar]
- 34.Jones J, Watt FM, Speight PM. J. Oral Pathol. Med. 1997;26:63. doi: 10.1111/j.1600-0714.1997.tb00023.x. [DOI] [PubMed] [Google Scholar]
- 35.Regezi J, Ramos D, Pytela R, Dekker N, Jordan R. Oral Oncol. 2002;38:332. doi: 10.1016/s1368-8375(01)00062-8. [DOI] [PubMed] [Google Scholar]
- 36.Thomas G, Nystrom M, Marshall J. J Oral Pathol Med. 2006;35 doi: 10.1111/j.1600-0714.2005.00374.x. [DOI] [PubMed] [Google Scholar]
- 37.Li S, Lin M, Liu Y-H, Sun X, McGuire MJ, Brown KC. Mol Cancer Ther. 2009;8:1239. doi: 10.1158/1535-7163.MCT-08-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deutsch HM, Glinski JA, Hernandez M, Haugwitz RD, Narayanan VL, Suffness M, Zalkow LH. J Med Chem. 1989;32:788. doi: 10.1021/jm00124a011. [DOI] [PubMed] [Google Scholar]
- 39.Mathew AE, Mejillano MR, Nath JP, Himes RH, Stella VJ. J Med Chem. 1992;35:145. doi: 10.1021/jm00079a019. [DOI] [PubMed] [Google Scholar]
- 40.Nicolaou K, Dai W-M, Gut R. Angew Chem Int Ed Engl. 1994;33:15. [Google Scholar]
- 41.Wang S, Zhelev NZ, Duff S, Fischer PM. Bioorg Med Chem Lett. 2006;16:2628. doi: 10.1016/j.bmcl.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 42.Kumar SK, Williams SA, Isaacs JT, Denmeade SR, Khan SR. Bioorg Med Chem. 2007;15:4973. doi: 10.1016/j.bmc.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 43.Ajaj KA, Biniossek ML, Kratz F. Bioconj Chem. 2009;20:390. doi: 10.1021/bc800429q. [DOI] [PubMed] [Google Scholar]
- 44.Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, Linnoila P, Matthews MJ, Bunn PA, Carney D, Minna JD, Mulshine JL. J. Cell. Biochem. 1996:32. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.