Abstract
We synthesized and characterized a new tagging reagent, (3R,4S)-1-(4-(aminomethyl)phenylsulfonyl)pyrrolidine-3,4-diol (APPD), for the selective fluorogenic derivatization of 3-nitrotyrosine (3-NT) residues in peptides (after reduction to 3-aminotyrosine) and affinity enrichment. The synthetic 3-NT-containing peptide, FSAY(3-NO2)LER, was employed as a model for method validation. Further, this derivatization protocol was successfully tested for analysis of 3-NT-containing proteins exposed to peroxynitrite in the total protein lysate of cultured C2C12 cells. The quantitation of 3-NT content in samples was achieved through either fluorescence spectrometry or boronate affinity chromatography with detection by specific fluorescence (excitation and emission wavelengths of 360 and 510 nm, respectively); the respective limits of detection were 95 and 68 nM (19 and 13 pmol total amount) of 3-NT. Importantly, the derivatized peptides show a strong retention on a synthetic boronate affinity column, containing sulfonamide-phenylboronic acid, under mild chromatographic conditions, affording a route to separate the derivatized peptides from large amounts (milligrams) of non-derivatized peptides, and to enrich them for fluorescent detection and MS identification. Tandem MS analysis identified chemical structures of peptide 3-NT fluorescent derivatives and revealed that the fluorescent derivatives undergo efficient backbone fragmentations, permitting sequence-specific identification of protein nitration at low concentrations of 3-NT in complex protein mixtures.
Keywords: 3-Nitrotyrosine; (3R, 4S)-1-(4-(aminomethyl)phenylsulfonyl) pyrrolidine-3,4-diol (APPD); fluorogenic derivatization; fluorescence; boronate affinity chromatography; mass spectrometry; proteomics
INTRODUCTION
Protein tyrosine (Tyr) nitration to 3-nitrotyrosine (3-NT) is a nitric oxide (NO)-dependent post-translational modification (PTM) associated with a number of diseases and biological aging [1–5]. In order to correlate protein Tyr nitration mechanistically with specific physiological and pathological conditions, it is important to identify protein target sequences and to quantify protein-3-NT residues on these proteins. Several proteomic studies have provided important information about protein targets for nitration in vivo [6–11]. However, it appears that most of these approaches only yielded data covering a small fraction of possible protein targets. Some strategies for the enrichment of 3-NT-containing peptides and proteins have been developed, based on the reduction of 3-NT to 3-aminotyrosine (3-AT) as a critical step, followed by either differential detection of 3-AT (instead of 3-NT) [11] or the reaction of 3-AT with N-hydroxysuccinimide esters coupled to biotin [12] or a functional group available for further derivatization [13]. However these derivatization approaches can suffer from side reactions of N-hydroxysuccinimide esters with other primary amines especially when the abundance of 3-NT is low. Moreover, the published methods [11–13] do not allow for quantification other than through integration of mass spectrometric signals. Therefore, we have developed a selective derivatization strategy of 3-AT with benzylamine, which yields a highly fluorescent benzoxazole product available for quantitative assessment by fluorescence spectrometry or HPLC with fluorescence detection [14]. This reagent does not form a fluorescent product with either peptide N-terminal or Lys amino groups, and, further, allows for relative quantification of original 3-NT-containing peptides through stable isotopic labeling using a couple of “light” and “heavy” reagents [14]. The chromatographic properties of benzylamine-tagged peptides can be controlled through substituents on the original benzylamine, as demonstrated in a comparison between benzylamine and 4-(aminomethyl)benzenesulfonic acid (ABS) [15]. We have now extended the derivatization strategy to incorporate a functional group for the affinity-enrichment of 3-NT-containing peptides, using a novel synthetic benzylamine derivative with a substituent suitable for boronate affinity chromatography. Boronate affinity separation is based on the pH-dependent formation of a pentacyclic complex of boronic acid with cis-diol compounds, which has been employed in the separation of biological molecules, such as catechols, nucleotides, nucleosides, carbohydrates, glycoproteins and some enzymes [16–20]. Recently, some tagging methods for specific residues have been reported for the separation of peptides/proteins by boronate affinity chromatography. For example, the guanidine group of arginine in peptides/proteins was covalently modified with 2,3-butanedione to yield cis-diol containing triazolidine derivatives amenable for separation by boronate affinity [21]. In this paper we describe the synthesis and application of (3R,4S)-1-(4-(aminomethyl)phenylsulfonyl) pyrrolidine-3,4-diol (APPD) for fluorogenic derivatization and boronate affinity enrichment of 3-NT in proteins and peptides. Our results suggest that the method is suitable for quantitative proteomic analysis of protein nitration.
Materials and methods
Reagents
All chemicals for the synthesis of APPD were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received unless otherwise stated. Solvents were of HPLC purity.
Synthesis and characterization of standard 3-NT-containing peptide
A 3-NT-containing peptide, representing the partial sequence of mouse muscle phosphorylase b, 545FSAY(NO2)LER551, was synthesized on an ACT 90 (Advanced ChemTech, Louisville, KY) instrument by means of a solid phase technique using Fmoc-protected amino acids and 3-NT purchased from Bachem Bioscience (King of Prussia, PA). The peptide was purified by semi-preparative HPLC performed on a Summit HPLC system (Dionex Corp., Sunnyvale, CA). Peptide purity was >99%, as characterized by ESI-MS and analytical reverse-phase HPLC analyses.
Synthesis of APPD tagging reagent
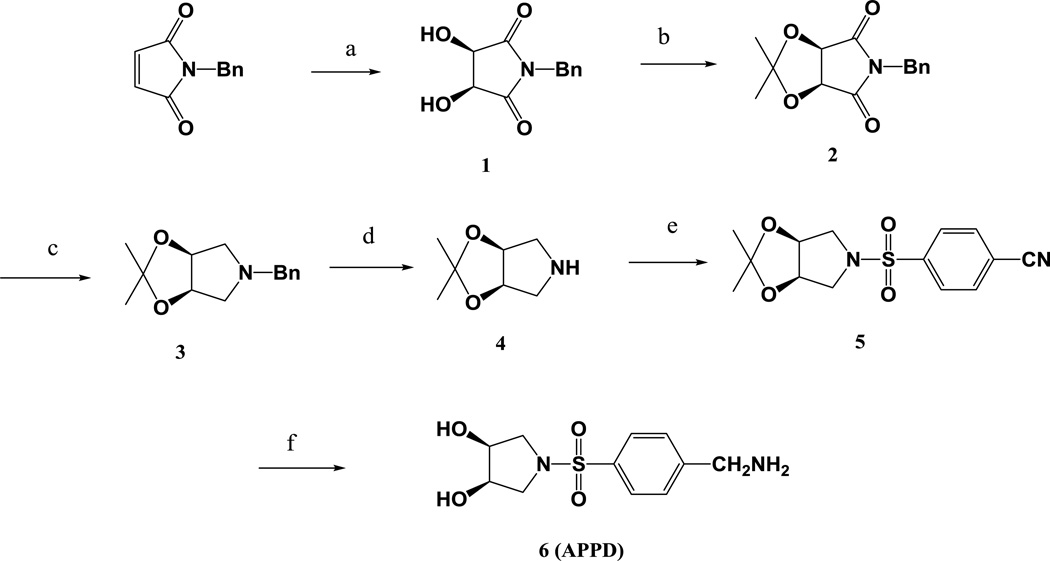
The synthetic steps for the synthesis of APPD are summarized in Figure 1.
Figure 1.
Synthesis of APPD. Reagents and conditions: (a) OsO4, NMO, H2O/acetone (5:1 v/v), 50 °C; (b) 2,2-dimethyloxypropane/TyOH; (c) LiAlH4, THF, 80 °C; (d) H2, Pt/C, MeOH; (e) 4-cyanobenzene-1-sulfonyl chloride, Et3N, MeCN; (f) H2, Pt/C, MeOH, 10% conc. HCl.
(3R,4S)-1-benzyl-3,4-dihydroxypyrrolidine-2,5-dione 1
N-benzylmaleimide was prepared by reaction of benzylamine with maleic anhydride followed by dehydration in acetic anhydride containing sodium acetate. The crude product was purified by dissolving in ethanol and subsequently precipitating with water (yield: 45%). To a stirred solution of 6.5 g of N-methylmorpholine N-oxide and 6 ml of osmium tetraoxide solution (2.5 wt% in tert-butyl alcohol) in a mixture of 50 ml of acetone and 10 ml of deionized water were added 10.0 g N-benzylmaleimide (osmium tetraoxide is toxic [22,23] and must be handled and disposed of with appropriate precautions). The mixture was stirred at 45°C for 24 h. After the reaction mixture was cooled to room temperature, 3.0 g Na2SO3 were added and stirred for 30 min, and filtered with celite through a sintered glass funnel. The filtrate was concentrated by evaporation under reduced pressure and extracted with 3 × 50 ml of ethyl acetate. The combined organic extract was dried over Na2SO4 and evaporated under reduced pressure, affording a deep brown gum which was allowed to granulate for 4 hours at room temperature. The resulting solid was washed with 30 ml of chloroform and dried in vacuum to give compound 1 (5.5 g, 46% yield) as white powder: mp 125–127°C; 1H NMR (DMSO-d6): δ 7.32 ~ 7.24 (m, 5H), 6.02 (d, 2H), 4.55 (s, 2H), 4.42 (d, 2H).
(3aR,6aS)-5-benzyl-2,2-dimethyl-3aH-[1,3]dioxolo[4,5-c]pyrrole-4,6(5H,6aH)-dione 2
To a slurry of 8.5 g of 1 in 40 ml of 2,2-dimethyloxypropane was added 0.02 g of p-toluenesulfonic acid and the mixture was stirred at room temperature for 10 h. The reaction mixture was then stirred with 0.5 g of sodium bicarbonate for 10 min, filtered, and concentrated by evaporation under reduced pressure. The residue was dissolved in 80 ml of chloroform and washed with 1 M Na2CO3 solution, dried over Na2SO4 and evaporated under reduced pressure. The solid that formed was washed with petroleum ether and dried under vacuum overnight, affording product 2 (9.6 g, 95% yield) as white powder. NMR (CDCl3): δ 7.41 ~ 7.28 (m, 5H), 4.86 (s, 2H), 4.69 (s, 2H), 1.43 (s, 3H), 1.30 (s, 3H).
(3aR,6aS)-5-benzyl-2,2-dimethyltetrahydro-3aH-[1,3]dioxolo[4,5-c]pyrrole 3
To a solution of 9.5 g of compound 2 in 20 ml of diethyl ether was added in portions a total of 5.0 g of LiAlH4 (LiAlH4 must be handled with appropriate precautions!). The slurry was heated to 80°C and refluxed for 24 h, and then cooled in an ice-bath. To the reaction mixture were slowly added 30 ml of ethyl acetate and then 20 ml of 30% NaOH solution under stirring. After 30 min, the mixture was filtered, and the filtration cake was washed twice with 30 ml of ethyl acetate. The combined organic layer was dried over Na2SO4 and evaporated under reduced pressure to give product 3 as pale yellow oil. Yield: 8.2 g, 92%. 1H NMR (CDCl3): δ 7.36 ~ 7.25 (m, 5H), 4.66 (d, 2H), 3.63 (s, 2H), 3.0 (dd, 2H), 2.1 (dd, 2H), 1.59 (s, 3H), 1.34 (s, 3H).
(3aR,6aS)-2,2-dimethyltetrahydro-3aH-[1,3]dioxolo[4,5-c]pyrrole 4
To a stirred solution of 8.0 g of compound 3 in 50 ml of methanol was carefully added 1.5 g of 20% Pd(OH)2/C catalyst under argon. The slurry mixture was hydrogenated at 20 psi for 18 h at room temperature, and filtered to remove the catalyst (the hydrogenation must be carried out with appropriate care), and the filtration cake was washed with 20 ml of methanol. The combined organic solutions were evaporated under reduced pressure. The residue was dissolved in 50 ml of diethyl ether, and 10 ml of 1.0 M HCl in ether were added. The crystalline precipitate that formed was collected by filtration, washed with ether and dried under vacuum to give compound 4 (5.42 g, 89%) as a white powder. 1H NMR (DMSO-d6): δ 4.65 (d, 2H), 3.08 (dd, 2H), 2.50 (dd, 2H), 1.44 (s, 3H), 1.31 (s, 3H).
4-(((3aR,6aS)-2,2-dimethyldihydro-3aH-[1,3]dioxolo[4,5-c]pyrrol-5(4H)-yl)sulfinyl)benzonitrile 5
To a solution of 3.0 g of 4 in 50 ml of acetonitrile were added 5.0 ml of triethylamine and 3.4 g of 4-cyanobenzene-1-sulfonyl chloride, and the mixture was stirred for 1 h and subsequently filtered. The filtrate was washed with 10 ml of a saturated aqueous solution of NaCl, and evaporated under reduced pressure to give product 5 (5.6 g, 89% yield) as a white solid. 1H NMR (DMSO-d6): δ 7.92 (d, 2H), 7.82 (d, 2H), 4.64 (s, 2H), 3.59 (dd, 2H), 2.97 (dd, 2H), 1.29 (s, 3H), 1.22 (s, 3H).
(3R,4S)-1-((4-(aminomethyl)phenyl)sulfonyl)pyrrolidine-3,4-diol (APPD) 6
To a solution of 5.0 g of compound 5 in 50 ml of methanol containing 1 ml of acetic acid was carefully added 0.5 g of 10% Pd/C catalyst. The mixture was hydrogenated at 40 psi for 18 h at room temperature, and subsequently filtered to remove the catalyst (the hydrogenation must be carried out with appropriate care); the filtration cake was washed with 20 ml of methanol. The combined organic solutions were evaporated under reduced pressure. To the residue was added 40 ml of 2 M HCl in aqueous solution, and the mixture was heated to 70°C and stirred for 2 hours. The solvent was removed by evaporation under reduced pressure to give product 6, which was washed with chloroform and ether, and dried under vacuum (3.4g, 68%). 1H-NMR (D2O): δ 7.94 (d, 2H), 7.71 (d, 2H), 4.31 (s, 2H), 4.14 (t, 2H), 3.53 (dd, 2H), 3.23 (dd, 2H). 13C-NMR (50% CD3OD/50% H2O): 139.58 (s, 1C), 137.22 (s, 1C), 130.83 (s, 2C), 129.16 (s, 2C), 71.47 (s, 2C), 52.68 (s, 2C), 43.62 (s, 1C). MS (ES+, 20 eV): m/z 273.06 ([M+H]+), Calculated for C11H16N2O4S: 272.08.
Preparation of peroxynitrite for protein nitration in vitro
Peroxynitrite (ONOO−) was prepared by the ozonolysis of sodium azide solutions as described earlier [24]; the ONOO− content in stock solutions containing 0.1% NaOH was quantified using ε302=1,670 M−1cm−1 [4].
Protein nitration in vitro
Prior to protein tyrosine nitration, 1 mg/mL (~10 µM) rabbit muscle phosphorylase b (Ph-b) (Prozyme, San Leandro, CA) in a buffer containing 1% (w/v) SDS, 50 mM NH4HCO3, pH 7.8, was reductively alkylated by incubation with 2 mM DTT for 30 min at 50°C followed by the addition of iodoacetic acid (IAA) at a final concentration of 5 mM and incubation for an additional 30 min at room temperature. The alkylated protein was separated from unreacted compounds and side products by precipitation in ten volumes of cold ethanol for 4 h at −20°C followed by centrifugation at 14,000 g for 2 min. This step was necessary to remove I−, which otherwise could react with ONOO− to form I2 leading to Tyr iodination as assessed through the formation of iodo-Tyr modified peptides by LC-MS/MS analysis (data not shown). Reconstituted 100-µL (100-µg) Ph-b aliquots in 0.1 M phosphate buffer containing 1% SDS were then nitrated by the addition of 10 µL alkaline stock solution of ONOO− (final concentration of 3 mM) while vortexing to yield ca. 100 µM protein 3-NT. The content of 3-NT on Ph-b was determined by UV-spectroscopy (ε430=4,400 M−1cm−1 at pH>9) using non-nitrated protein samples for background control [4]. Ph-b samples containing desired concentrations of 3-NT were prepared by mixing of nitrated and control protein at specific ratios.
Protein nitration in cell culture
C2C12 murine myoblasts were grown in Petri dishes as monolayers to ca. 50% confluence in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and penicillin/streptomycin at 37°C and 5% CO2. The resulting monolayers were washed with PBS, trypsinized, washed with DMEM and PBS, and collected by centrifugation at 500 g for 2 min. A cell suspension in PBS containing 1.5 mg/ml protein was exposed to peroxynitrite (final concentration 3 mM) under vortexing for 5 seconds in 1.5-mL Eppendorf tubes. Cells were lysed by the addition of 1% SDS to PBS buffer and sonication for 30 sec. at 10% power output of the probe sonicator Sonic Dismembrator 500 (Fisher Scientific, Pittsburgh, PA). The supernatant was used for the analysis of protein nitration.
Preparation of protein tryptic digests
Following the nitration, C2C12 cell proteins were subjected to reductive alkylation of Cys residues (in the presence of 1% SDS) and cold ethanol precipitation as described above for phosphorylase b sample preparation. After centrifugation, protein pellets were collected and reconstituted in digestion buffer containing 50 mM NH4HCO3 (pH 7.8), 10% (v/v) acetonitrile, and trypsin (sequence grade from Promega, Madison, WI) at a 1:20 (w/w) ratio to proteins. Digestion was carried out overnight at 37°C.
Derivatization of 3-NT peptides with APPD
Prior to APPD tagging, 3-NT peptides were reduced to 3-AT peptides by incubation with 10 mM sodium dithionite (Na2S2O4) for 2 min at room temperature and the resultant 3-AT-containing peptides immediately separated from the reagent by reverse-phase HPLC (see below). Tagging with APPD was carried out in 0.1 M sodium phosphate buffer (pH 9.0) in the presence of K3Fe(CN)6. Concentrations of the reagents were experimentally evaluated (see below). A typical procedure for the derivatization would be as follows: to 0.25 ml of peptide solution in 0.1 M PBS (pH 9.0) 10 mM APPD and 100 µM K3Fe(CN)6 are added and the reaction mixture is incubated at room temperature for 1 h. The reaction was monitored by fluorescence spectrometry and reversed-phase HPLC with UV and fluorescence detection.
Fluorescence measurements
After derivatization, fluorescence spectra were measured in 0.5-mL 1-cm light-pass fluorescence cuvettes (Hellma, Plainview, NY) using a Shimadzu RF-5000U fluorescence spectrophotometer with excitation and emission band widths set at 5 nm. Spectra were then digitized and fluorescence intensities were calculated from the spectra.
Reverse-phase HPLC
Synthetic and tryptic peptides, and their derivatives, were separated by reverse-phase HPLC using Vydac C18 columns and acetonitrile (ACN) gradients. Desalting of peptides after 3-NT reduction to 3-AT was achieved on guard columns (4.6 mm ID × 10 mm length) equilibrated with 0.01% aqueous trifluoroacetic acid (TFA) (mobile phase A), and after washing for at least 5 min under control of the baseline with UV detection, peptides were eluted by a steep linear gradient increasing ACN in 0.1% TFA from 0 to 80% (v/v) within 5 min., followed by an additional 5 min. elution with 80% (v/v) ACN in 0.1% aqueous TFA. Fractions containing peptides (from 6 to 10 min.) were collected and dried in a speed-vac before reconstitution in the media compatible with the next analysis or derivatization step.
For analytical HPLC of APPD-tagged FSAY*LER (here Y* denominates various derivatives of the tyrosine residue, such as 3-NT, 3-AT, and derivatization products) samples were injected onto a Vydac C18 column (4.6 mm ID × 250 mm length) equilibrated with 10%ACN/90%H2O/0.1%TFA (v/v/v), and peptides were separated by a linear increase of ACN in the mobile phase (1%/min) followed by a washing step for 10 min at 80% ACN. UV and fluorescence detection were achieved using a photodiode array (PDA) detector SPD-M10AVP and fluorescence detector RF-10AXL (both from Shimadzu USA Manufacturing, Canby, OR), respectively.
Boronate affinity chromatography
Sulfonamide-phenylboronic acid-functionalized (0.45 mmol/g) silica gel (Nucleosil, 5 µm, 500 Å, Marcherey-Nagel, Düren) was prepared as described [25] and used as boronate affinity solid phase in this work. The solid phase was slurry packed (0.5 g silica phase in 50 ml methanol) into an empty stainless steel chromatography column (4.6 mm i.d. × 50 mm) under a pressure of 8000 p.s.i. with an air-driven fluid pump (Haskel, Burbank, CA). Boronate affinity chromatography was conducted using 50 mM NH4HCO3 buffer (pH 7.8) as mobile phase for binding of the APPD-tagged peptides and water (pH~5.8) for elution, in the presence or in the absence of 30% (v/v) ACN. The flow rate was 1.0 mL/min, and the effluent was monitored with UV and fluorescence detection. A TSK-Gel Boronate-5PW column (1000Å, 10µ, 7.5 mm i.d. × 75 mm length) was purchased from Tosoh Biosciences, Montgomeryville, PA.
MALDI-TOF Mass Spectrometry
For MALDI-TOF MS analysis, either desalted (using C18 ZipTips, Millipore Co., Bedford, MA) or non-treated samples were directly applied onto the stainless steel sample plate with 2 µl of 50% (v/v) acetonitrile/0.1% TFA saturated with α-cyano-4-hydroxycinnamic acid. Mass spectra were acquired in the mass range between 500 and 3000 amu on a Voyager-DE STR mass spectrometer (PerSeptive Biosystems, Framingham, MA) equipped with a 337 nm nitrogen laser. The instrument was operated in the positive reflector mode with the following parameters: accelerating voltage: 20,000 V; grid voltage: 70–75%; mirror voltage ratio: 1.12; guide wire: 0.02%; extraction delay time: 150 nsec.
Capillary liquid chromatography-mass spectrometry analysis for protein mapping
Samples were desalted using Oasis™ HLB columns (Waters Corporation, Milford, MA) as suggested by the manufacturer, and introduced into an LTQ-FT hybrid linear quadrupole ion trap Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (ThermoFinnigan, Bremen, Germany) via capillary liquid chromatography (capLC), analogous to a published procedure [26]. In brief, peptides were separated on a reverse-phase LC Packings PepMap C18 column (0.300 × 150 mm) at a flow rate of 10 µL/min with a linear gradient rising from 0 to 65% acetonitrile in 0.06% aqueous formic acid over a period of 55 min using an LC Packing Ultimate Chromatograph (Dionex). LC-MS experiments were performed in a data-dependent acquisition manner using the Xcalibur 2.0 software (Thermo Scientific). The five most intensive precursor ions in a survey MS1 spectrum acquired in the Fourier transform-ion cyclotron resonance over a mass range of 300–2000 m/z were selected and fragmented in the linear ion trap by collision-induced dissociation. The ion selection threshold was 500 counts.
Raw experimental files were processed by TurboSequest search using BioWorks 3.2 (ThermoFinnigan). The resulted MS/MS peak lists in dta format were combined within each experiment using an in-house written Perl script and submitted for peptide/protein identification to the Mascot 2.2 (Matrix Science, London, UK) database-searching program using a most recent Swiss-Prot or IPI mouse database with a fragment ion mass tolerance of 0.20 a.m.u. and a parent ion tolerance of 10 ppm. Trypsin specificity and up to 2 missed cleavage sites were used in the search parameters. In order to find all possible products (delta masses) of derivatization reaction between APPD and nitropeptides we used the P-Mod software ([27] to map experimental data. Based on those data, variable protein modifications for Sequest and Mascot searches were set to be +16 on M (oxidation), +15 on Y (aminotyrosine), +45 on Y (nitration), +264 on Y, +281 on Y, +536 on Y (APPD derivatives). An additional shift of +80 on Y for all the above Y modifications was considered to take into account a potential sulfation of tyrosine residues during the reduction of 3-nitrotyrosine to 3-aminotyrosine with sodium dithionite. Carboxymethylation of cysteine residues was considered a fixed modification.
Sequest and Mascot results were imported into a Scaffold 3.0 software (Proteome Software Inc., Potland, OR; free viewer is available at http://www.proteomesoftware.com) for analyzing with the X!Tandem search algorithm and statistical validation of peptide/protein identities. Peptide identifications were accepted if they could be established at greater than 95% probability as specified by the Peptide Prophet algorithm as described [28]. Protein identifications were accepted if they could be established at >95% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [29]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Statistical analysis
For quantitative analysis, all measurements were performed in triplicate, and the data presented as mean ± standard deviation. The limits of detection were calculated from concentration dependencies including at least 7 data points over a limited concentration range as LOD=3sB/b, where sB (the standard error of blank) was obtained from the linear regression analysis (y=a+bx) as a standard deviation of y, and b is the slope of the best linear fit line from the linear regression [30].
Results
Synthesis of Tagging Reagent (APPD)
The synthetic scheme for the preparation of APPD is shown in Fig.1. The key intermediate, cis-diol pyrrolidine, was derived from (3R,4S)-1-benzyl-3,4-dihydroxypyrrolidine-2,5-dione 1, which was prepared by the OsO4-catalyzed enantioselective dihydroxylation of N-benzyl maleimide [31]. 1 was protected with 2,2-dimethyloxypropane, which was sequentially reduced by LiAlH4 at 80°C and by hydrogenation over 20% Pd hydroxide/C catalyst to remove the benzyl group, affording the protected pyrrole 4. The crude product 4 was purified by precipitating its hydrochloride salt in ether and the overall yield of the first four steps was 33%. Product 4 was treated with 4-cyanobenzene-1-sulfonyl chloride to give 5 in 87% yield. The conversion of 5 to the target molecule 6 was achieved by the hydrogenation over 10% Pd/C catalyst in methanol, containing 2% of acetic acid. The deprotection of cis-diol with 2.0 M HCl solution afforded the target compound 6.
Fluorogenic derivatization of 3-NT containing peptides with APPD
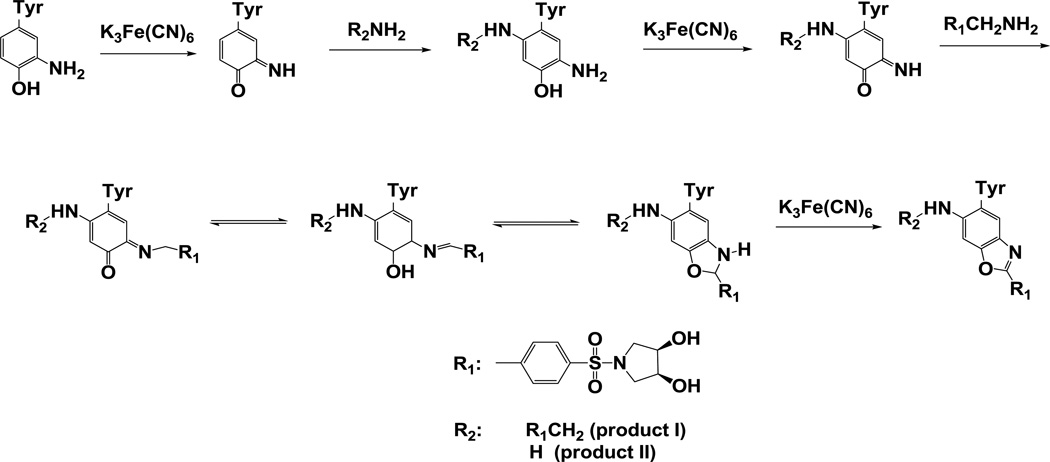
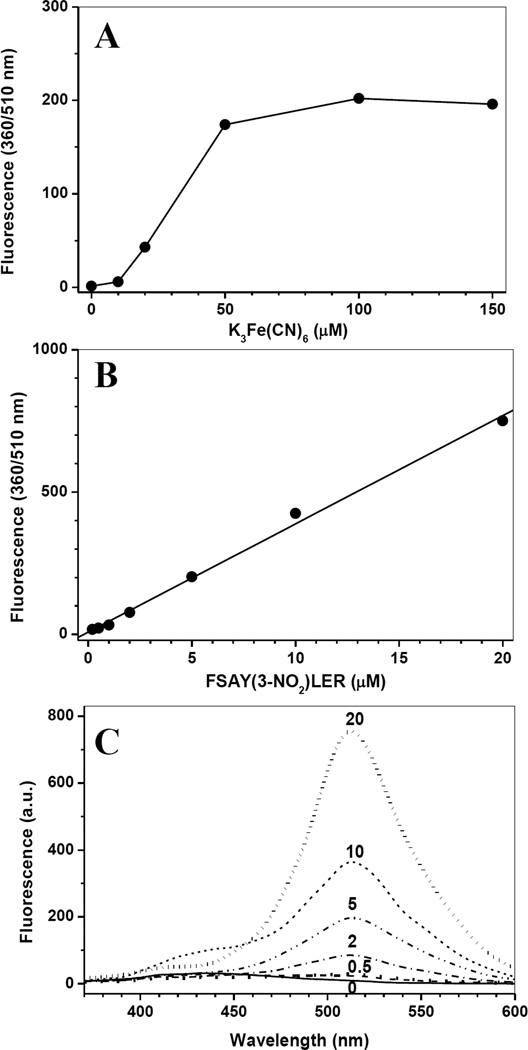
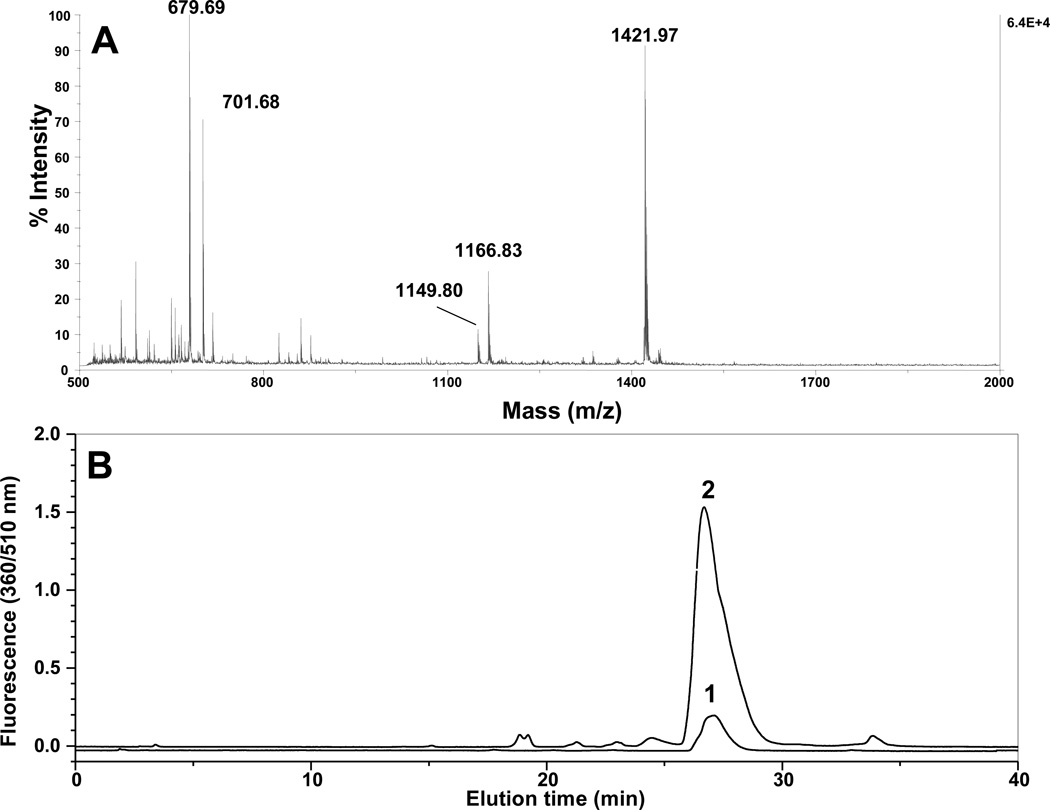
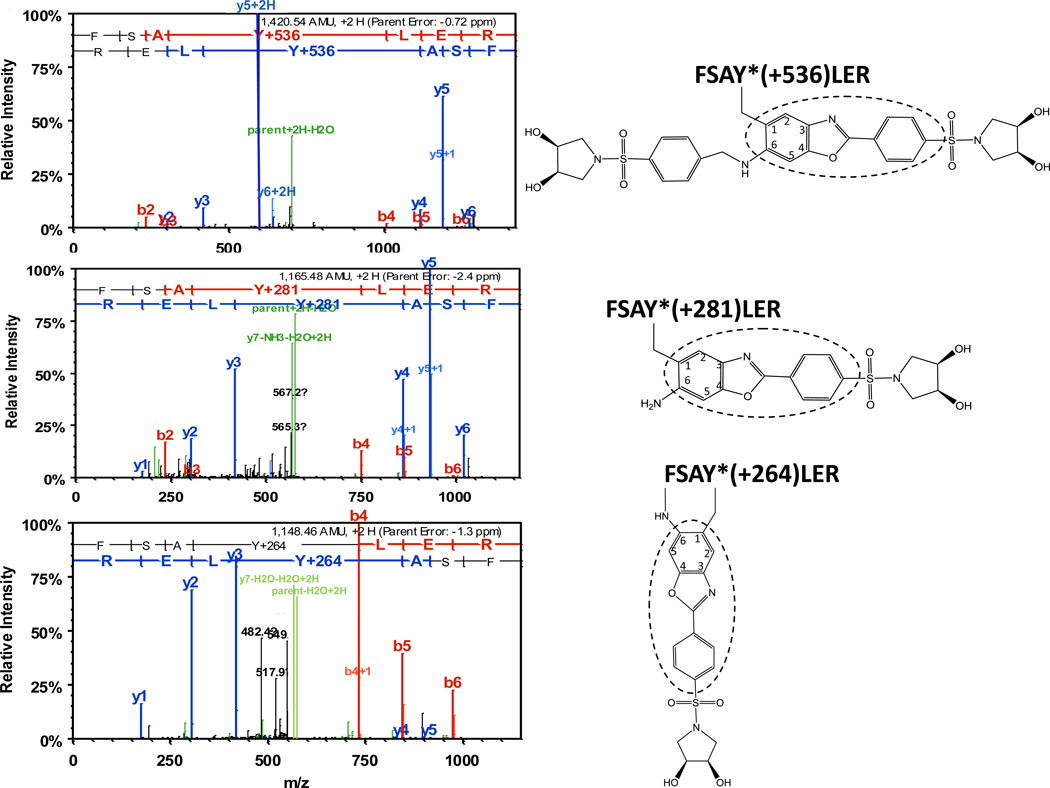
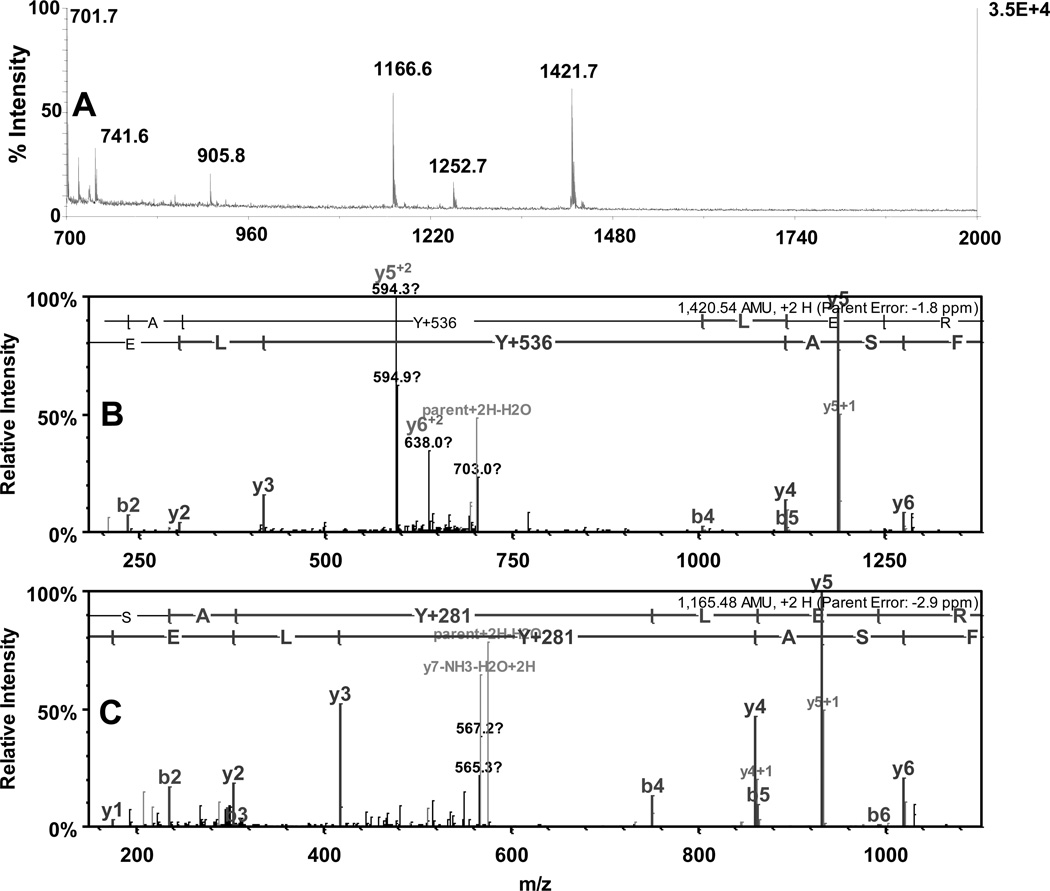
The reaction sequence for the fluorogenic derivatization of a 3-AT containing peptide with APPD is shown in Figure 2, in accord with the published reaction scheme for benzylamine [15]. Prior to fluorescent tagging, 3-NT-containing peptides (mass shift of +45 amu relative to Tyr) are completely reduced by 10 mM sodium dithionite to respective 3-AT-containing sequences (mass shift of +15 relative to Tyr) as confirmed by MS analysis (data not shown). Concentrations of reagents for fluorogenic derivatization were optimized based on yields of fluorescent products assessed by fluorescence spectrometry. For a 3-NT concentration of 5 µM, an increase of APPD concentration over 10 mM (i.e. at more than 1000-fold molar excess of the reagent) did not significantly affect the yield of a fluorescent product with maximum emission at 510 nm and excitation maximum at 360 nm (data not presented). A concentration dependence of the fluorescence intensity at 510 nm on the concentration of oxidant, K3Fe(CN)6, showed a saturation behavior at ca. 100 µM (Figure 3A). Tagging FSAY(3-NH2)LER with 10 mM APPD and 100 µM K3Fe(CN)6 resulted in a nearly linear dependence of the fluorescence intensity at 510 nm on the peptide concentration between 0 and 20 µM (Fig. 3, B and C). MALDI-TOF MS analysis (Fig. 4A) shows that APPD tagging produced two major benzoxazole products of the peptide with m/z 1421.9 (product I) and 1166.8 (product II), which correspond to peptide derivatives of +536 and +281 amu, respectively. The respective structures for these products are displayed in the inserts of Figures 5A and 5B. LC-MS/MS analysis confirmed that in the sequence of tagged peptides these characteristic mass shifts located on the Tyr residue, Tyr+536 and Tyr+281, respectively, based on the mass difference between the y4 and y3 fragments in the spectra (Fig.5, A and B). Here, the addition of APPD and ammonia, respectively, to the 6-position of the benzoxazole ring is suggested based on previous structural analysis of benzylamine reaction with 4-methylcatechol [14]. Importantly, no significant remaining amounts of FSAY(3-NH2)LER (m/z 900.5) or FSAY(3-NO2)LER (m/z 930.4) were observed after the reaction. A small yield (ca. 7%) of a product with m/z 1149.8 (product III) was observed in the MALDI-TOF MS spectrum (Fig. 4A). This mass is consistent with the intramolecular addition of the N-terminal amino group to position 6 in the final benzoxazole, a hypothetical structure of which is shown in the insert of Fig.5C, and based on the observed MS/MS fragmentation pattern. Analogous products have been observed upon derivatization of 3-AT-containing peptides with 4-(aminomethyl)benzenesulfonic acid (ABS) [32]. Importantly, product III will contribute to the total fluorescence at 510 nm and nearly co-elutes with products I and II in the reverse-phase chromatogram (HPLC fractions collected between 26 and 29 min in Fig. 4B), confirmed by capLC-FT-ICR-MS analysis (data not shown).
Figure 2.
Derivatization pathway of peptide 3-AT with APPD.
Figure 3.
The dependences of fluorescence intensity after tagging of FSAY(3-NO2)LER with APPD on the concentration of K3Fe(CN)6 (A) or 3-NT peptide (B) and fluorescence spectra at different concentrations of 3-NT (C). A - FSAY(3-NO2)LER (5 µM) was reduced to FSAY(3-NH2)LER and incubated with 10 mM APPD and different concentrations of K3Fe(CN)6 in 0.1 M sodium phosphate buffer (pH 9.0) for 1 h. B and C - Fluorescence intensities and spectra, respectively, at different concentrations of the 3-NT peptide tagged with 10 mM APPD and 100 µM K3Fe(CN)6 as described above. Fluorescence was measured in a 0.5-mL (1-cm) cuvette using excitation and emission wavelengths of 360 and 510 nm, respectively.
Figure 4.
Analysis by MALDI-TOF MS (A) and reverse-phase HPLC detected by fluorescence (B) of APPD-tagged FSAY(3-NO2)LER. After reduction of 3-NT to 3-AT the peptide (10 µM) was incubated for 1 h with 10 mM APPD and 20 µM (curve 1) or 100 µM (curve 2) of K3Fe(CN)6 in 0.1 M sodium phosphate buffer (pH 9.0) at room temperature. Chromatograms were detected using excitation and fluorescence wavelengths of 360 and 510 nm, respectively.
Figure 5.
ESI-MS/MS identification of different benzoxazole products after nitro-to-amino reduction of FSAY(3-NO2)LER (10 µM) and tagging with 10 mM APPD and 100 µM K3Fe(CN)6. The structures shown in inserts contain different substitutions in position 6 of the original 3-NT ring (the fluorescent 2-phenylbenzoxasole core is circled).
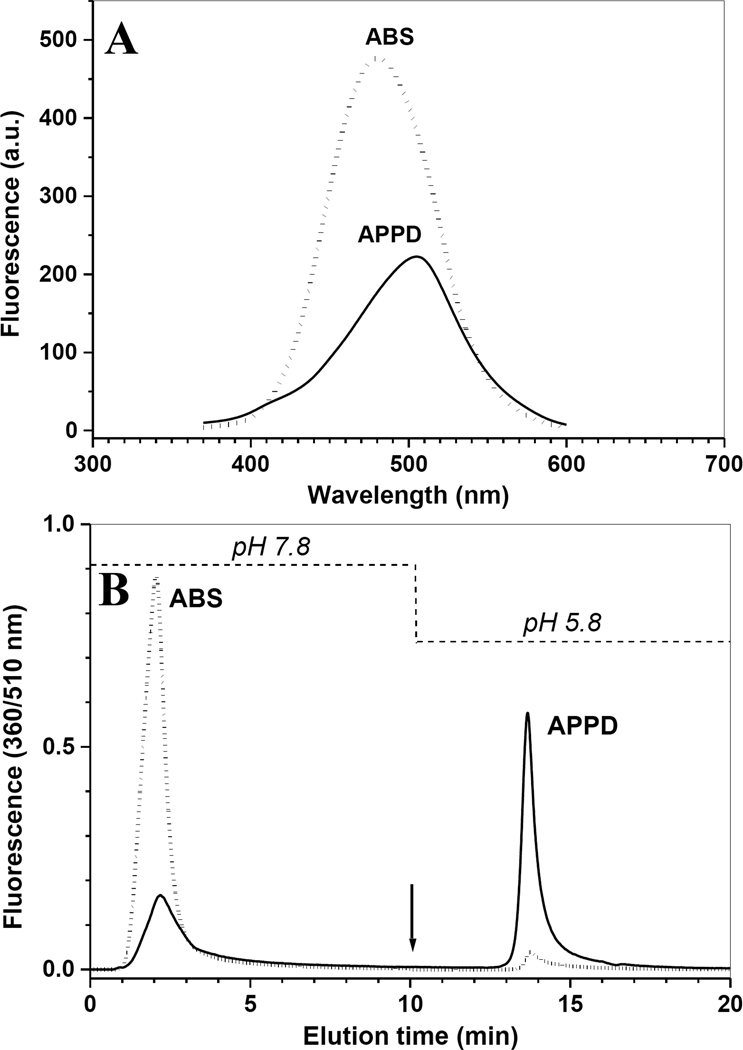
In addition to the 510-nm fluorescence (benzoxazole), fluorescence spectrometry detected the formation of another fluorescent species with an emission maximum at ca. 430 nm, which is also present in the control samples (reagents without peptide; spectra in Fig. 3C). We did not focus on the identification of fluorescent side products in this study; however, reverse-phase HPLC shows the presence of several fluorescent peaks besides the major peak with a retention time of ca. 27 min., which appear at increasing concentrations of the oxidant, K3Fe(CN)6 (compare chromatograms 1 and 2 in Fig.4B). This is the reason to use relatively low concentrations of the oxidant (not more than 100 µM; see Fig.3A) particularly at 3-NT concentrations below 1 µM. These results are in accord with our previous study on 3-AT tagging with an other benzylamine derivative, ABS, where we have determined that maximum yields of benzoxazole product can be achieved at a ratio of K3Fe(CN)6 to 3-AT of ca. 5 to 1 and that a further increase in the oxidant concentration results mostly in fluorescent side product formation [32]. The background fluorescence after tagging with ABS was attributed to the reagent background and, potentially, to the formation of protein oxidation products during the course of the derivatization procedure. Irrespective of its nature, the relative contribution of this background fluorescence is higher at lower concentrations of nitropeptides, which defines a detection limit of about 0.1 µM 3-NT by fluorescence spectrometry [32]. However, specifically for APPD tagging, we have the unique opportunity to separate the APPD tagged peptides from fluorescence side products by boronic acid affinity chromatography, and to monitor them by their specific fluorescence at 510 nm.
Boronate affinity separation and enrichment of a fluorescent APPD-tagged synthetic model peptide
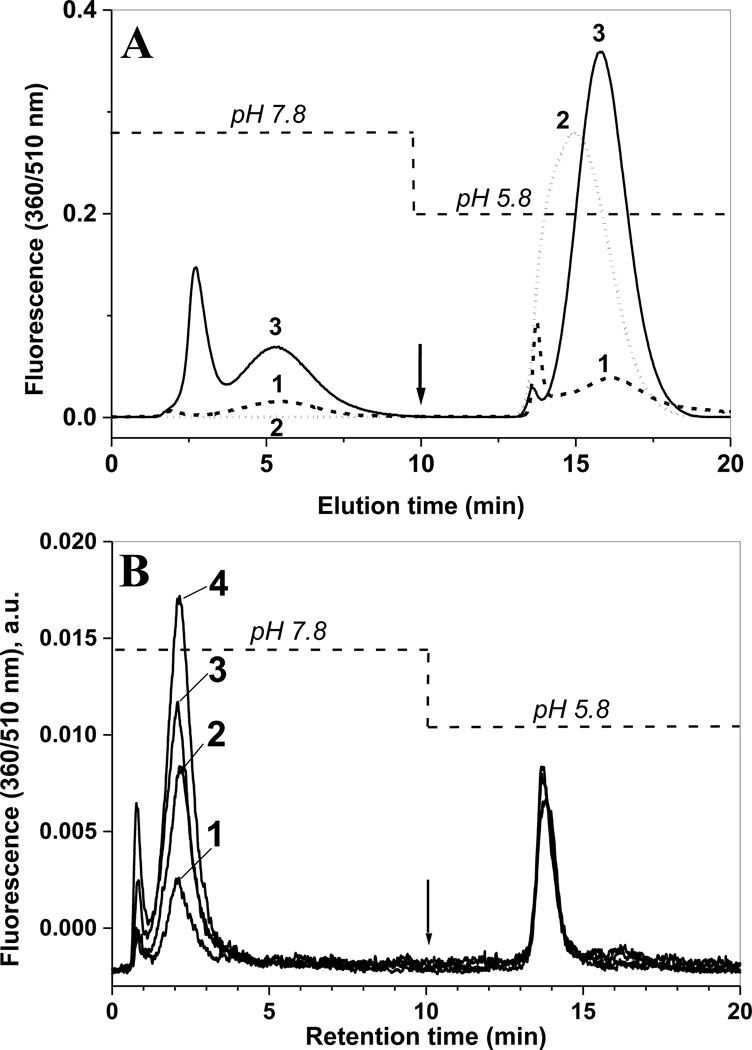
The synthetic APPD reagent contains two separate functional groups: the benzylamine group for formation of fluorescent benzoxazole and the cis-diol group, which associates in a pH-dependent manner with boronate anion. The latter properties can be utilized for enrichment of the APPD-derivatized 3-NT peptides by boronate affinity chromatography. We first tested the boronate affinity enrichment of an APPD-tagged model peptide on a modified silica solid phase containing a sulfonamide-substituted phenylboronic acid with a boronate pKa of 7.5 [25]. This solid phase displays an enhanced boronate binding of alkyl- and aryl-cis-diol compounds under neutral conditions. Fig. 6A shows the boronate affinity HPLC with fluorescence detection at 510 nm of APPD-tagged FSAY(3-NO2)LER using a column (4.6 i.d. × 50 mm length) packed with the synthetic sulfonamide-substituted phenylboronic acid. The APPD-tagged peptide was bound to the column in 50 mM NH4HCO3 buffer (pH 7.8) and washed for 10 min (at 1 ml/min) with this buffer. Subsequently, the tagged peptide was eluted with water (pH~5.8) during the next 10 min (trace 1 in Fig.6A). Further acidification of the elution buffer is not necessary, since it does not yield greater amounts of APPD-tagged peptides but requires additional efforts to clean up samples for subsequent MS analysis. To account for non-specific binding to the column material due to hydrophobic interactions, increasing concentrations of ACN were added to both binding and elution buffers. We found that the addition of 30% (v/v) ACN effectively blocks non-specific affinity during both washing and elution steps (Fig. 6A, trace 3). Without organic solvent a significant accumulation of the fluorescent peptide occurs on the column, which can be washed out by 30% ACN in the mobile phases during a subsequent blank run (trace 2 in Fig.6A). The fluorescent fractions eluting after acidification (between 13 and 18 min) were collected and analyzed by MALDI-TOF MS (Fig. 7A) and capLC-FT-ICR-MS (Fig.7, B and C), which confirmed that the eluate contained predominantly APPD-tagged FSAY*LER in the form of products I (m/z 1421.97) and II (m/z 1166.83) (the respective structures are shown in the inserts of Fig. 5A and 5B). It appears that both APPD derivatives of FSAY*LER, containing either one or two cis-diol groups, have a comparable affinity to the column based on a comparable abundance of the respective peptide ions in the MALDI mass spectrum (Fig. 7A).
Figure 6.
Boronic affinity chromatography of APPD tagged synthetic peptide (A) and of tryptic digest of nitrated rabbit phosphorylase b mixed with proteins from lysate of C2C12 cultured cells (B) detected by fluorescence (excitation and emission wavelengths are 360 and 510 nm, respectively). In panel A, 200 µL of 10 µM FSAY(3-NO2)LER after reduction of 3-NT to 3-AT and incubation with 10 mM APPD and 100 µM K3Fe(CN)6 for 1 h was injected onto phenylboronic column equilibrated with buffer A containing 50 mM NH4HCO3 (pH 7.8) in the absence (1) or presence (2 and 3) of 30% (v/v) acetonitrile. The arrow indicates switching from buffer A to water in the mobile phase without changing the percentage of organic solvent. Trace 2 represents a blank injection (washing run) after the separation performed in the absence of organic solvent (trace 1). Panel B: Boronic affinity separation of tryptic peptides from nitrated Ph-b (1 µg protein containing 100 pmol 3-NT) only (trace 1) and in the presence of C2C12 cell proteins (trace 2 – 0.2, trace 3 – 0.5, and trace 4 – 1 mg total protein) reduced and tagged in the same conditions.
Figure 7.
Analysis of fluorescent boronate affinity HPLC fractions (13–18 min. in chromatogram 3, Fig.6A) by MALDI-TOF (A) and capLC-FT-ICR MS (B and C). B and C represent tandem MS spectra for APPD-tagged FSAY*LER ions with mass shifts of Y+536 (m/z 1421.5) and Y+281 (m/z 1166.6), respectively, present in panel A.
Qualitative enrichment of nitrated peptides from a biological matrix
It is well recognized that the presence of biological matrix may interfere with and even completely block both the specific chemical derivatization of low-abundant analytes and subsequent LC and MS analysis [33]. We tested our APPD derivatization approach using small aliquots of in vitro nitrated protein, Ph-b, mixed with 100–1000 fold excess of proteins derived from C2C12 cultured cells to obtain levels of 100 to 2000 pmol protein-bound 3-NT per mg total protein in the mixture. In terms of molar ratios, the amount of 100 pmol 3-NT/mg protein would represent average Tyr nitration levels of 0.005 mol 3-NT per mol of protein with an average molecular weight of 50 kDa. That is, statistically in these mixtures only one out of 200 proteins would contain a nitrated Tyr residue, and each nitrated protein would yield only one nitropeptide but, on average, 50–100 non-nitrated peptides upon tryptic digestion. Fig. 6B shows that a protein digest containing 1 µg (~10 pmol) of nitrated Ph-b and 100 pmol of 3-NT can be derivatized with APPD to yield a fluorescent product with a 510 nm emission maximum, which is successfully enriched by boronic affinity chromatography and detected by its specific fluorescence. Importantly, the presence of an excess amount of non-nitrated peptides (trypsin digests from 0.2 to 1 mg C2C12 proteins) did not reduce the yield of fluorescent benzoxazole products based on the 510-nm fluorescence.
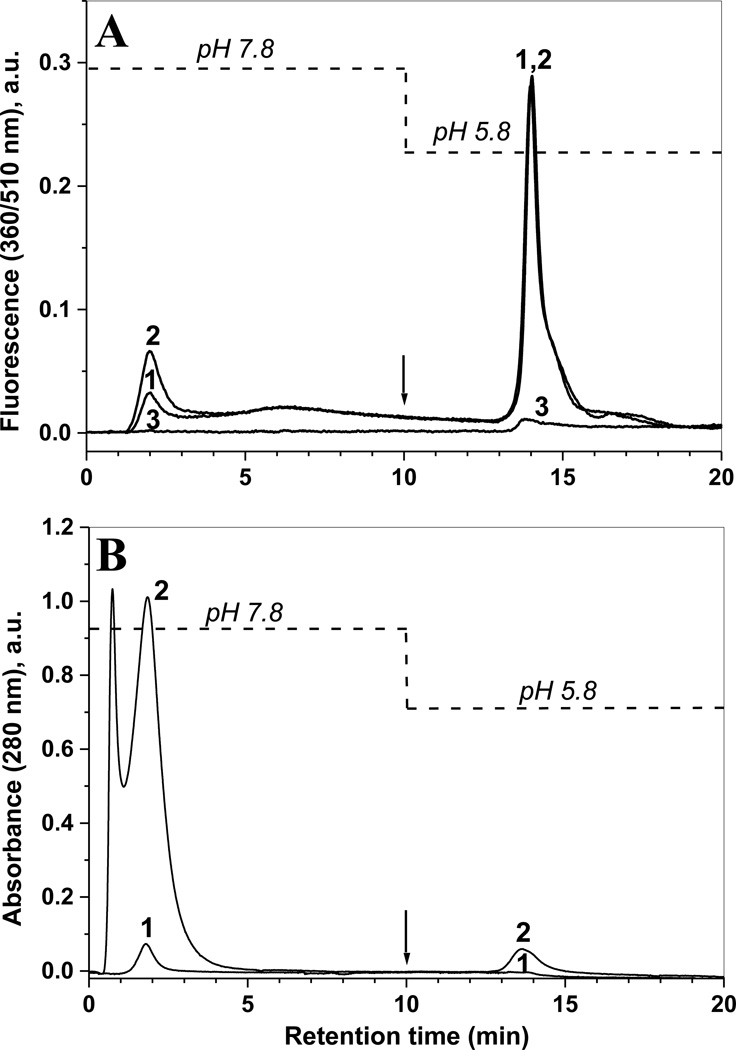
Additional proof for the specific binding of 510-nm fluorescent products (benzoxazole) to the boronate column was obtained through a proteomic analysis of non-retained peptides (HPLC fractions from 1 to 10 min), and more tightly retained peptides (13–18 min) as shown in Fig. 8. In this experiment, slightly higher amounts of nitrated Ph-b (5 µg) were used to obtain a better coverage of Ph-b peptides. The boronate affinity chromatograms after APPD derivatization of nitrated Ph-b digest alone and in the presence of 0.2 mg protein digest from C2C12 cells show significant differences in matrix peptide content in both fractions based on 280-nm absorbance (traces 1 and 2 in Fig.8B, respectively), but no changes in the yield of fluorescent products in the boronate affinity fractions based on specific 510-nm fluorescence detection (traces 1 and 2 in Fig. 8A). CapLC-FT-ICR-MS analysis followed by Sequest search against mouse and rabbit protein databases, identified native peptides from more than 200 mouse proteins (not shown) and multiple native but no APPD tagged rabbit Ph-b peptides in the flow-through (1–10 min) fractions (Fig. 9A). In contrast, the fluorescent boronate affinity chromatographic fractions contained only APPD-tagged peptides from the rabbit Ph-b sequence (Fig.9B).
Figure 8.
Boronate affinity chromatography of model nitrated protein, Ph-b, mixed with high amounts of C2C12 cell lysate protein, through APPD tagging of digested proteins and detected by specific fluorescence (A) or 280-nm absorbance (B). Nitrated Ph-b (5 µg) was analyzed either alone (trace 1) or in the presence of 0.2 mg C2C12 proteins (trace 2); trace 3 represents a reagent control (APPD tagging in the absence of 3-NT). 3-NT containing peptides were reduced to corresponding 3-AT peptides and tagged with 10 mM APPD and 50 µM K3Fe(CN)6 in 0.2 M NH4HCO3 (ABC) buffer at pH 9, bound to boronate affinity column in 0.1 M ABC buffer at pH 7.8 (0–10 min) and eluted with H2O at pH 5.8 (10–20 min) at a flow rate of 1 ml/min. Boronate affinity (13–18 min) and flow-through fractions (1–10 min) were collected for LC-MS analysis.
Figure 9.
Sequence coverage for Ph-b in flow-through (A) and boronate affinity (B) fractions collected during chromatographic run shown in Fig.8 (trace 2). In panel B, APPD derivatization products detected in the sample (Y+536, Y+ 264, and Y+281) are shown in bold, italic and underlined font, respectively.
The specific enrichment of APPD-tagged peptides via boronate affinity chromatography can be demonstrated by comparative analysis of the chromatograms obtained for nitropeptides tagged with either APPD or ABS, which differs from APPD by the lack of the cis-diol pyrrolidine ring. Tagging with ABS results in the formation of a fluorescent derivative with maximum emission at 480 nm and ca. two-fold higher chemical and/or fluorescence quantum yields as compared with APPD (Fig. 10A), in accord with our previous results [32]. The ABS derivatives did not bind to the column and eluted during the washing step immediately after injection unlike the APPD derivatives (Fig. 10B). These results clearly demonstrate the significance of the cis-diol group in the APPD reagent for boronate affinity enrichment. For comparison of our synthetic sulfonamide-substituted phenylboronic acid, the boronate affinity chromatography of the APPD-tagged peptides was conducted on a commercially available column (TSK-Gel Boronate-5PW 1000A, 10µ, 7.5 mm i.d. × 75 mm length) containing immobilized m-aminophenylboronic acid, under the same HPLC conditions. This column displayed a similar affinity for the APPD-tagged peptides from C2C12 protein digests as our synthetic column (data not presented) providing additional evidence for the role of pH-dependent complex formation between the APPD cis-diol function and the boronate anion of the stationary phase.
Figure 10.
Comparison of fluorescence spectra (A) and boronate affinity chromatographic runs detected by fluorescence (B) of nitrated C2C12 cultured cell protein digests (250 µg total protein containing 500 nmol 3-NT), reduced with 20 mM sodium dithionite and tagged with 10 mM of either APPD (solid lines) or ABS (dotted lines) and 100 µM K3Fe(CN)6 in 100 mM PBS (pH 9.0). Chromatographic conditions were as in the legend to Fig.8.
Quantitative analysis of APPD-tagged 3-NT in complex peptide mixtures from proteins nitrated in vitro by boronate affinity chromatography with fluorescence detection
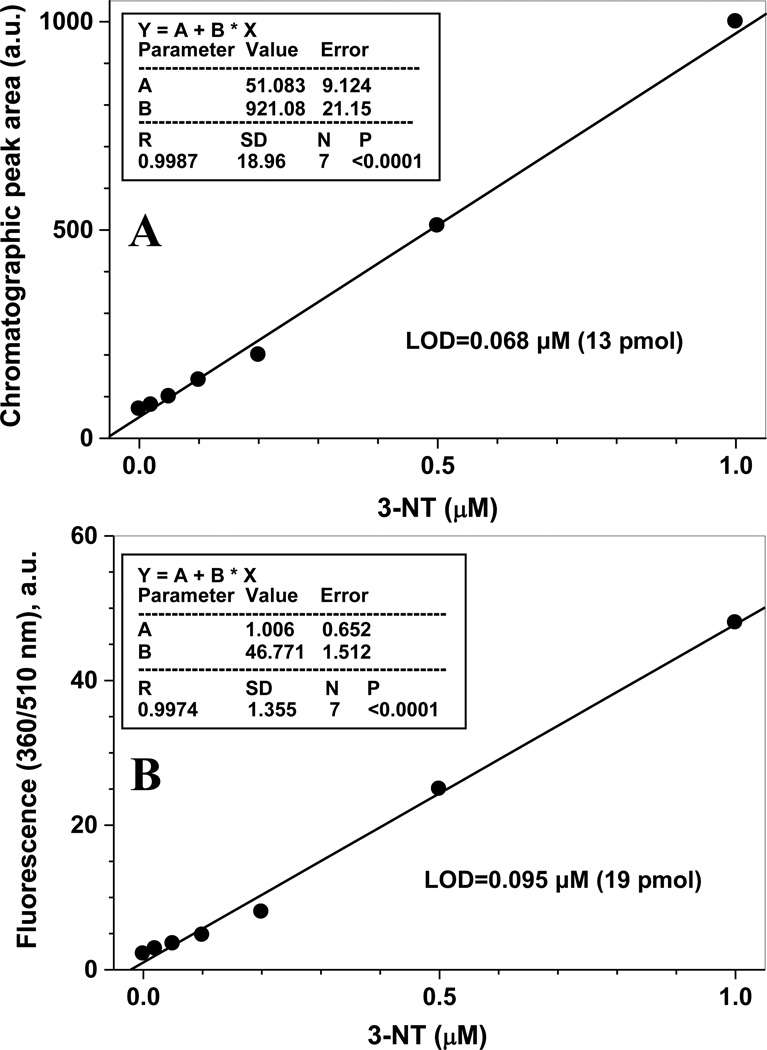
In the previous section, we had tested the qualitative enrichment of APPD-tagged peptides from a complex biological matrix. To test whether boronate affinity chromatography of APPD-tagged nitropeptides may represent a quantitative method to analyze low levels of protein tyrosine nitration in complex peptide mixtures, we nitrated Ph-b by peroxynitrite in vitro and mixed with an excess of proteins from lysate of C2C12 cultured cells. 3-NT concentrations were quantified through specific 430-nm absorbance as described in the experimental section. After trypsin digestion, reduction of 3-NT with sodium dithionite, and derivatization with APPD as described above, samples were separated by boronate affinity chromatography and detected through the specific fluorescence of APPD-tagged peptides (excitation and emission wavelengths of 360 and 510 nm, respectively). The dependence of the peak areas on the original concentration of 3-NT is shown in Fig. 11A. Importantly, the results are presented for relatively low, biologically relevant 3-NT concentrations (below 1 µM) in the presence of highly abundant non-nitrated peptides (digest of 1 mg total protein from C2C12 cultured cell lysate). A linear regression analysis gave the limit of detection (LOD) of ca. 68 nM of 3-NT, corresponding to 13 pmol of 3-NT per injection. This value is lower than the LOD for fluorescence spectrometry analysis alone, 95 nM of 3-NT (19 pmol per sample) (Fig. 11B). Although the calculated values for APPD tagging are comparable to those reported for ABS tagging (0.09 µM and 40 pmol 3-NT [32]), quantitation through APPD tagging/boronate affinity chromatography can be performed in the presence of at least 10-fold larger amounts of biological matrix (1 mg in the case of APPD vs 100 µg for ABS). This feature offers an opportunity of enrichment and accumulation of APPD-tagged peptides from large amounts of biological material that is very important, particularly, for sequence-specific analysis of low abundant protein nitration in tissues.
Figure 11.
Validation of the fluorogenic APPD derivatization as quantitative method for peptide 3-NT determination by boronate affinity chromatography with fluorescence detection (A) and by fluorescence spectrometry (B). 3-NT concentration in samples (0–1 µM) was achieved through addition of different amounts of nitrated Ph-b, containing ca. 10 mol 3-NT/mol protein (1 µM 3-NT/2.5 µg Ph-b), to 0.5 mg of C2C12 cell lysate protein. Samples were digested with trypsin, reduced by 10 mm sodium dithionite and derivatized with 2 mM APPD and 20 µM K3Fe(CN)6 in 50 mM ammonium bicarbonate buffer (pH 9.0). Boronate affinity chromatography conditions were as in the legend to Fig. 8.
Sequence-specific characterization of protein Tyr nitration by fluorogenic tagging with APPD and boronate affinity chromatography enrichment of tagged peptides
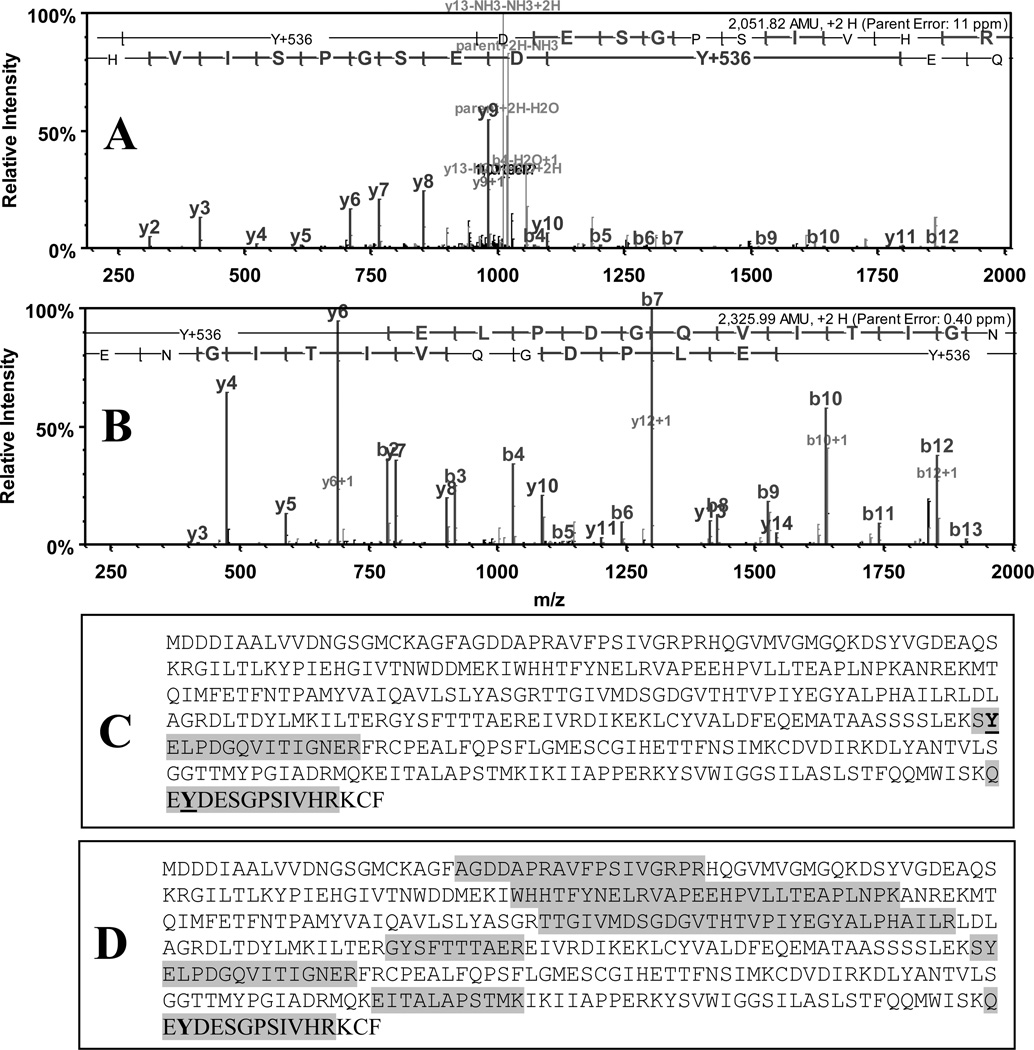
To test whether low-level protein tyrosine nitration in complex protein mixtures can be assessed in a sequence-specific matter, we analyzed by capLC-FT-ICR-MS the fluorescent boronate affinity fractions of APPD-tagged peptides from peroxynitrite exposed C2C12 cells. Protein nitration was achieved at a level of ca. 1 nmol 3-NT per 0.5 mg total protein. The efficiency of APPD tagging/boronate affinity enrichment was addressed through comparison of capLC-FT-ICR-MS data with those for non-tagged or ABS-tagged samples prepared from the same amount of material in similar conditions except for affinity separation of tagged peptides. Analysis of non-tagged or ABS-tagged nitrated C2C12 protein digests identified >100 proteins at highest probability settings; no ABS-tagged peptides were revealed (data not shown). In contrast, APPD-tagging/boronate affinity enrichment resulted in the detection of two APPD tagged peptides (Tyr +536 amu), indicating two tyrosine nitration sites in the sequence of mouse actin 1 (Fig.12, A and B). Importantly, no non-tagged peptides from actin 1 (Fig. 12, C) or other proteins (data not shown) can be identified in the samples after boronate affinity enrichment, in contrast to non-purified sample (Fig. 12, D). Since the yields of fluorescent benzoxazole products are higher in the ABS tagged samples (Fig. 10A), our data suggest that for successful sequence-specific characterization of tagged 3-NT the absolute amounts of an analyte, which is tagged 3-NT, are less important than the relative abundance in the sample. Therefore, our approach of APPD tagging/boronate affinity enrichment of nitropeptides, in addition to accumulation of APPD-tagged peptides from larger amounts of biological material, offers the option of a removal of non-tagged matrix peptides, which suppress the sequence-specific MS analysis of low abundant protein tyrosine nitration.
Figure 12.
Representative capLC-FT-ICR tandem MS spectra of APPD-tagged peptides in the protein tryptic digest from peroxynitrite-exposed C2C12 cells, obtained after boronate affinity enrichment of a sample contained 1 nmol 3-NT in 500 µg of protein. A and B represent tandem MS spectra of two peptides, SY*ELPDGQVITIGNER and QEY*DESGPSIVHR (mass shift on both of +536 relative to Tyr), from the sequence of mouse cytoplasmic actin 1 (gene name: actb). Protein sequence coverage is shown in panel C; for comparison, panel D shows sequence coverage for this protein observed without APPD tagging and boronate affinity enrichment.
Discussion
In this study, a novel chemical tagging reagent, APPD, was synthesized and applied to fluorogenic derivatization and boronate affinity enrichment of 3-NT-containing peptides. Based on the derivatization of a synthetic peptide, FSAY(3-NO2)LER, we established a tagging protocol optimized for maximum yields of fluorescence benzoxazole derivatives from 3-NT. Though different fluorescent products of APPD-tagged 3-NT peptides are detected by MS analysis, with different substitutions in position 6 of the benzoxazole, they all contain the 2-benzoxazole core and contribute to the measured fluorescence with emission maximum at 510 nm and excitation maximum at 360 nm. Fluorescent 2-benzoxazole derivatives are formed via condensation with the benzylamine moiety of APPD specifically from protein/peptide 3-AT, and, potentially, from 3-hydroxytyrosine (DOPA) [14,15] and 5-hydroxytryptophan [32], two another important biomarkers of oxidative protein modification. The latter two species do not require a reduction step to form benzoxazole upon tagging with benzylamines [14], and can be fluorometrically assessed separately from 3-NT provided that the respective non-reduced controls are included. In our experiments with C2C12 cells exposed to peroxynitrite we did not observe significant amounts of fluorescence products emitting at 510 nm in non-reduced samples. However, unidentified fluorescent side products with an emission maximum at 430 nm may also form from APPD irrespective of the presence of 3-NT that causes detection limits of 95 and 68 nM, respectively, for fluorescence spectroscopy and boronate affinity HPLC-FL analysis of APPD tagged 3-NT. These numbers are better than those previously reported for fluorogenic 3-NT tagging with ABS, which lacks the affinity functionality [32]. The critical advantage of APPD tagging/boronate affinity enrichment is that APPD-tagged peptides will be separated from non-tagged material and eluted virtually in a single peak. Therefore, our approach offers a possibility for the accumulation of APPD tagged 3-NT peptides for both quantitative fluorescence spectrometry and sequence-specific MS analysis provided that the amount of material is not limited (this is a common situation in studies of tissue biomarkers) suggesting its potential application in the characterization of protein nitration in complex biological systems.
Acknowledgements
This research was supported by the NIH (AG23551, AG25350) and a grant from Pfizer. The authors thank Dr. D. Yakovlev (Biochemical Research Service Laboratory, University of Kansas) for the synthesis and purification of the nitropeptide FSAY(3-NO2)LER.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turko IV, Murad F. Protein nitration in cardiovascular diseases. Pharmacol. Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds MR, Berry RW, Binder L. Nitration in neurodegeneration: deciphering the “Hows” “nYs”. Biochemistry. 2007;46:7325–7336. doi: 10.1021/bi700430y. [DOI] [PubMed] [Google Scholar]
- 3.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 2002;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 4.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. USA. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc. Natl. Acad. Sci. USA. 2001;98:12056–12061. doi: 10.1073/pnas.221269198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanski J, Hong S-J, Schöneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J. Biol. Chem. 2005;280:24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- 8.Kanski J, Behring A, Pelling J, Schöneich C. Proteomic identification of 3-nitrotyrosine-containing rat cardiac proteins: Effects of biological aging. Am. J. Physiol. 2005;288:H371–H381. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- 9.Kanski J, Alterman MA, Schöneich C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radic. Biol. Med. 2003;35:1229–1239. doi: 10.1016/s0891-5849(03)00500-8. [DOI] [PubMed] [Google Scholar]
- 10.Sacksteder CA, Qian W-J, Knyushko TV, Wang H, Chin MH, Lacan G, Melega WP, Camp DG, Smith RD, Smith DJ, Squier TC, Bigelow DJ. Endogenously nitrated proteins in mouse brain: Links to neurodegenerative disease. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 11.Ghesquiere B, Colaert N, Hensens K, Dejager L, Vanhaute C, Verleysen K, Kas K, Timmerman E, Goethals M, Libert C, Vandekerckhove J, Gevaert K. In vitro and in vivo protein-bound tyrosine nitration characterized by diagonal chromatography. Mol. Cell Proteom. 2009;8:2642–2652. doi: 10.1074/mcp.M900259-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikov G, Bhat V, Wishnok JS, Tannenbaum SR. Analysis of nitrated proteins by nitrotyrosine-specific affinity probes and mass spectrometry. Anal. Biochem. 2003;320:214–222. doi: 10.1016/s0003-2697(03)00359-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Qian W, Knyuschko TV, Clauss TRW, Purvine SO, Moore RJ, Sacksteder CA, Chin MH, Smith DJ, Camp DG, II, Bigelow DJ, Smith RD. A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. J. Proteome Res. 2007;6:2257–2268. doi: 10.1021/pr0606934. [DOI] [PubMed] [Google Scholar]
- 14.Pennington J, Schöneich C, Stobaugh JF. Selective fluorogenic derivatization with isotope coding of catechols and 2-aminophenol with benzylamine: a chemical basis for the relative determination of 3-hydroxytyrosine and 3- nitrotyrosine peptides. Chromatograhia. 2007;66:649–659. [Google Scholar]
- 15.Sharov VS, Dremina ES, Pennington J, Killmer J, Asmus C, Thorson M, Hong SJ, Li X-B, Stobaugh JF, Schöneich C. Selective fluorogenic derivatization of 3-nitrotyrosine and 3,4-dihydroxyphenylalanine (DOPA) in peptides: a method designed for quantitative proteomic analysis. Meth. Enzymol. 2008;441:19–32. doi: 10.1016/S0076-6879(08)01202-0. [DOI] [PubMed] [Google Scholar]
- 16.Bergold A, Scouten WH. Borate chromatography. Chem. Anal. 1983;66:149–187. [Google Scholar]
- 17.Liu X-C, Scouten WH. Miscellaneous methods in affinity chromatography. Part I: boronic acids as selective ligands for affinity chromatography. Biochromatogr. 2002:307–317. [Google Scholar]
- 18.Wilson NL, Karlsson NG, Packer NH. Separation Methods in Proteomics. CRC Press LLC; 2006. Enrichment and analysis of glycoproteins in the proteome. [Google Scholar]
- 19.Hage DS. Clinical applications of high-performance affinity chromatography. Adv. Chromatogr. 2003;42:377–420. [PubMed] [Google Scholar]
- 20.Singhal RP, DeSilva S, Shyamali M. Boronate affinity chromatography. Adv. Chromatogr. 1992;31:293–335. [PubMed] [Google Scholar]
- 21.Foettinger A, Leitner A, Lindner W, Christian D. Solid-phase capture and release of arginine peptides by selective tagging and boronate affinity chromatography. J. Chromatogr. A. 2005;1079:187–196. doi: 10.1016/j.chroma.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 22.Smith IC, Carson BL, Ferguson TL. Osmium: an appraisal of environmental exposure. Environ. Health Perspect. 1974;8:201–213. doi: 10.1289/ehp.748201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najrana T, Saito Y, Uraki F, Kubo K, Yamamoto K. Spontaneous and osmiumtetroxide-induced mutagenesis in an Escherichia coli strain deficient in both endonuclease III and endonuclease VIII. Mutagenesis. 2000;15:121–125. doi: 10.1093/mutage/15.2.121. [DOI] [PubMed] [Google Scholar]
- 24.Pryor WA, Cueto R, Jin X, Koppenol WH, Ngu-Schwemlein M, Squadrito GL, Uppu PL, Uppu RM. A practical method for preparing peroxynitrite solutions of low ionic strength and free of hydrogen peroxide. Free Radic. Biol. Med. 1995;18:75–83. doi: 10.1016/0891-5849(94)00105-s. [DOI] [PubMed] [Google Scholar]
- 25.Li X-B, Pennington J, Stobaugh JF, Schöneich C. Synthesis of sulfonamide- and sulfonyl-phenylboronic acid modified silica phases for boronate affinity chromatography at physiological pH. Anal. Biochem. 2008;372:227–236. doi: 10.1016/j.ab.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikehata K, Duzhak TG, Galeva NA, Ji T, Koen YM, Hanzlik RP. Protein targets of reactive metabolites of thiobenzamide in rat liver in vivo. Chem. Res. Toxicol. 2008;21:1432–1442. doi: 10.1021/tx800093k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen BT, Davey SW, Ham AJ, Liebler DC. P-Mod: an algorithm and software to map modifications to peptide sequences using tandem MS data. J. Proteome Res. 2005;4:358–368. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 30.Miller JC, Miller JN. Ellis Horwood. London: 1992. Statistics for analytical chemistry; pp. 115–117. [Google Scholar]
- 31.Mitsugu A, Kyozo S. Characterization of a model compound for the lysine tyrosylquinone cofactor of lysyl oxidase. Biochem. Byophys. Res. Commun. 2001;218:193–199. doi: 10.1006/bbrc.2001.4315. [DOI] [PubMed] [Google Scholar]
- 32.Sharov VS, Dremina ES, Galeva NA, Gerstenecker GS, Li X-B, Dobrowsky RT, Stobaugh JF, Schöneich C. Fluorogenic tagging of protein 3-nitrotyrosine with 4-(aminomethyl) benzenesulfonic acid for quantitative analysis of protein tyrosine nitration. Chromatographia. 2010;71:37–53. doi: 10.1365/s10337-009-1409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuriel T, Deeb RS, Hajjar DP, Gross SS. Protein 3-nitrotyrosine in complex biological samples: quantitation by high-pressure liquid chromatography/electrochemical detection and emergence of proteomic approaches for unbiased identification of modification sites. Methods Enzymol. 2008;441:1–17. doi: 10.1016/S0076-6879(08)01201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]