Abstract
The Notch family of receptors plays essential roles in many phases of development, and dysregulation of Notch activity is increasingly recognized as a player in many diseases. O-Glycosylation of the Notch extracellular domain is essential for Notch activity, and tissue-specific alterations in the glycan structures are known to regulate activity. Here we review recent advances in identification and characterization of the enzymes responsible for glycosylating Notch and molecular mechanisms for how these O-glycans affect Notch activity.
Introduction
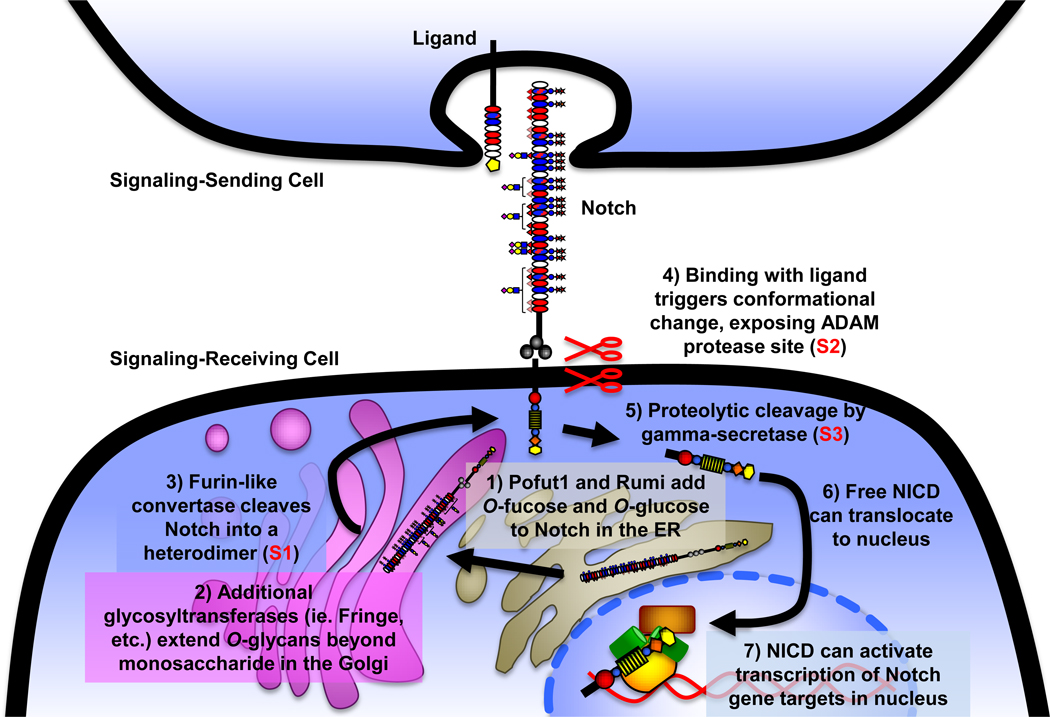
The Notch protein plays an important role as a transmembrane signaling receptor in a wide variety of developmental pathways [1,2]. Notch is conserved across all metazoans, and there are four mammalian homologs (Notch1–4). Loss of individual mouse Notch homologs 1 or 2 results in embryonic lethality, and mutations of Notch or downstream signaling components within the pathway have been implicated in a multitude of disease states in humans, including T-cell Acute Lymphoblastic Leukemia, CADASIL (Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy), Alagille Syndrome, Spondylocostal Dysostosis, Multiple Sclerosis, several heart defects, and Breast Cancer (reviewed in [1–4]). The Notch locus encodes a large (~300 kDa) single-pass Type I transmembrane receptor, comprised of a large extracellular domain (ECD) with multiple tandem Epidermal Growth Factor-like (EGF) repeats and negative regulatory region (NRR), followed by a transmembrane region and a large intracellular domain (ICD) involved in downstream signaling events (Figure 1A) [1]. The Notch signaling pathway is activated upon binding of the Notch ECD to one of its ligands presented on an apposing cell (Figure 2).
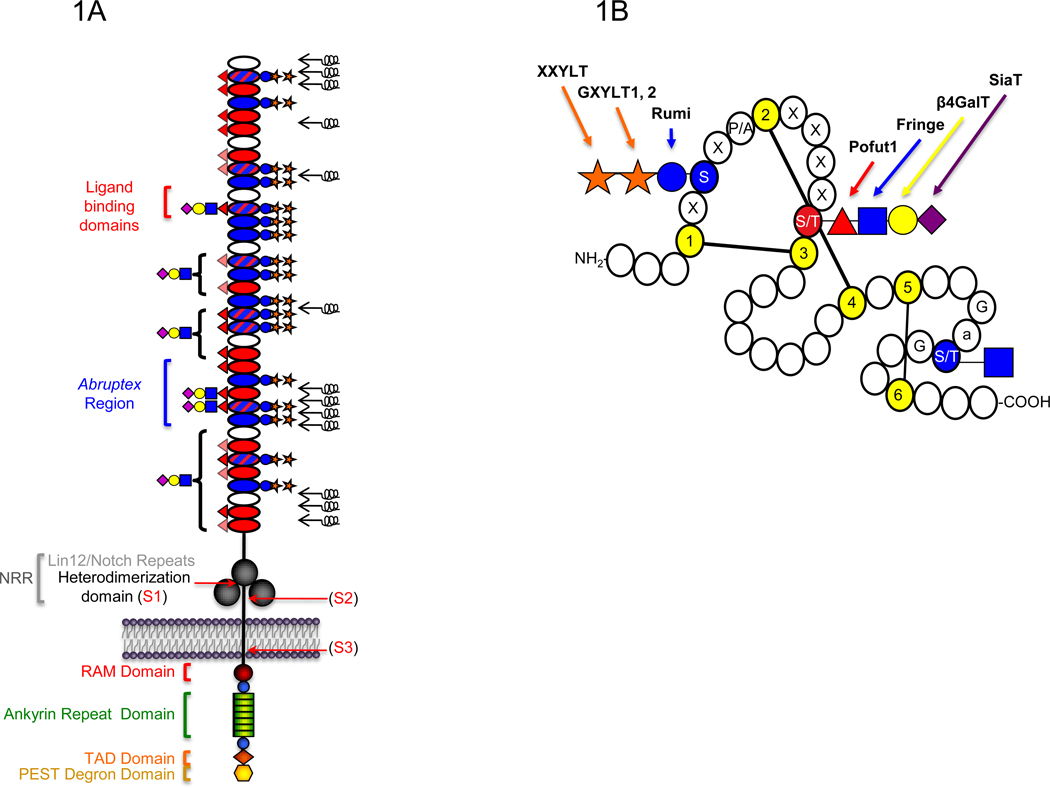
Figure 1. Schematic representation of the domain structure of the Notch receptor and an Epidermal Growth Factor-like (EGF) repeat.
A. The extracellular domain (ECD) of mouse Notch1 consists of 36 tandem EGF repeats (ovals) followed C-terminally by three Lin12/Notch repeats (LNRs) (large grey circles) and a heterodimerization domain (black line) [1]. EGF repeats containing the consensus sequence for O-glucosylation, O-fucosylation, or both, are filled with blue, red, or blue and red hatched lines, respectively. Mapped O-fucose and O-glucose glycans are indicated on each EGF repeat based on published data [19–21] (lightly shaded red triangles indicate O-fucose sites not yet confirmed). Results suggest that all O-glucose sites are modified with O-glucose trisaccharide, although some O-glucose monosaccharide form also exists at lower levels (not shown) [19]. Elongation of the saccharide beyond the O-fucose is based on work from [20,21]. Brackets indicate that elongation occurs on some or all of the O-fucose sites in that region. Symbols are based on Consortium for Functional Glycomics guidelines: glucose, blue circle; xylose, orange star; fucose, red triangle; GlcNAc, blue square; Galactose, yellow circle; Sialic Acid, purple diamond. Non-calcium-binding EGF repeats are unable to coordinate calcium ions to their N-terminal linker region, and are predicted to be flexible regions of the ECD (black springs) [47]. Ligands are known to physically interact with EGF11–12 [44,45]. The Abruptex region is defined by a series of mutations in EGF 24–29 of Drosophila Notch known to hyperactivate Notch and be refractory to Fringe [43]. The LNRs and heterodimerization domain together form the Negative Regulatory Region (NRR). Recent structural studies suggest that in the resting state, the LNRs mask the heterodimerization domain, preventing proteolysis [49,50]. Ligand binding is believed to result in a conformational change exposing the S2 protease site (see Figure 2). Sites of receptor cleavage events (S1-S3) are indicated with red arrows. The intracellular domain (ICD) is rich in several regulatory motifs as indicated, including a RAM (Regulation of Amino Acid Metabolism) domain, immediately followed by a Nuclear Localization Signal (NLS) (blue circles) and seven tandem ankyrin repeats (green rectangles). Following these motifs are a second NLS (blue circle), a Transcriptional Activation Domain (TAD) (orange diamond), and a PEST (Pro, Glu, Ser, Thr-rich) degron sequence involved in turnover of the ICD at the C-terminus (yellow hexagon). B. Short 40 amino acid motifs known as EGF repeats are defined by the presence of six conserved Cys (yellow) that form three disulfide bridges. Potential sites of O-glucosylation (C1-x-S-x-P/A-C2), O-fucosylation (C2-x-x-x-x-S/T-C3), and O-GlcNAc modification (no consensus sequence defined, but observed between Cys 5 and Cys 6 on Ser or Thr following Gly and an aromatic residue (a), and preceding Gly) are indicated in red or blue. Glycosyltransferases that carry out addition of each sugar are indicated with arrows.
Work in a number of laboratories over the past decade has demonstrated that O-fucosylation and O-glucosylation of the EGF repeats in the Notch ECD are essential for its function [3–9]. Elimination of the enzyme responsible for addition of O-fucose to EGF repeats (Protein O-fucosyltransferase 1, Pofut1 in mice, Ofut1 in Drosophila) reveals that O-fucosylation is universally required for all Notch signaling [10–12]. Elongation at the O-fucose by the β1–3N-acetylglucosaminyl transferase, Fringe, on EGF repeats modulates Notch activity in a number of tissue specific contexts and serves as a paradigm for how alterations in the glycosylation status of a receptor affect activity [13,14]. Genetic studies on the biological role of O-glucosylation have not been as extensive as for O-fucosylation, but elimination of the enzyme responsible for addition of O-glucose to EGF repeats (Protein O-glucosyltransferase, Poglut, gene name Rumi) also results in severe Notch-like phenotypes in flies or mice [15,16]. In this review we will focus on advances in our understanding of Notch glycosylation since it was last reviewed in this series [7], with a focus on what we have learned about the biochemistry of Notch O-glycosylation and current models for how the glycans affect Notch function.
Biochemistry of Notch O-glycosylation
The O-fucose and O-glucose glycans on Notch occur at specific consensus sequences within the context of EGF repeats, which make up the majority of the Notch ECD (Figure 1). Recently, O-GlcNAc modification, a third form of O-glycosylation was identified on EGF repeats as well, occurring on hydroxy amino acids between the fifth and sixth conserved Cys of an EGF repeat [17]. This sequence context has been sufficient to identify predicted O-GlcNAc sites on other EGF repeat-containing proteins, including the Notch ligand Delta [17]. Although no function for O-GlcNAc on Notch has yet been found, it is added to Notch by an enzymatic activity distinct from the well-known nuclear/cytoplasmic O-GlcNAc transferase, OGT [8,17,18].
While database searches with the O-fucose and O-glucose consensus sequences reveal nearly 100 mammalian proteins are predicted to be O-fucosylated [4], and over 40 proteins are potentially O-glucosylated [16], the Notch family of receptors contain the highest incidence of each consensus sequence and are predicted to be the most widely modified (Figure 1) [4,16]. Recent site-mapping work in our laboratory has revealed that O-glucose also exists on EGF 9 of mouse Notch1 at an unconventional sequence: C1ASAAC2, suggesting that Ala can replace Pro N-terminal to Cys 2 [19]. Database searches with the revised O-glucose consensus sequence (C1-x-S-x-A/P-C2) reveal additional new predicted sites on mouse Notch1 and 3, as well as sites on several novel proteins [19].
The fully extended forms of O-fucose and O-glucose glycans are shown diagrammatically in Figure 1B. O-Fucose on EGF repeats (including those from Notch [13,20–22]) can be elongated to a tetrasaccharide in mammals: Siaα2–3/6Galβ1–4GlcNAcβ1–3Fuc (Figure 1B) [23,24], whereas elongation beyond the disaccharide GlcNAcβ1–3Fuc has not been detected on Drosophila Notch protein isolated from S2 cells [25]. Recently, Aoki et al. carried out studies to define the O-glycome of Drosophila melanogaster by mass spectrometry, using β-elimination to release the O-linked sugar modifications from total protein extracts of fly embryos [26]. They discovered a novel glucuronic acid-containing O-linked fucose trisaccharide: GlcAβ1–4(GlcNAcβ1–3)Fucitol. Given the similarity to the GlcNAcβ1–3Fuc on the EGF repeats of Notch, the trisaccharide was proposed to be derived from Notch, although further work needs to be done to determine whether this trisaccharide exists on Notch or other proteins in vivo. O-Glucose is typically found elongated to a trisaccharide on EGF repeats (Xylα1-3Xylα1–3Glc, Figure 1B) [23], including on mouse Notch1 [19,22,27]. In their O-glycome analysis of Drosophila, Aoki and co-workers also detected O-glucose mono- (Glucitol) and disaccharide (Xylα1–3Glucitol), but no trisaccharide among the reductively released saccharides [26]. Consistent with these results, mass spectral O-glucose site mapping on Drosophila Notch has confirmed the presence of mono- and disaccharide forms of O-glucose at specific sites ([15], Rana et al., unpublished).
Nearly all of the enzymes responsible for addition of O-fucose and O-glucose glycans to EGF repeats have been identified (Figure 1B). Pofut1 is a soluble, ER-localized enzyme that adds fucose to Ser or Thr in the C2-x-x-x-x-(S/T)-C3 consensus sequence [28,29]. Fringe is a Golgi-localized β1–3N-acetylglucosaminyltransferase that extends O-fucose to the disaccharide [13,14,30]. While three Fringe homologs exist in mammals (Lunatic, Manic, and Radical Fringe), a single Fringe exists in flies [31]. The structure GlcNAcβ1–3Fucα1-O-Ser/Thr can be further elongated in mammals to the tetrasaccharide by sequential action of a β1-4galactosyltransferase and an α2–3 or α2–6sialyltransferase.
Rumi was recently identified as the gene encoding Poglut, a soluble, ER-localized enzyme responsible for addition of glucose to Ser in the C1-x-S-x-A/P-C2 consensus sequence [15,16,32]. Two human glucoside α1–3xylosyltransferases (GXYLT1 and GXYLT2) have been recently identified that extend O-glucose to a disaccharide [33]. Both are predicted to be Type II membrane glycoproteins, similar to most Golgi-localized glycosyltransferases [34]. The only enzyme required for O-glucose trisaccharide biosynthesis not yet identified is the xyloside α1-3xylosyltransferase (XXYLT) that adds a second xylose to generate the Xylα1–3Xylα1–3Glcβ1-O-Ser trisaccharide found in mammals. Recent glycoproteomic studies on O-glucose site occupancy show O-glucose trisaccharide occurs at high stoichiometries at most sites, but some site and cell-specific underglucosylation has been observed on mouse Notch1 and 2 (Figure 1A, [16,19,35]). These results suggest the extent of modification at some sites may be more sensitive to expression levels of Rumi than others. Interestingly, recent results suggest that Notch signaling is sensitive to the levels of Rumi expressed in a tissue [16].
Models for how O-glycans affect Notch function
O-Fucose and Fringe modification affect ligand binding
Elimination of Pofut1 in mice has a profound effect on ligand binding in both embryonic stem cells [36] and lymphoid cells [37]. A chaperone-like activity has been reported for Drosophila Ofut1 that is required for cell-surface expression of Notch in flies [38], but this chaperone activity has not been clearly seen in the mouse system, as cells lacking Pofut1 have Notch proteins on their surfaces [36,37]. This discrepancy may be explained by differences in species, or in cell-dependent expression of other chaperones. A small decrease in cell surface expression of Notch proteins is seen in embryonic stem cells lacking Pofut1 [36] and in somites from mice with a hypomorphic allele of Pofut1, cax [39]. More detailed analysis of the chaperone activity of Pofut1/Ofut1 has been reviewed elsewhere [3,5,8].
Similar to O-fucosylation, the major effect of Fringe-mediated O-fucose elongation appears to be modulation of Notch-ligand binding, whereby Delta activation of Notch is potentiated, while signaling via Serrate is inhibited. In flies, data indicate that GlcNAc is the terminal sugar added to O-fucose residues on Drosophila Notch, and that the disaccharide is sufficient for observing a Fringe effect. In vitro extension to trisaccharide causes no change in in vitro ligand binding as assessed in vitro [25]. While the mammalian system is more complicated (4 receptors, 5 ligands, and 3 Fringes), the majority of data suggest mammalian Fringe modification also alters ligand binding [3,5,8,9]. Elongation beyond GlcNAc to the trisaccharide (Galβ1–4GlcNAcβ1–3-Fucoseα1-O-Ser/Thr) is necessary to see a Fringe effect in a mammalian cell system [40], an important departure from requirements in the fly system. Consistent with these findings, β4-galactosyltransferase-1 knockout mice exhibit a minor Notch phenotype [41].
O-Glucose affects a step after ligand binding but prior to proteolysis
To examine what roles Rumi and O-glucose play in proper Notch function in Drosophila, Acar et al. examined steps of the canonical signaling cascade where Rumi may be exerting regulation [15]. Notch is present at the cell surface in cells lacking Rumi, indicating that O-glucosylation is not required for cell surface presentation of the receptor, and that Rumi does not have any chaperone-like activity. Defects in ligand binding were ruled out as the cause for the rumi phenotype, as Notch generated in rumi knockdown cells binds Delta as strongly as protein generated from control cells. Interestingly, analysis of protein extracts from various tissues of rumi mutant flies showed significantly decreased Notch proteolysis (S2 or S3 cleavage, Figure 2). As proteolysis requires conformational changes in the ECD (Figure 2), this suggests O-glucosylation may affect the structure of the ECD. Similar results have been obtained in mammalian cells where rumi has been knocked down using RNAi [16]. The temperature sensitivity of the phenotype in flies is consistent with destabilization of the Notch ECD due to the absence of O-glucose [15]. This lack of stability could in turn interfere with the ability of the receptor to couple ligand binding to requisite conformational changes necessary for proteolysis (Figure 2).
Figure 2. Notch activation pathway.
Notch is O-fucosylated and O-glucosylated in the ER (1), and elongated by additional glycosyltransferases in the Golgi (2). Notch is also cleaved by a furin-like convertase in the Golgi (S1 cleavage, 3), generating the mature heterodimeric receptor that is subsequently expressed at the cell surface. Following interaction with a DSL (Delta, Serrate, Lag2) ligand presented on an apposing cell, Notch ECD undergoes a conformational change exposing the protease site for an ADAM protease (Kuzbanian in Drosophila) at the membrane interface (S2 cleavage, 4), and is subsequently cleaved by gamma-secretase just inside the cell membrane (S3 cleavage, 5), causing ICD release. Upon S3 cleavage, the ICD is free to translocate to the nucleus (6), bind to a member of the CSL (CBF/SuH/LAG-1) family of transcription factors via its RAM domain, and this binary complex serves as a platform for Mastermind-like (MAML)-family cofactors, which in turn can act as scaffolding for transcriptional complex formation. The assembled transcriptional activation complex can then activate expression of Notch target genes (7), including Hairy-Enhancer of Split (Hes) and Hairy-Enhancer of Split related with YRPW motif (Hey) family members [1].
O-Glycosylation sites important for Notch activity
Several studies have examined whether specific O-fucose or O-glucose sites on Notch are important for activity. Eliminating any of three highly conserved O-fucose sites at EGF 12, 26, or 27 within mouse Notch1 alters activity in cell-based Notch signaling assays [21]. EGF 12 is part of the ligand-binding region of Notch (Figure 1A). Ge and Stanley generated a mouse line carrying a point mutation in the O-fucosylation site of EGF 12 in endogenous Notch1 [42]. Loss of this site resulted in a mild Notch phenotype with defects in T cell development. This result shows that O-fucosylation of EGF 12 plays an important role in Notch1 function, but suggests that additional O-fucose sites, such as those on EGF 26 or 27, are likely contributing to Notch activity. Interestingly, EGF 26 and 27 are in the Abruptex region of the Notch receptor (Figure 1A), named for a group of mutations in Drosophila Notch that result in hyperactivated Notch that is refractory to Fringe [43].
We have also performed cell-based signaling assays to examine the role of each O-glucosylation site in mouse Notch1. Only elimination of O-glucose at EGF 28 results in significant decreases in Delta-like1-mediated Notch1 signaling [19]. This is a surprising result, as there are 17 sites of O-glucosylation on Notch1, and suggests that individual mutations may not explain the major effects of Rumi. Of interest is the fact that like EGF repeats 26 and 27, EGF 28 also maps to the Abruptex region of the receptor (Figure 1A).
Structural analysis of Notch signaling
Structural studies of the Notch family of receptors are required to fully understand the molecular mechanisms by which O-fucose and O-glucose glycans affect Notch function. The structural implications of O-glycosylation on Notch-ligand interactions have yet to be defined, but recent structural studies provide insights into Notch ECD global structure, and the possible impact of O-glycosylation on altering that structure. Several studies have focused on the structure of EGF 11–12, the ligand-binding region of Notch [44,45]. Recently, the structures of unmodified, O-fucosylated, and Fringe elongated mouse Notch1 EGF 12 were compared by NMR [46]. Interestingly, the fucose moiety appears to contribute to stability of the anti-parallel β-sheet in the EGF repeat, and Fringe elongation to the GlcNAc-β1,3-Fucose causes a significant conformational shift of several residues within the O-fucose consensus region. This may provide a mechanism for how Fringe modification indirectly exerts its effects on Notch activity at EGF 12.
Although the Notch ECD is traditionally schematically represented as a linear and rigid structure (Figure 1A), several recent studies indicate that this is unlikely to be an accurate depiction of the global structure of the receptor. Some EGF repeats are capable of coordinating calcium via conserved acidic/polar residues at the N-terminal linker region between two adjoining EGFs (calcium binding EGF, or cbEGF). Using EGF 11–13 of human Notch1, Hambleton and co-workers observed that in the presence of calcium, the calcium-binding motifs in the linker between two adjacent EGF repeats impart rigidity to this region [47]. The flexibility of non-cbEGF linkers, which do not coordinate calcium, is supported by the observation that consecutive non-cbEGF repeats from Plasmodium falciparum merozoite surface protein-1 assume a U-shaped structure in which the non-cbEGF domains share a hydrophobic interface. Based on these results, Hambleton and co-workers proposed that the Notch ECD consists of both rigid and flexible regions dependent on the presence or absence of cbEGFs. Of special interest is the fact that the organization of calcium-binding sites within the Notch ECD is highly conserved [45], suggesting these regions of structural rigidity and flexibility are essential for Notch ECD function.
Support for this model of regions of flexibility and rigidity comes from electron microscopy (EM) studies by Kelly et al. [48]. Using the ECD of both Drosophila and human Notch1, their images depict structures that appear to have globular structures where the ECD is folded up on itself, consistent with the flexible regions predicted to exist in both proteins based on positioning of cbEGFs in these receptors. Further work remains to be performed to address whether these structures are biologically active, or whether O-fucose or O-glucose glycans affect this structural conformation.
Conclusions
Just over 10 years ago, Notch activity was shown to be regulated by Fringe-mediated elongation of O-fucose on its ECD [13,14]. Since then most of the enzymes responsible for addition of O-fucose and O-glucose glycans have been identified, and their importance for Notch activity has been confirmed. We now know that the predicted consensus sites are modified, usually at high stoichiometries, at least in Notch protein overproduced in tissue culture systems. Future studies need to examine the site-specific modification of Notch proteins isolated from physiologically relevant in vivo sources. We have also learned through mutagenesis of the O-fucose site in the ligand-binding domain (EGF 12) and individual O-fucose or O-glucose sites in the Abruptex region (EGF 26, 27, and 28), that O-glycosylation of these sites plays important roles in Notch1 function. Elimination of the O-fucose site at EGF 12 decreases Notch1 activity in vivo [42]. Future studies need to examine the in vivo importance of other sites (e.g. O-fucose on EGF 26 or 27, O-glucose on EGF 28). Finally, structural studies on the Notch ECD need to be performed on protein with different modifications (e.g. with or without Fringe or GXYLT elongation), to better understand how these glycans modulate Notch function.
Highlights.
Notch is modified at multiple sites with O-fucose and O-glucose glycans
Elongation of O-fucose glycans by the glycosyltransferase Fringe regulates Notch-ligand binding
Rumi encodes the protein O-glucosyltransferase responsible for adding O-glucose to Notch
O-Glucose may stabilize Notch to allow proper receptor activation
Multiple O-glycan sites contribute to Notch ligand-binding and activity
O-Glycosylation may alter the flexibility of the Notch extracellular domain
Acknowledgements
The authors would like to thank members of the Haltiwanger laboratory for critical reading of the manuscript and helpful discussions. Original work was supported by NIH grant GM061126 and training grant NCI T32 CA009176.
Abbreviations
- ICD
Intracellular Domain
- ECD
Extracellular Domain
- Poglut
Protein O-Glucosyltransferase
- Pofut1/Ofut1
Protein O-Fucosyltransferase 1/O-fucosyltransferase 1
- EGF repeat
Epidermal Growth Factor-like Repeat
- cbEGF
Calcium-binding EGF repeat
- OGT
O-GlcNAc Transferase
- LNR
Lin-12/Notch Repeat
- RAM
Regulation of Amino Acid Metabolism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of interest to highlight:
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Developmental cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Jafar-Nejad H, Leonardi J, Fernandez-Valdivia R. Role of glycans and glycosyltransferases in the regulation of Notch signaling. Glycobiology. 2010;20:931–949. doi: 10.1093/glycob/cwq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rampal R, Luther KB, Haltiwanger RS. Notch signaling in normal and disease States: possible therapies related to glycosylation. Current molecular medicine. 2007;7:427–445. doi: 10.2174/156652407780831593. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi H, Haltiwanger RS. Role of glycosylation of Notch in development. Seminars in cell & developmental biology. 2010;21:638–645. doi: 10.1016/j.semcdb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanley P, Guidos CJ. Regulation of Notch signaling during T- and B-cell development by O-fucose glycans. Immunological reviews. 2009;230:201–215. doi: 10.1111/j.1600-065X.2009.00791.x. [DOI] [PubMed] [Google Scholar]
- 7.Stanley P. Regulation of Notch signaling by glycosylation. Current opinion in structural biology. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Current topics in developmental biology. 2010;92:131–164. doi: 10.1016/S0070-2153(10)92004-8. [DOI] [PubMed] [Google Scholar]
- 9.Luther KB, Haltiwanger RS. Role of unusual O-glycans in intercellular signaling. The international journal of biochemistry & cell biology. 2009;41:1011–1024. doi: 10.1016/j.biocel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okajima T, Irvine KD. Regulation of notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 11.Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M, Kitagawa M, Harigaya K, Spana E, Bilder D, et al. neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development. 2003;130:4785–4795. doi: 10.1242/dev.00679. [DOI] [PubMed] [Google Scholar]
- 13.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 14.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 15. Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. The rumi gene was identified during a mutant screen for modifiers of Notch activity in flies. Sequence analysis revealed it to be a soluble, ER-localized protein with a CAP10 glycosyltransferase-like domain, consistent with previous studies on Poglut activity in cells. Expressed and purified Rumi protein was then demonstrated to contain Poglut activity.
- 16. Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS, Jafar-Nejad H. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138:1925–1934. doi: 10.1242/dev.060020. Elimination of the mouse homologue of Rumi results in a severe embryonic lethal phenotype with somewhat more severe defects than expected from loss of Notch function. These studies suggest Rumi is required for Notch activity in mammalian cells, but that additional targets may also be affected.
- 17.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. The Journal of biological chemistry. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 18.Sakaidani Y, Furukawa K, Okajima T. O-GlcNAc modification of the extracellular domain of Notch receptors. Methods in enzymology. 2010;480:355–373. doi: 10.1016/S0076-6879(10)80016-3. [DOI] [PubMed] [Google Scholar]
- 19. Rana NA, Nita-Lazar A, Takeuchi H, Kakuda S, Luther KB, Haltiwanger RS. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. The Journal of biological chemistry. 2011 doi: 10.1074/jbc.M111.268243. IN PRESS. This paper provides the first comprehensive site mapping of predicted O-glucose sites on any of the Notch proteins using highly sensitive mass spectral methods. O-Glucose trisaccharide was found at all predicted sites, and at a novel site within EGF 9. Stoichiometry of modification was high at most sites, but a few showed variability. Mutatagenesis studies revealed that elimination of the O-glucose site at EGF 28 results in a significant decrease in Dll1-mediated Notch1 activation using cell-based assays.
- 20.Shao L, Moloney DJ, Haltiwanger R. Fringe modifies O-fucose on mouse Notch1 at epidermal growth factor-like repeats within the ligand-binding site and the Abruptex region. The Journal of biological chemistry. 2003;278:7775–7782. doi: 10.1074/jbc.M212221200. [DOI] [PubMed] [Google Scholar]
- 21.Rampal R, Arboleda-Velasquez JF, Nita-Lazar A, Kosik KS, Haltiwanger RS. Highly conserved O-fucose sites have distinct effects on Notch1 function. The Journal of biological chemistry. 2005;280:32133–32140. doi: 10.1074/jbc.M506104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, Haltiwanger RS. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. The Journal of biological chemistry. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 23.Harris RJ, Spellman MW. O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology. 1993;3:219–224. doi: 10.1093/glycob/3.3.219. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura H, Takao T, Hase S, Shimonishi Y, Iwanaga S. Human factor IX has a tetrasaccharide O-glycosidically linked to serine 61 through the fucose residue. The Journal of biological chemistry. 1992;267:17520–17525. [PubMed] [Google Scholar]
- 25.Xu A, Haines N, Dlugosz M, Rana NA, Takeuchi H, Haltiwanger RS, Irvine KD. In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. The Journal of biological chemistry. 2007;282:35153–35162. doi: 10.1074/jbc.M707040200. [DOI] [PubMed] [Google Scholar]
- 26. Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. The Journal of biological chemistry. 2008;283:30385–30400. doi: 10.1074/jbc.M804925200. Here the authors performed a total O-glycome analysis by mass spectral analysis of O-glycans released from proteins extracted from Drosophila embryos. Surprisingly, one of the most abundant structures detected was a novel O-fucose trisaccharide: GlcAβ1-4(GlcNAcβ1-3)Fucitol. Detailed analysis showed this structure more abundant in Fringe-expressing cells, suggesting it may be present on Notch. These same studies showed the presence of O-glucose monosaccharide (Glucitol) and disaccharide (Xylα1-3Glucitol), but no O-glucose trisaccharide (Xylα1-3Xylα1-3Glucitol) nor O-fucose monosaccharide (Fucitol) or disaccharide (GlcNAcβ1-3Fucitol).
- 27. Whitworth GE, Zandberg WF, Clark T, Vocadlo DJ. Mammalian Notch is modified by D-Xyl-alpha1-3-D-Xyl-alpha1-3-D-Glc-beta1-O-Ser: implementation of a method to study O-glucosylation. Glycobiology. 2010;20:287–299. doi: 10.1093/glycob/cwp173. Using chemically synthesized Xylα1-3Xylα1-3Glucose as a standard, these authors confirmed that the O-glucose trisaccharide on Notch (previously reported, but for which a structure had not been determined) has the same structure as that previously found on EGF repeats from human clotting factors 7 and 9.
- 28.Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, Haltiwanger RS. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. The Journal of biological chemistry. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 29.Luo Y, Haltiwanger RS. O-fucosylation of notch occurs in the endoplasmic reticulum. The Journal of biological chemistry. 2005;280:11289–11294. doi: 10.1074/jbc.M414574200. [DOI] [PubMed] [Google Scholar]
- 30.Luther KB, Schindelin H, Haltiwanger RS. Structural and mechanistic insights into lunatic fringe from a kinetic analysis of enzyme mutants. The Journal of biological chemistry. 2009;284:3294–3305. doi: 10.1074/jbc.M805502200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 32.Lee TV, Takeuchi H, Jafar-Nejad H. Regulation of notch signaling via O-glucosylation insights from Drosophila studies. Methods in enzymology. 2010;480:375–398. doi: 10.1016/S0076-6879(10)80017-5. [DOI] [PubMed] [Google Scholar]
- 33. Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. The Journal of biological chemistry. 2010;285:1582–1586. doi: 10.1074/jbc.C109.065409. These studies identified the xylosyltransferase responsible for adding α1-3linked xylose to O-glucose on EGF repeats. Two genes encoding this activity were identified: GXYLT1 and GXYLT2. Both were identified based on their similarity to the UDP-glucose: glycoprotein glucosyltransferase (UGGT) involved in folding of N-glycosylated proteins in the ER.
- 34.Paulson JC, Colley KJ. Glycosyltransferases. Structure, localization, and control of cell type-specific glycosylation. The Journal of biological chemistry. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 35.Bakker H, Oka T, Ashikov A, Yadav A, Berger M, Rana NA, Bai X, Jigami Y, Haltiwanger RS, Esko JD, et al. Functional UDP-xylose transport across the endoplasmic reticulum/Golgi membrane in a Chinese hamster ovary cell mutant defective in UDP-xylose Synthase. The Journal of biological chemistry. 2009;284:2576–2583. doi: 10.1074/jbc.M804394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P. Roles of Pofut1 and O-fucose in mammalian Notch signaling. The Journal of biological chemistry. 2008;283:13638–13651. doi: 10.1074/jbc.M802027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, Xin W, Gerson S, Stanley P, Lowe JB, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117:5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005;307:1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- 39.Schuster-Gossler K, Harris B, Johnson KR, Serth J, Gossler A. Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development. BMC developmental biology. 2009;9:6. doi: 10.1186/1471-213X-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Moloney DJ, Stanley P. Fringe modulation of Jagged1-induced Notch signaling requires the action of beta 4galactosyltransferase-1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13716–13721. doi: 10.1073/pnas.241398098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Lu L, Shi S, Stanley P. Expression of Notch signaling pathway genes in mouse embryos lacking beta4galactosyltransferase-1. Gene expression patterns : GEP. 2006;6:376–382. doi: 10.1016/j.modgep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 42. Ge C, Stanley P. The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1539–1544. doi: 10.1073/pnas.0702846105. The O-fucose site in EGF 12 of mouse Notch1 was mutated in the endogenous locus. Homozygotes showed a hypomorphic Notch phenotype, specifically in T cell development. These studies clearly demonstrate the importance of O-fucose on EGF12, but the lack of a full Notch phenotype (as seen in Pofut1 null mice) suggests that O-fucose on other EGF repeats must also play important roles in Notch1 function.
- 43.de Celis JF, Bray SJ. The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development. 2000;127:1291–1302. doi: 10.1242/dev.127.6.1291. [DOI] [PubMed] [Google Scholar]
- 44.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 45.Xu A, Lei L, Irvine KD. Regions of Drosophila Notch that contribute to ligand binding and the modulatory influence of Fringe. The Journal of biological chemistry. 2005;280:30158–30165. doi: 10.1074/jbc.M505569200. [DOI] [PubMed] [Google Scholar]
- 46. Hiruma-Shimizu K, Hosoguchi K, Liu Y, Fujitani N, Ohta T, Hinou H, Matsushita T, Shimizu H, Feizi T, Nishimura S. Chemical synthesis, folding, and structural insights into O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor. Journal of the American Chemical Society. 2010;132:14857–14865. doi: 10.1021/ja105216u. Here the authors chemically synthesized EGF repeat 12 from mouse Notch1 with O-fucose, or GlcNAcβ1-3Fucose, attached. Comparison of the structures by NMR revealed that the presence of the GlcNAc caused a significant, local, conformational change in residues near the O-fucose glycosylation site.
- 47.Hambleton S, Valeyev NV, Muranyi A, Knott V, Werner JM, McMichael AJ, Handford PA, Downing AK. Structural and functional properties of the human notch-1 ligand binding region. Structure. 2004;12:2173–2183. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 48.Kelly DF, Lake RJ, Middelkoop TC, Fan HY, Artavanis-Tsakonas S, Walz T. Molecular structure and dimeric organization of the Notch extracellular domain as revealed by electron microscopy. PloS one. 2010;5:e10532. doi: 10.1371/journal.pone.0010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, Huang L, Vitelli S, Vo KT, Haytko P, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PloS one. 2010;5:e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]