Abstract
S-nitrosothiols (RSNO) present in nanomolar concentrations in cells and body fluids play an important role in vasodilation, in preventing platelet aggregation, leukocyte adhesion and for cellular signaling. However, because of the low levels of RSNO and interference with other nitric oxide species, reliable assays that measure both high molecular -weight and low molecular weight RSNO in plasma and RBCs have been difficult to develop and there is no consensus of reported values. We have previously developed a sensitive method using Cu(II)-ascorbic acid at neutral pH, which was specific for RSNO without interference of nitrite or other NOx species. However, due to neutral pH foaming, this method was not suitable for determinations in plasma or RBCs with high protein content. This method has now been modified by using CuCl2 and ascorbic acid in glacial acetic acid. The low pH solves the foaming problem. However, protonation of under acidic conditions facilitates the formation of RSNO. For this method to specifically measure RSNO in the sample, the unreacted thiols are blocked by reacting with N-ethylmaleimide [NEM] and nitrite is blocked by reacting with acidified sulfanilamide before being analyzed by chemiluminescence. Using this method, S-nitrosothiols have been determined in the range of 2 nM to 26 nM (mean ±SE = 10.18±2.1) in plasma and 0 to 88.1nM (mean ±SE = 51.27± 10.5) in red blood cells.
Keywords: S-nitrosothiols, nitric oxide, ozone based chemiluminescence assay, plasma, red blood cells
1. Introduction
S-nitrosothiols (RSNO) are adducts of nitric oxide with thiol groups. Nitric oxide does not directly react with thiols to form RSNOs at physiological pH (1). However, oxidation of NO by oxygen forms dinitrogen trioxide, which can react with thiols to form RSNO (2). High molecular weight and low molecular weight RSNO are present in nanomolar concentrations in vivo in cells and body fluids. It has been proposed that RSNOs in the plasma play a role in vasodilation, anti-platelet aggregation activity and anti-leukocyte adhesion properties (3;(4). They also serve as intracellular signaling molecules (5). Blood RSNO levels have been shown to vary under various pathological conditions and, therefore, have potential clinical relevance (6).
Determination of total or individual S-nitrosylated proteins is very challenging. Several methods have been developed to determine the total RSNO in plasma, red blood cells and whole blood (7). The most popular and simple method for the determination of RSNO is the Saville reaction involving the treatment of RSNO with mercuric chloride, which releases NO+ that then reacts with Griess reagents to form an azo dye that can be detected calorimetrically. Fluorescence methods have also been developed for the detection of RSNOs using 4,5 diaminofluorescein dyes. These methods are, however, not sensitive enough to measure the physiological levels of RSNO that are expected to be < 0.1μM. The ozone based chemiluminescence method using Sievers NOA 280i instrument, is one of the highly sensitive methods to determine nanomolar concentrations of NO. The details of the instrument and techniques for measurement of NO have been reviewed (8;9;10). The bond between sulfur and NO must be cleaved in order to determine RSNOs by this method. Several methods are available in the literature for this purpose. They include photolysis (11), Cu(I)-Iodide (12) CuCl2 + cysteine (13), tri-iodide (7) and Cu(II) + ascorbate (10;14) to cleave the RSNO bond at neutral pH or acidic pH. The KI/I2 reductive reagent has been widely used to determine nitrite and RSNO in biological fluids (10;15). It has been shown that this reagent measures the nitrite and N-nitrosamines in addition to RSNO (16). Marley et al. have employed pretreatment of the sample with acidified sulfanilamide to remove the nitrite interference (12). We developed a method to determine the RSNO using Cu+2- ascorbic acid at neutral pH without interference of nitrite (14). This method has a high sensitivity to measure both high and low molecular weight RSNOs including S-nitrosohemoglobin and albumin-SNO. However, this method is not suitable for high protein content samples due to formation of foam at neutral pH in the reaction vessel. Since the RSNO levels in biological samples are in low nanomolar concentrations, a large volume, which contains a high level of protein, needs to be injected to see a signal. Therefore, this method has been modified by using Cu(II)Cl2 and ascorbate in strong acidic acid instead of neutral pH. We found that Cu(II) in the presence of ascorbate readily cleaves the RSNO bond to stiochiometrically releases NO. This reagent does not release NO from nitrated lipids, proteins and N-nitrosamines.
The released NO in the reaction vessel is carried by inert gas to the detector where it reacts with ozone to produce a chemiluminescence signal proportional to the concentration. This method also detects nitrite in addition to RSNO. This nitrite interference is, however, eliminated by pre-treating the sample with acidified sulfanilamide, which forms a diazonium complex with nitrite that does not generate a chemiluminescence signal.
2. Materials
2. 1. Specific Equipment
Sievers Nitric Oxide Analyzer Model 280 (GE Analytical Instruments, Boulder, CO) (see ref 8 for more details)
Liquid program (3.21)
Origin program
Oxygen and inert gases (argon or nitrogen)
Hamilton syringes 50, 100, 250 μl, with 5 inch long needles
Portable centrifuge
2. 2. Reagents
Water: Ultra pure water (UPH2O) (at 18.2 MΩ-cm) from PURELAB Plus or Millipore water Milli Q-Gard 2 Purification Pack or doubled distilled water (See Note 6).
50 mM diethylenetriaminepentaacetic acid (DTPA) solution: Suspended 983.4 mg DTPA in 25 ml of UPH2O water. Add 1 M NaOH drop wise until it dissolved. Adjust the pH to 7.0. Make up the final volume to 50 ml with UPH2O. This solution is stable at room temperature.
5% Acidified sulfanilamide: Dissolve 5 g sulfanilamide in 100ml 2M HCl.
-
Preparation of S-nitrosoglutathione (GSNO) standard solution.
Stock solution: 10 mM glutathione solution (Sigma-Aldrich) is prepared by dissolving 30.732 mg of glutathione in 10 ml of 1M HCl. Similarly, 10 mM nitrite solution is prepared by dissolving 6.9 mg nitrite in 10 ml of UPH2O containing 0. 2mM DTPA solution (40 μl of 50 mM DTPA diluted to 10ml with UPH2O). 5ml of glutathione solution is mixed with 5 ml of nitrite solution to form GSNO. This solution should be capped, protected from light and kept cold (4°C). The concentration of this solution is determined by diluting 0.25 ml of GSNO stock solution to 2.5 ml with UPH2O in a cuvette and an absorption spectrum is run from 250 nm to 450 nm. The maximum absorbance at 335 nm is recorded. The concentration of GSNO is determined by using the mM extinction coefficient of 0.980 at 335 nm. Calculation= absorbance at 335/0.980 × 10 (dilution factor). This value is in the range of 4 to 5mM. The concentration of GSNO is adjusted to 1 mM by appropriate dilution with 0.5 M HCl.
Working Standard solution (1μM): 10 μl of stock solution (1mM) is diluted in 10 ml of sulfanilamide solution (9:1V/V, UPH2O: 5% acidified sulfanilamide). This 1μM working standard is serially diluted with acidified sulfanilamide (9:1V/V, UPH2O: 5% acidified sulfanilamide) at a ratio of 1:2, 1:4 … down to a concentration of 7.8 nM (Table 1). (See notes 1, 8 & 9)
250 mM N-ethylmaleimide (NEM) solution: Dissolve 313 mg in 10 ml PBS. This reagent should be prepared fresh every day.
100 mM potassium ferricyanide: Dissolve 329 g in 10 ml UPH2O.
SNO preservation solution: 9.75 ml potassium ferricyanide mixed with 0.1 ml of triton X100, 0.1ml of 250 mM NEM and 50 μl of 50mM DTPA.
100 mM cupric chloride (CuCl2): Dissolve 170.48 mg cupric chloride dihydrate in 10 ml UPH2O.
125 mM ascorbic acid solution: Dissolve 220.15 mg in 10 ml of UPH2O. Prepare fresh reagent every day.
Table 1.
Preparation of GSNO standards.
| Ratio | Volume of 1 μM GSNO std (ml) | Volume of acidified sulfanilamide (ml) | Total volume (ml) | Final nitrite concentration (nM) |
|---|---|---|---|---|
| 1:0 | 10 | 0 | 10 | 1000 |
| 1:2 | 5 | 5 | 10 | 500 |
| 1:4 | 2.5 | 7.5 | 10 | 250 |
| 1:8 | 1.25 | 8.75 | 10 | 125 |
| 1:16 | 0.625 | 9.375 | 10 | 62.5 |
| 1:32 | 0.3125 | 9.6875 | 10 | 31.25 |
| 1:64 | 0.156 | 9.844 | 10 | 15.625 |
| 1:128 | 0.078 | 9.922 | 10 | 7.812 |
3. Methods
3.1 Calibration curve for S-nitrosothiols
Add 7.5 ml glacial acetic acid, 200 μl of 100 mM Cu(II)Cl2 and 200 μl of 125 mM ascorbic acid into the purge vessel.
Gas bubbler/sodium hydroxide trap is connected between the purge vessel and the detector.
Water bath temperature for circulating water around water-jacked purge vessel is set at 37°C. Cold water circulation for condenser is not required.
Liquid program is used to acquire data. The reaction mixture is continuously purged with inert gas at constant cell pressure by adjusting the gas flow. The data is acquired every ¼ sec. Once the baseline is stabilized, 100μl of working standard solutions from 7.812, 15.62, 31.25, 62.5, 125, 250, 500 and 1000 nm GSNO is injected into the purge vessel using a 5 inch long needle(see note 9). Chemiluminescence signals are recorded. Duplicate measurements are made for each sample.
Origin program is used to analyze the data: It can be purchased from OriginLab Corporation, One Roundhouse Plaza, Suite 303, Northampton, MA 01060 USA, www.originlab.com.
The data from the liquid program is imported into the Origin program in an ASCII format. Chemiluminescence signals were regenerated by plotting the time (X axis with each row equivalent to 0.00417 min) against mV values (Y axis) (Fig.1A).
The area under the curve of each signal is calculated after determination of the baseline and finding the peaks using the tool menu of the program (Table 2).
Standard Curve: The standard curve is constructed from the average area of the peaks versus concentration. The slope and intercept of the standard curve is determined (Fig 1B).
Figure 1.

(A) Chemiluminescence signals of GSNO standards: GSNO (7.812 -1000nM; 100μl) is injected into the purge vessel of Nitric Oxide Analyzer that contains 7.5 ml glacial acetic acid 200μl of 100mM Cu(II)Cl2 and 200μl of ascorbic acid. (B) Calibration curve for S-nitrosothiols is obtained by integrating the area under the curve.
Table 2.
Average area under the curve of each signal.
| Nitrite concentration (nM) | Signal area under curve (AU) |
|---|---|
| 7.812 | 0.150 |
| 15.625 | 0.32 |
| 31.25 | 0.467 |
| 62.5 | 0.975 |
| 125 | 1.835 |
| 250 | 4.035 |
| 500 | 7.89 |
| 1000 | 16.1 |
The area of the blank value (water) 0.201 is subtracted from the area of all the nitrite standards. A.U. Arbitrary Units
3.2. Blood collection
Venous blood is drawn from fasting human subjects into 5 ml heparin vacutainers. 6.5mM NEM (26μl:250mM NEM/ ml blood) and 0.2mM DTPA (5 μl: 50mM DTPA/ml blood) are immediately mixed with whole blood and after 1 min, centrifuged at 4500 RCF for 5 min. NEM and DTPA prevent the decay of plasma S-nitrosothiols to nitrite (See notes 1 & 2). Process the plasma and red blood cells as soon as possible after separation from the blood.
-
Processing of plasma:
0.9 ml plasma that should be free of hemolysis is immediately transferred into light protected (dark colored) microtubes that contained 0.1 ml of 5% acidified sulfanilamide (See note 3). Plasma is immediately frozen on dry ice and stored at -85°C or -150°C for future analysis or measured immediately.
-
Processing of RBCs:
1 ml of RBCs (pipette from the bottom of tube) is mixed with 1ml of preservation solution and incubated for 5 min to lyse the cells and oxidize the hemoglobin.(see note 5)
0.25 ml of hemolysate is passed through a G-25 Sephadex column, which is pre-equilibrated with 0.1mM DTPA-PBS buffer, and the hemoglobin fraction is collected.
This column separation is carried out under subdued light preferably in a cold room (at 4 °C).
-
The concentration of methemoglobin is determined using the mM extinction coefficient of 3.7 at 630nm.
Concentration of heme (mM) = Absorbance at 630nm/3.7×dilution factor.
The concentration of the heme will be around 2.0 mM.
0.9ml of this solution is mixed with 0.1ml of 5% acidified sulfanilamide in 1 M HCl. This sample can be frozen at -85 or -150 for later analysis or measured immediately.
3.3. Measurement of plasma RSNOs
Thaw the plasma under subdued light just prior to determination. Thawed plasma samples are kept on ice until injected. (See note 8)
1 ml plasma sample is divided into two aliquots. One aliquot is incubated with 5mM HgCl2 (25μl:100mM HgCl2) for 5 min and to the second aliquot 25 μl UPH2O is added.
100 μl plasma is injected using a Hamilton syringe into the bottom of the purge vessel that contains 7.5 ml glacial acetic acid, 200 μl of 100 mM CuCl2 and 200 μl of 125 mM ascorbic acid (See note 9).
Each sample is injected in triplicate. The reagents are changed after every three or four injections.
Data is transferred to the Origin program and the chemiluminescence signals are regenerated as mentioned for the nitrite standard curve (see above).
The signals are smoothed by averaging the data of 20 points to eliminate the noise and for baseline correction (Fig.2).
Open the smoothed data worksheet and the chemiluminescence signals are regenerated. The area under the curve is determined as described for the standard curve. An example of raw data before and after being smoothened is shown in Figures 2A & 2B.
The RSNO values are calculated by taking the difference in the area of the chemiluminescence signals for samples in the presence and absence of HgCl2 (Fig.3).
Figure 2.
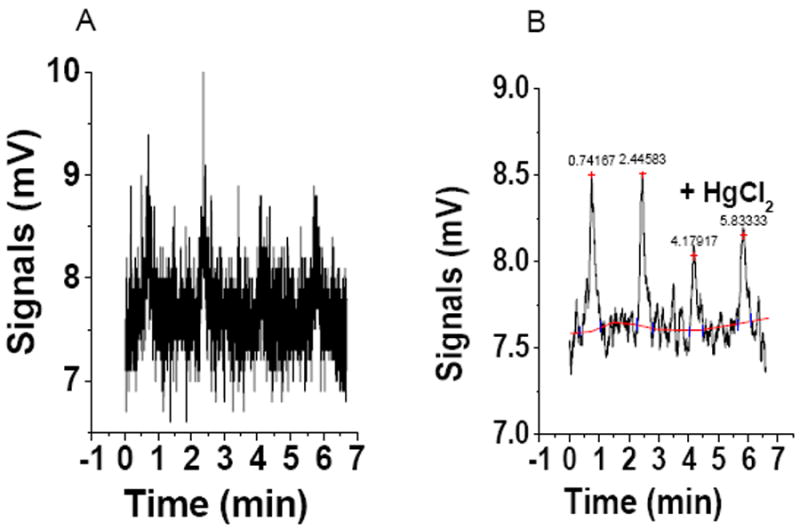
Chemiluminescence signal of plasma S-nitrosothiols: 100ul of plasma treated with 6.5 mM NEM, 0.2 mM DTPA and 5% sulfanilamidein 2M HCl as mentioned in methods section is injected into the purge vessel. (A) Chemiluminescence signals from the raw data, and (B) Chemiluminscence signals from the smoothed data using origin program for calculating the area under the curve.
Figure 3.

S-nitrosothiols values in plasma and red blood cells.(!) Plasma samples ; (,) Red blood cells.
Calculation:
An example for a sample of plasma S-nitrosothiols (nM):
3.4. Measurement of RBC RSNO
Thaw the RBC lysate under subdued light just prior to determination. Thawed lysates are kept on ice until injected.
1 ml of freshly processed RBC sample or thawed sample is divided into two aliquots. 5 mM HgCl2 (25 μl:100 mM) is added to one aliquot and 25 μl UPH2O is added to the second aliquot and incubated for 5 min.
100 μl to 200 μl sample is injected using a Hamilton syringe into the bottom of the purge vessel that contains 7.5 ml of glacial acetic acid, 200 μl of 100 mM CuCl2 and 200 μl of 125 mM ascorbic acid.
Reagents are changed every two or three injections.
The signals are processed as mentioned in 3.3 for plasma samples (see above)
The RSNO values are calculated by taking the difference in the area of the chemiluminescence signals for samples in the presence and absence of HgCl2 (Fig.3).
Calculation:
An example for a sample of RBC high molecular weight S-nitrosothiols (nM):
3.5. Determination of RSNO in cells
After completion of the incubation period for cells, the formation of RSNO- is stopped by adding 5 to 10 fold excess of NEM and allowing 5 to 10 min to block all the thiol groups. Cells are lysed by sonication and the protein content was determined. Treat the cell lysate with acidified sulfanilamide (9:1V/V, cell lysate: 5% acidified sulfanilamide) to remove the nitrite and to stabilize the RSNO. Inject 1 to 5 mg protein samples into the purge vessel depended on the RSNOs content.
Acknowledgments
Research Support: This research was supported by the Intramural Research Program of the NIH, National Institute on Aging
Footnotes
Always use fresh high-quality water (UPH2O with resistivity = 18.2 MΩ-cm containing metal chelator preferably 0.1 mM DTPA to prepare the solutions.
RSNO are light and temperature sensitive. Samples should be protected from light by carrying out all steps under subdued light and/or using dark color tubes. Keep samples on ice or in a cold room.
S-nitrosothiols are unstable in presence of free thiol groups and metal contaminants. In order to preserve the RSNO, blood should be collected in NEM and DTPA containing vacutainer tubes or NEM and DTPA should be added to blood as soon as it is collected. Allow 1 or 2 min to react NEM with thiols and centrifuge the blood immediately.
RSNO are more stable under acidic conditions. Therefore, mix the plasma with acidified sulfanilamide solution (9:1V/V, plasma: 5% acidified sulfanilamide) immediately after separation from the blood. This reagent removes the nitrite and stabilizes the RSNO in room temperature and frozen storage conditions.
Make sure that free thiols are blocked with NEM before addition of acidified sulfanilamide to plasma to remove the nitrite. Otherwise free thiols react with nitrite under acidic conditions to generate RSNO. Some investigators have used NEM together with acidified sulfanilamide to block the thiols and remove the nitrite. But we observed that nitrite reacts with thiols to form RSNO under acidic conditions before it complexes with sulfanilamide.
Thiols groups also should be blocked with NEM before addition of potassium ferricyanide (preservation solution) to RBCs to oxidize the hemoglobin. Otherwise oxidation of iron nitrosylhemoglobin by potassium ferricyanide generates S-nitrosohemoglobin. Addition of NEM together with potassium ferricyanide does not completely prevent the generation S-nitrosohemoglobin.
Always use fresh high quality water (UPH2O with resistivity = 18.2 MΩ-cm) containing a metal chelator preferably 0.1mM DTPA to prepare the solutions.
Always cap the reagent bottles tightly; otherwise atmospheric nitric oxide species dissolve in liquids over time.
Make sure that there is no gas leak in the purge system. Maintain constant instrument cell pressure by adjusting the inert gas flow. Run the system at least 30 min for stabilization prior to initiating sample analysis.
We strongly suggest the use of the Origin program for integration of peaks. It can be purchased from Origin Lab Corporation, One Roundhouse Plaza, Suite 303, Northampton, MA 01060 USA, www.originlab.com.
Thaw the plasma samples just prior to use, while protecting from light. GSNO standards also should be prepared just prior to use. Complete the analysis within 2-3 hrs.
Inject the samples at the bottom of the purge vessel using a syringe with a 5-inch long needle for good reproducibility. Syringe needle should be rinsed with water and wiped after every injection. Otherwise sample will be contaminated with Cu(I) reagent, which degrades RSNO.
Make sure that free thiols are blocked with NEM before addition of acidified sulfanilamide to the plasma to remove the nitrite. Otherwise free thiols react with nitrite under acidic conditions generating RSNOs. Some investigators have used NEM together with acidified sulfanilamide to block the thiols and remove the nitrite. But we observed that nitrite reacts with thiols to form RSNOs under acidic conditions before it complexes with sulfanilamide.
When high protein samples (plasma) are injecting, leave a sufficient gap between injections until the gas bubbles are regenerated in the purge vessel.
Reference List
- 1.Hogg N. The biochemistry and physiology of S-nitrosothiols. Annu Rev Pharmacol Toxicol. 2002;42:585–600. doi: 10.1146/annurev.pharmtox.42.092501.104328. [DOI] [PubMed] [Google Scholar]
- 2.Kharitonov VG, Sundquist AR, Sharma VS. Kinetics of nitrosation of thiols by nitric oxide in the presence of oxygen. J Biol Chem. 1995;270:28158–28164. doi: 10.1074/jbc.270.47.28158. [DOI] [PubMed] [Google Scholar]
- 3.Ignarro LJ, Edwards JC, Gruetter DY, Barry BK, Gruetter CA. Possible involvement of S-nitrosothiols in the activation of guanylate cyclase by nitroso compounds. FEBS Lett. 1980;110:275–278. doi: 10.1016/0014-5793(80)80091-3. [DOI] [PubMed] [Google Scholar]
- 4.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 5.Gaston BM, Carver J, Doctor A, Palmer LA. S-nitrosylation signaling in cell biology. Mol Interv. 2003;3:253–263. doi: 10.1124/mi.3.5.253. [DOI] [PubMed] [Google Scholar]
- 6.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 7.MacArthur PH, Shiva S, Gladwin MT. Measurement of circulating nitrite and S-nitrosothiols by reductive chemiluminescence. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:93–105. doi: 10.1016/j.jchromb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Dunham AJ, Barkley RM, Sievers RE. Aqueous nitrite ion determination by selective reduction and gas phase nitric oxide chemiluminescence. Anal Chem. 1995;67:220–224. doi: 10.1021/ac00097a033. [DOI] [PubMed] [Google Scholar]
- 9.Nagababu E, Rifkind JM. Measurement of plasma nitrite by chemiluminescence without interference of S-, N-nitroso and nitrated species. Free Radic Biol Med. 2007;42:1146–1154. doi: 10.1016/j.freeradbiomed.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu S, Wang X, Gladwin MT, Kim-Shapiro DB. Chemiluminescent detection of S-nitrosated proteins: comparison of tri-iodide, copper/CO/cysteine, and modified copper/cysteine methods. Methods Enzymol. 2008;440:137–156. doi: 10.1016/S0076-6879(07)00808-7. [DOI] [PubMed] [Google Scholar]
- 11.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marley R, Feelisch M, Holt S, Moore K. A chemiluminescense-based assay for S-nitrosoalbumin and other plasma S-nitrosothiols. Free Radic Res. 2000;32:1–9. doi: 10.1080/10715760000300011. [DOI] [PubMed] [Google Scholar]
- 13.Fang K, Ragsdale NV, Carey RM, MacDonald T, Gaston B. Reductive assays for S-nitrosothiols: implications for measurements in biological systems. Biochem Biophys Res Commun. 1998;252:535–540. doi: 10.1006/bbrc.1998.9688. [DOI] [PubMed] [Google Scholar]
- 14.Nagababu E, Ramasamy S, Rifkind JM. S-nitrosohemoglobin: a mechanism for its formation in conjunction with nitrite reduction by deoxyhemoglobin. Nitric Oxide. 2006;15:20–29. doi: 10.1016/j.niox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Yang BK, Vivas EX, Reiter CD, Gladwin MT. Methodologies for the sensitive and specific measurement of S-nitrosothiols, iron-nitrosyls, and nitrite in biological samples. Free Radic Res. 2003;37:1–10. doi: 10.1080/1071576021000033112. [DOI] [PubMed] [Google Scholar]
- 16.Rassaf T, Bryan NS, Kelm M, Feelisch M. Concomitant presence of N-nitroso and S-nitroso proteins in human plasma. Free Radic Biol Med. 2002;33:1590–1596. doi: 10.1016/s0891-5849(02)01183-8. [DOI] [PubMed] [Google Scholar]


