Abstract
The replication system of phage T7 provides a model for DNA replication. Biochemical, structural, and single-molecule analyses together provide insight into replisome mechanics. A complex of polymerase, a processivity factor, and helicase mediates leading strand synthesis. Establishment of the complex requires an interaction of the C-terminal tail of the helicase with the polymerase. During synthesis the complex is stabilized by other interactions to provide for a processivity of 5 kilobase (kb). The C-terminal tail also interacts with a distinct region of the polymerase to capture dissociating polymerase to increase the processivity to >17 kb. The lagging strand is synthesized discontinuously within a loop that forms and resolves during each cycle of Okazaki fragment synthesis. The synthesis of a primer as well as the termination of a fragment signal loop resolution.
Introduction
Bacteriophage T7 is a lytic phage that infects Escherichia coli. Its genome of 40 kilobase (kb) encodes approximately 50 proteins of which three, along with one host protein, can constitute a functioning replisome. The T7 replisome mediates coordinated DNA synthesis in a manner that mimics the fundamental steps found in higher organisms [1,2]. The helicase domain of the hexameric gene 4 protein (gp4) unwinds dsDNA to provide a ssDNA template for gene 5 DNA polymerase (gp5) (Figure 1). The leading strand is synthesized continuously by gp5 in complex with its processivity factor E. coli thioredoxin (trx). The lagging strand is synthesized discontinuously to yield Okazaki fragments that are processed to form a continuous strand. The synthesis of each Okazaki fragment is initiated by the extension of tetraribonucleotides synthesized by the primase domain of gp4. The nascent Okazaki fragments are found within a replication loop that undergoes multiple rounds of formation and resolution during each cycle of synthesis of an Okazaki fragment. The gene 2.5 ssDNA-binding protein (gp2.5) is essential for protection of ssDNA produced during replication and for coordination of the synthesis of both strands [3]. The T7 replisome is a dynamic entity wherein each step is coordinated and proteins continuously enter and exit.
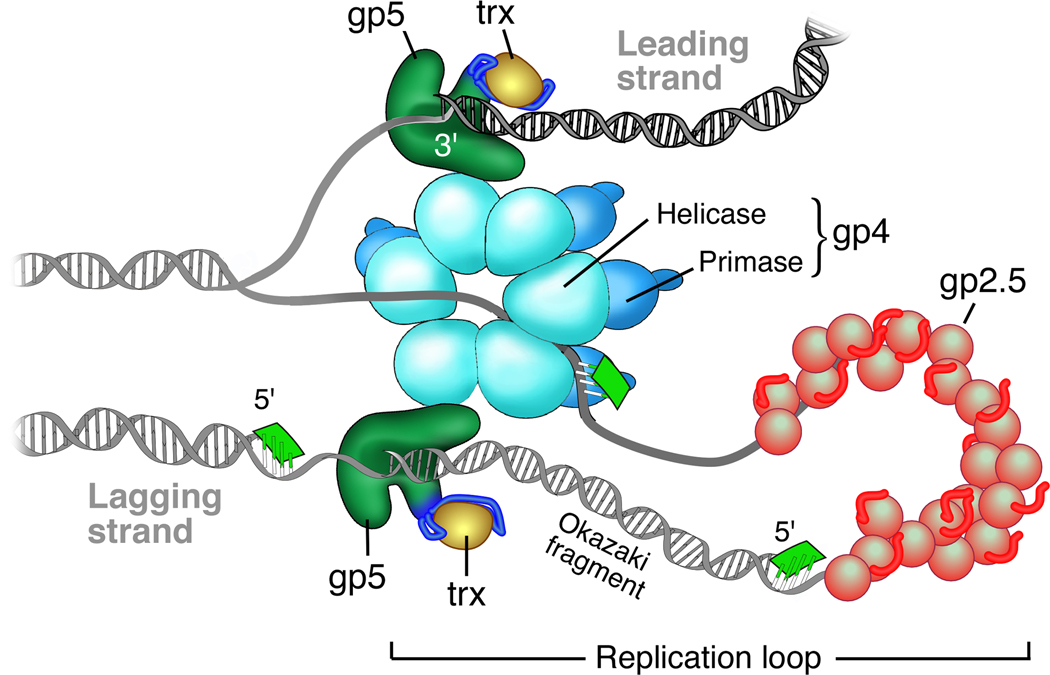
Figure 1.
Model of the T7 replisome. The hexameric helicase domain of gp4, encircling the lagging strand, unwinds the duplex DNA to provide two single-stranded DNA templates. Gp5/trx bound to the helicase catalyzes the polymerization of nucleotides on the leading strand. The ssDNA extruded by the translocating helicase forms a replication loop in which the primase domain of gp4 catalyzes the synthesis of a tetraribonucleotide (green) to serve as a primer for the lagging strand gp5/trx that is also bound to the helicase. Gp2.5 coats the exposed ssDNA.
Other DNA replication systems are generally more complicated than the T7 system. Helicase and primase are found in separate proteins although they must physically interact to properly function. Additional proteins in these systems include processivity clamps, loading proteins for the helicase and processivity factor, and proofreading exonucleases. How does the T7 replisome function with only four proteins whereas others require far more? It is clear that the four proteins of the T7 replisome have usurped many of these functions. The limited number of proteins has made possible: (i) reconstitution of a replisome [2], (ii) determination of the crystal structures of the proteins [4–6], and (iii) visualization of active replisomes by single-molecule techniques [7,8]. A more detailed review of T7 DNA replication is available [2].
Processivity of DNA polymerase
The binding of trx to a flexible loop (thioredoxin binding domain, TBD) located at the tip of the thumb of gp5 increases the rate of dNTP incorporation by 20 to 80-fold to yield a processivity of approximately 800 nucleotides (nt) per binding event [9–12]. Unlike ring-shaped processivity factors that encircle DNA, binding of trx to the TBD in gp5 reorganizes this flexible region to better grasp the DNA. Comparative analysis of gp5 and gp5/trx by small angular X-ray scattering and exonuclease footprinting show that the binding of trx increases protein surface interactions with the duplex portion of the primer-template [13]. The extended surface renders gp5/trx more resistant to salt and facilitates sliding of the DNA in the binding cleft. These adjustments induced by trx binding most probably create a closed conformation of the binding crevice rather than the open complex shown in the crystallographic structure [4]. Single-molecule analyses reveal the enhancement of gp5 sliding on DNA by trx [14]. While gp5 alone frequently dissociates from dsDNA and rebinds, the binding of trx to gp5 suppresses such microscopic hopping so that gp5/trx can slide on the DNA in search for a 3’-terminus. The processivity of 800 nt resulting from the binding of trx does not explain the high processivity of >17 kb observed during movement of the replisome. As discussed below this enhanced processivity derives from two modes of gp5/trx-helicase interaction.
DNA polymerase-helicase interactions for leading strand synthesis
T7 DNA polymerase interacts with DNA helicase, an essential interaction to coordinate nucleotide polymerization with unwinding of the DNA. T7 gp4 is present as a mixture of hexamers and heptamers in the absence of DNA but assembles onto ssDNA as a hexamer, raising the possibility that loading onto DNA involves the loss of a subunit [15,16]. In other replication systems a helicase loading protein is required for the assembly of the helicase. Once loaded onto ssDNA the helicase translocates 5’ to 3’ using the energy of hydrolysis of dTTP. Upon encountering duplex DNA its continued movement unwinds the duplex. Structural determinants in the nucleotide and the nucleotide binding pocket of gp4 responsible for the dTTP requirement have been identified [17,18].
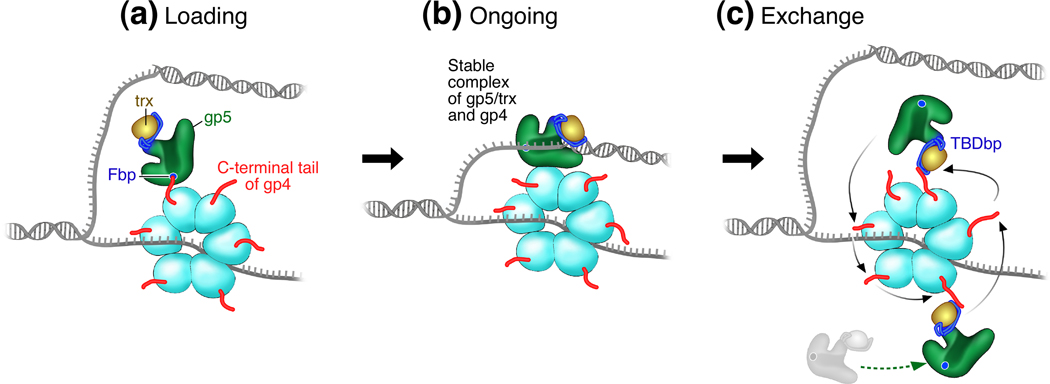
Gp4 has an acidic C-terminal tail that has multiple interactions with gp5/trx. One of these interactions is essential for the initiation of leading strand synthesis together with gp5/trx. A solvent exposed patch consisting of four basic amino acids is located on the surface of gp5 facing the duplex region of the DNA at a replication fork [4]. This basic patch is adjacent to the template strand as it exits the active site of gp5. Neutralization of the basic charges eliminates the ability of the polymerase to mediate strand-displacement synthesis with gp4 [19]. Binding studies show that altered polymerase is defective in binding to gp4 and that the acidic C-terminal tail of gp4 is involved in this interaction. However, the altered polymerase is able to replace wild-type gp5 once DNA synthesis is underway for strand-displacement synthesis. The ability of DNA polymerase in solution to exchange with the replicating DNA polymerase is medicated by interaction between the C-terminal tail of gp4 and the TBD as discussed below. Since the basic patch in the TBD is retained in the altered polymerase, it can bind to the C-terminal tail of gp4 and then exchange with the replicating DNA polymerase. The results suggest that the interaction between the C-terminal tail of the helicase and the basic patch of DNA polymerase is crucial for loading of gp5/trx onto the replication fork to initiate strand-displacement synthesis (Figure 2a).
Figure 2.
Three modes of interactions between gp4 and gp5/trx during leading strand synthesis. (a) Gp5/trx is loaded onto the hexameric gp4 through an interaction of the acidic C-terminal tail of gp4 with a basic patch at the front side (Fbp) of gp5. (b) A high affinity complex between the two proteins coordinates their activities during DNA synthesis. (c) Gp5/trx temporarily dissociated from gp4 or present in solution is retained with gp4 through an interaction between two basic loops in the trx binding domain (TBDbp) of gp5 and the acidic C-terminal tail of gp4.
The activities of both polymerase and helicase are reciprocally enhanced by their interactions. Unwinding of dsDNA by gp4 is 10-fold slower than translocation on ssDNA as measured by pre-steady state kinetics [20]. DNA synthesis catalyzed by gp5/trx at the replication fork increases the rate of unwinding to 114 base pairs per second, equivalent to that observed for translocation of gp4 on ssDNA [21]. Gp5/trx not only destabilizes the duplex to facilitate forward Brownian push of the helicase but it also prevents backward slipping of the helicase by synthesizing the complementary strand [21,22]. Gp4 variants defective in ssDNA binding as well as unwinding can mediate efficient strand-displacement synthesis in the presence of gp5/trx, thus confirming the supporting role of gp5/trx [23].
Interactions with gp4 enable gp5/trx to increase its processivity from incorporation of about 800 nt on ssDNA template per binding event to >17 kb during leading strand DNA synthesis on duplex DNA [7]. A major source for the increased processivity is the interaction that couples helicase unwinding to the polymerization of nucleotides by gp5/trx (Figure 2b). This interaction occurs only when gp5/trx and gp4 are functioning together during leading strand DNA synthesis. This stable interaction can be observed by surface plasmon resonance when gp5/trx is bound to a primer-template in the presence of the dideoxynucleoside 5’-triphosphate corresponding to the next incoming dNTP [24]. The sites of this interaction have not been identified but it does not involve the acidic C-terminal tail of gp4. This stable interaction provides for a processivity of around 5 kb [25].
In addition to the two modes of binding of gp5/trx and gp4 aforementioned– a mode for loading and the one for ongoing DNA synthesis – a third mode of interaction allows for exchange of DNA polymerases during DNA synthesis (Figure 2c). The C-terminal tail of gp4 interacts not only with the basic patch adjacent to the template, as in the loading mode, but also with two basic loops located within the TBD of gp5 [24]. These two solvent exposed loops are partially configured by the binding of trx to the TBD. In addition, one lysine in trx located in proximity to one of the two loops also contributes to the interaction [26].
The C-terminal tail of gp4 binds to these two loops and provides for backup DNA polymerase in the event the replicating gp5/trx dissociates. Elimination of the TBD in gp5 decreases the binding affinity to gp4 by 90-fold [24]. This interaction also allows for the capture of the replicating gp5/trx upon dissociation and its return to the primer-template. The ability of the hexameric gp4 to retain the dissociating replicating polymerase increases the processivity from 5 kb to >17 kb [25]. Abolishment of the charges in the two loops or removal of the C-terminal tail of gp4 reduces the normal high processivity to 5 kb as measured by single-molecule techniques [25]. Gp4 allows for the exchange of gp5/trx in solution with the replicating gp5/trx at the replication fork without affecting processivity. In the cell there is most probably an abundance of DNA polymerase thus assuring that there is always a polymerase bound to the helicase ready for exchange in the event the replicating polymerase dissociates. Exchange of gp5/trx in the T7 replisome was initially demonstrated in ensemble experiments [27] and later confirmed quantitatively using single-molecule measurements [28].
Synthesis and delivery of primers during lagging strand synthesis
Synthesis of the lagging strand requires the synthesis of oligoribonucleotides by the primase domain of gp4. These oligoribonucleotides are then used as primers to initiate the synthesis of Okazaki fragments. Like other prokaryotic primases, T7 primase catalyzes the synthesis of short oligoribonucleotides from specific sequences (primase recognition sites) on the template [29]. At the basic recognition sequence 5’-GTC-3’ T7 primase synthesizes the diribonucleotide 5’-pppAC-3’ that is then extended to the functional tetraribonucleotide 5’-pppACCA, pppACCC, or pppACAC-3’ provided the complementary sequence is present in the template [30] (Figure 3a). A stable complex between the primase and its recognition sequence is formed only when the ribonucleotides necessary for tetraribonucleotide formation are present [31]. Consistent with previous kinetic data [32], the binding affinity is relatively weak (Kd ~20 µM). Although the primase binds to its recognition sequence in the presence of preformed tetraribonucleotides, its binding is less stringent than with ribonucleotides. The results suggest that conformational changes that occur during synthesis of functional primers enable the primase to form a specific complex with DNA.
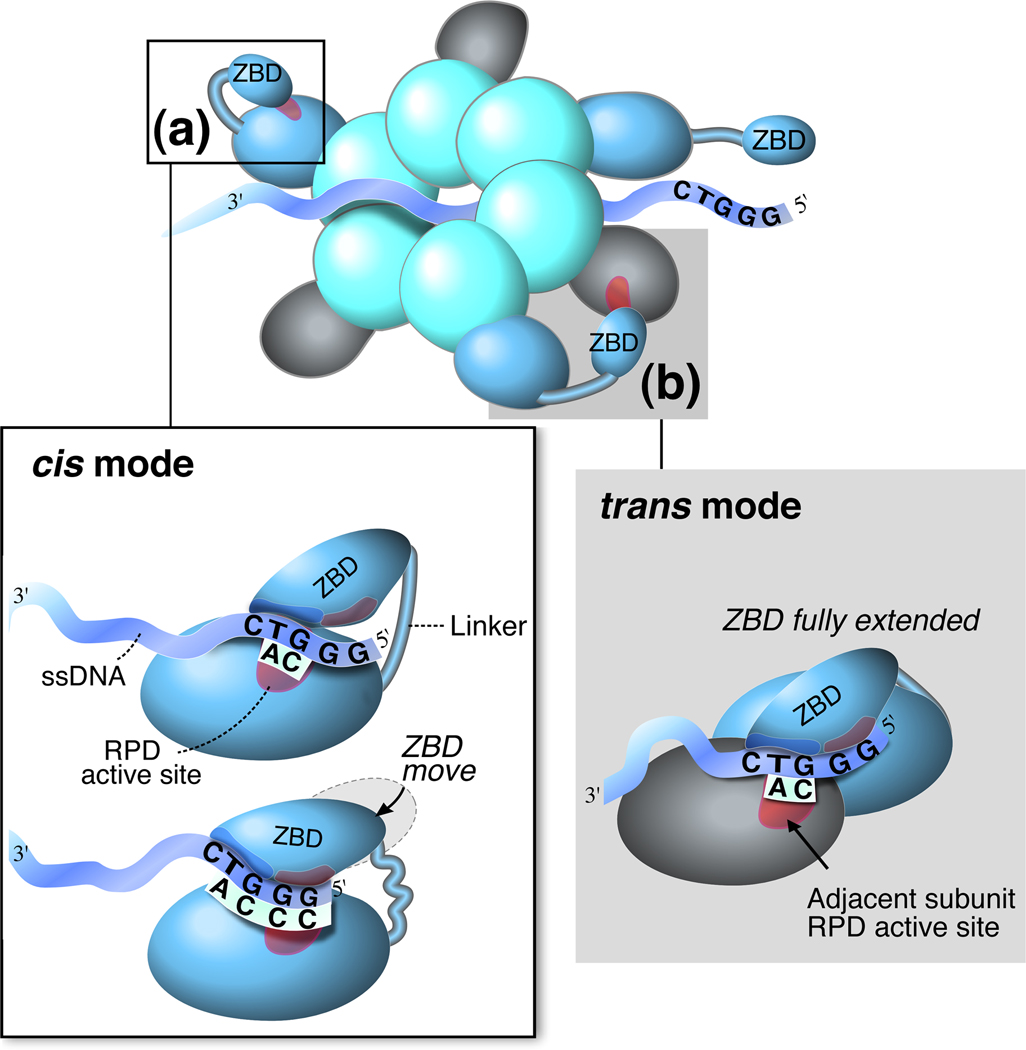
Figure 3.
Two distinct modes involved in primer synthesis by the primase domain of gp4. Synthesis of tetraribonucleotides at the primase recognition sequence requires interactions between the zinc-binding domain (ZBD) and RNA polymerase domain (RPD) of the primase domain of gp4. (a) Diribonucleotide pppAC is synthesized from the 5’-GTC-3’ basic recognition sequence through interactions between the ZBD and RPD in the same subunit (cis mode). Extension of the diribonucleotide to the functional tetraribonucleotide is facilitated by shortening the linker between the ZBD and RPD. (b) In the hexameric gp4 structure, the ZBD in one subunit interacts with the RPD in an adjacent subunit to catalyze the synthesis of a primer (trans mode).
The interaction between the primase and the hexameric helicase results in a structure in which the primase domain of one subunit interacts with that of an adjacent subunit (Figure 3b). This arrangement allows the zinc-binding motif of one subunit of gp4 to interact with the RNA polymerase catalytic site on another subunit to catalyze primer synthesis in a “trans mode” [33]. Results obtained by modifying the length of the linker connecting the RNA polymerase domain and the zinc-binding domain suggest a model in which the primase catalyzes the synthesis of pppAC in a cis mode and the extension occurs in a trans mode [34]. The trans mode of primer synthesis could function as a brake to stop helicase activity and hence leading strand DNA synthesis [7]. This pause would prevent leading strand synthesis from outpacing lagging strand synthesis.
Once primers are synthesized they are delivered to gp5/trx to initiate the synthesis of an Okazaki fragment. Tryptophan 69, located in the N-terminal region of the RNA polymerase domain of the primase plays a critical role in the delivery of primers to gp5/trx [35]. Substitution of lysine for Trp69 results in a primase that can synthesize but fails to stabilize the tetraribonucleotide.
Gp4 exists in two molecular weight forms in vivo, a 56-kDa and a 63-kDa form [36]. The 56-kDa form arises from an internal initiation codon and ribosome binding site located within the 63-kDa form [37]. The 56-kDa gp4 lacks the N-terminal 63 residues that comprise the zinc-binding domain and therefore cannot catalyze the de novo synthesis of tetraribonucleotides. However it can deliver a wide range of preformed oligonucleotides, including tRNA, to gp5/trx for use as primers [35]. Suppressor phages grown with the 56-kDa gp4 reveal alteration in gene 5.5 that encodes a protein that potentially regulates transcription of both host and phage [38], suggesting alterative mechanisms to acquire primers from a RNA transcript (B.Zhu et al., unpublished).
Regulation of lagging strand synthesis
Lagging strand synthesis is considerably more complex than leading strand synthesis yet both proceed at the same rate with a reconstituted T7 replisome [3]. The association of both the leading and lagging strand DNA polymerase with the gp4 must play a critical role in this coordination. For the parallel progression with the leading strand, the lagging strand polymerase forms a replication loop (Figure 1). The nascent Okazaki fragment is located within the loop and the eventual length of the Okazaki fragment is, on average, 0.8 kb [8]. The mechanisms regulating loop size, the use of primase recognition sites, and the length of Okazaki fragments have been difficult to dissect. Visualization of the formation and release of the replication loop by single-molecule analysis has provided insight into this complex process [8]. Analysis of the distributions of loop sizes and lag times between loops reveals that initiation of primer synthesis (signaling model) and the completion of an Okazaki fragment (collision model) each serve as a trigger for loop release. The presence of two triggers may represent a fail-safe mechanism ensuring the timely reset of the replisome after the synthesis of every Okazaki fragment.
The primase of gp4, like the helicase, is stimulated by interaction with gp5/trx. Primer synthesis during leading strand synthesis is approximately 10-fold greater relative to gp4 alone on ssDNA [39]. Single-molecule experiments show that in the presence of ATP and CTP, precursors for primer synthesis, leading strand synthesis periodically halts for approximately 6 seconds [7]. This cessation of DNA synthesis is the time, determined by kinetic studies, required to initiate and synthesize a primer by gp4 [32]. These results are compatible with a model in which the initiation of primer synthesis halts the movement of the helicase to stop leading strand synthesis during the lengthy period of primer synthesis. However, in ensemble experiments the rate of leading strand synthesis is not affected by the presence of a primase recognition site on the lagging strand [40]. Single-molecule FRET studies suggest that gp4 maintains contact with the primase recognition sequence and the primer, resulting in a priming loop [39]. In these studies no pause was observed during primer synthesis with leading synthesis slowed by the rate of helicase movement. The basis for the difference in these studies is not known. It is possible that different phases in primer synthesis and loop formation are being observed with a priming loop preceding the larger replication loop.
Role of gene 2.5 ssDNA-binding protein
T7 single-stranded DNA-binding protein (gp2.5) has multiple roles. Like gp4 it has an acidic C-terminal tail that is essential for T7 DNA replication [41]. In a reconstituted replication system it is essential for the coordination of leading and lagging strand synthesis [3]. The C-terminal tails of gp4 and gp2.5 are similar in length and composition and chimeric proteins in which the tails have been switched support the growth of T7 [42,43]. Like other ssDNA-binding proteins gp2.5 has an OB-fold consisting of anti-parallel β-sheets that form a barrel with a well-defined cleft [6]. Structural and mutagenesis data show that ssDNA binds within the cleft via stacking and electrostatic interactions [44]. Cross-linking studies show that in the absence of ssDNA the C-terminal tail resides within the DNA binding cleft most likely to protect from random binding of charged molecules and to coordinate the DNA and protein interaction of gp2.5.
Not surprisingly, gp2.5 physically binds to gp5/trx through interactions of the acidic C-terminal tail with both the basic patches located in the TBD [24] and on the front of gp5 [19]. Despite the similarity of the C-terminal tails, sequential bindings of gp4 and gp2.5 to gp5/trx in a polymerizing mode suggest that they interact with gp5/trx in a distinct manner [45]. Gp2.5 enables gp5/trx to catalyze strand-displacement DNA synthesis at a nick in DNA, a process that involves the C-terminal tail [46]. It has been postulated that the binding of gp2.5 to the ssDNA extruded by the helicase results in condensation of the DNA with the increasing mass leading to resolution of the replication loop [3].
Interestingly, the C-terminal tails of both gp4 and gp2.5 bear a phenylalanine at the last position. A screening for suppressors of gp2.5 lacking the C-terminal phenylalanine identified residues that map in proximity to aromatic residues and to residues in contact with DNA in the crystal structure of gp5/trx bound to DNA [47]. Gp2.5 lacking the C-terminal phenylalanine has a lower affinity for gp5/trx relative to the wild-type gp2.5 and this affinity is partially restored by the suppressor mutations in gp5 [46].
Additional factors contributing to T7 DNA replication
T7 phage derives most of the nucleotides needed for DNA replication from breakdown of host DNA to deoxynucleoside 5’-monophosphates. Host enzymes can convert these nucleoside monophosphates to the di and triphosphate nucleotides but may not be sufficient to meet the demand for the rapid DNA synthesis that occurs in phage-infected cells. A screening for host proteins essential for phage growth identified E. coli CMP kinase that also phosphorylates dCMP as an essential protein [48]. Gene 1.7 encodes a nucleotide kinase that phosphorylates dTMP and dGMP to dTDP and dGDP and subsequently, albeit at a slower rate, to the nucleoside triphosphate [49]. The ability of gp1.7 to phosphorylate dideoxythymidylate explains the sensitivity of E. coli harboring gene 1.7 to dideoxythymidine; E. coli thymidylate kinase cannot phosphorylate dideoxythymidylate [50]. Increased dTTP is beneficial for DNA synthesis mediated by gp5/trx and unwinding by gp4 helicase.
Conclusions
Protein interactions occur within the T7 replisome in a coordinated manner. Binding of trx reconfigures the TBD in gp5 to enhance contact with DNA and with gp4 and gp2.5. Interactions of gp5/trx with the helicase domain of gp4 involve loading of the helicase, coordination of polymerization and unwinding, and exchange of gp5/trx during leading strand synthesis. Reciprocal stimulation of the polymerase and helicase results in highly processive synthesis. Synthesis of oligonucleotides by the primase domain of gp4 not only provides primers for the synthesis of Okazaki fragments but also signals the formation and resolution of replication loops on the lagging strand. The C-terminus of gp2.5 modulates ssDNA binding and its interaction with gp5/trx contributes to coordinated DNA synthesis. Besides these four key components, other proteins also contribute to the efficiency of replication. These proteins include those necessary for the processing of Okazaki fragments and for supplying nucleotides to the replisome. Structural information on the intact replisome as well as its subassemblies would improve greatly our understanding of a functional replisome.
Highlights.
A reconstituted replisome with four proteins
Coordinated synthesis
A replication loop
Acknowledgements
The authors are grateful to Steve Moskowitz for preparing the figures. This work was supported by National Institutes of Health Public Health Service Grant GM 54397.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Richardson CC. Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell. 1983;33:315–317. doi: 10.1016/0092-8674(83)90411-7. [DOI] [PubMed] [Google Scholar]
- 2. Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. This review provides a comprehensive description on the interactions that occur during DNA replication with emphasis on the bacteriophage T7 system. Principles governing interactions between components of the replisome and mechanisms for coordinated synthesis of leading and lagging strands are broadly discussed.
- 3.Lee J, Chastain PD, 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 4.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 5.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 6.Hollis T, Stattel JM, Walther DS, Richardson CC, Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc Natl Acad Sci U S A. 2001;98:9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. This study uses single-molecule techniques to render real time observation of kinetics of the T7 replication complex. The high processivity of leading strand synthesis, the pausing of the replicating complex during primer synthesis, and the formation and resolution of a replication loop are observed.
- 8. Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457:336–339. doi: 10.1038/nature07512. The formation and release of replication loops is examined by single-molecule techniques. Analyses of events indicate that both primer synthesis and completion of an Okazaki fragment regulate the dynamic replication loop.
- 9.Huber HE, Tabor S, Richardson CC. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 10.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 11.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 12.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 13. Akabayov B, Akabayov SR, Lee SJ, Tabor S, Kulczyk AW, Richardson CC. Conformational dynamics of bacteriophage T7 DNA polymerase and its processivity factor, Escherichia coli thioredoxin. Proc Natl Acad Sci U S A. 2010;107:15033–15038. doi: 10.1073/pnas.1010141107. T7 DNA polymerase and its complex with E. coli thioredoxin are examined by structural and biochemical analyses. Evidence is presented that conformational changes induced by the binding of thioredoxin increase interactions with DNA to enhance processivity of the polymerase.
- 14.Etson CM, Hamdan SM, Richardson CC, van Oijen AM. Thioredoxin suppresses microscopic hopping of T7 DNA polymerase on duplex DNA. Proc Natl Acad Sci U S A. 2010;107:1900–1905. doi: 10.1073/pnas.0912664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19:3418–3427. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crampton DJ, Ohi M, Qimron U, Walz T, Richardson CC. Oligomeric states of bacteriophage T7 gene 4 primase/helicase. J Mol Biol. 2006;360:667–677. doi: 10.1016/j.jmb.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 17.Satapathy AK, Crampton DJ, Beauchamp BB, Richardson CC. Promiscuous usage of nucleotides by the DNA helicase of bacteriophage T7: determinants of nucleotide specificity. J Biol Chem. 2009;284:14286–14295. doi: 10.1074/jbc.M900557200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SJ, Richardson CC. Molecular basis for recognition of nucleoside triphosphate by gene 4 helicase of bacteriophage T7. J Biol Chem. 2010;285:31462–31471. doi: 10.1074/jbc.M110.156067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Lee SJ, Zhu B, Tran NQ, Tabor S, Richardson CC. Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc Natl Acad Sci U S A. 108:9372–9377. doi: 10.1073/pnas.1106678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci U S A. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satapathy AK, Kochaniak AB, Mukherjee S, Crampton DJ, van Oijen A, Richardson CC. Residues in the central beta-hairpin of the DNA helicase of bacteriophage T7 are important in DNA unwinding. Proc Natl Acad Sci U S A. 2010;107:6782–6787. doi: 10.1073/pnas.1002734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamdan SM, Marintcheva B, Cook T, Lee SJ, Tabor S, Richardson CC. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci U S A. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamdan SM, Johnson DE, Tanner NA, Lee JB, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. Interactions between T7 DNA polymerase and helicase in leading strand synthesis are dissected. Highly processive polymerase activity results from the coordinated interaction between helicase and polymerase and is further increased by electrostatic interactions between the two proteins.
- 26.Ghosh S, Hamdan SM, Cook TE, Richardson CC. Interactions of Escherichia coli thioredoxin, the processivity factor, with bacteriophage T7 DNA polymerase and helicase. J Biol Chem. 2008;283:32077–32084. doi: 10.1074/jbc.M805062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson DE, Takahashi M, Hamdan SM, Lee SJ, Richardson CC. Exchange of DNA polymerases at the replication fork of bacteriophage T7. Proc Natl Acad Sci U S A. 2007;104:5312–5317. doi: 10.1073/pnas.0701062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loparo JJ, Kulczyk AW, Richardson CC, van Oijen AM. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc Natl Acad Sci U S A. 2011;108:3584–3589. doi: 10.1073/pnas.1018824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 30.Tabor S, Richardson CC. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc Natl Acad Sci U S A. 1981;78:205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee SJ, Zhu B, Hamdan SM, Richardson CC. Mechanism of sequence-specific template binding by the DNA primase of bacteriophage T7. Nucleic Acids Res. 2010;38:4372–4383. doi: 10.1093/nar/gkq205. The physical binding between T7 DNA primase and template DNA is examined by surface plasmon resonance. Conformational changes in the primase during functional primer synthesis lead to a stable complex of the primase with its recognition sequence
- 32.Frick DN, Richardson CC. Interaction of bacteriophage T7 gene 4 primase with its template recognition site. J Biol Chem. 1999;274:35889–35898. doi: 10.1074/jbc.274.50.35889. [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Richardson CC. Interaction of adjacent primase domains within the hexameric gene 4 helicase-primase of bacteriophage T7. Proc Natl Acad Sci U S A. 2002;99:12703–12708. doi: 10.1073/pnas.202471499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qimron U, Lee SJ, Hamdan SM, Richardson CC. Primer initiation and extension by T7 DNA primase. EMBO J. 2006;25:2199–2208. doi: 10.1038/sj.emboj.7601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu B, Lee SJ, Richardson CC. Direct role for the RNA polymerase domain of T7 primase in primer delivery. Proc Natl Acad Sci U S A. 2010;107:9099–9104. doi: 10.1073/pnas.1004220107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 37.Mendelman LV, Notarnicola SM, Richardson CC. Roles of bacteriophage T7 gene 4 proteins in providing primase and helicase functions in vivo. Proc Natl Acad Sci U S A. 1992;89:10638–10642. doi: 10.1073/pnas.89.22.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q, Richardson CC. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:1761–1765. doi: 10.1073/pnas.90.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2009;462:940–943. doi: 10.1038/nature08611. Pre-steady state kinetics together with fluorescence resonance energy transfer analysis demonstrates DNA synthesis is coordinated by various protein interactions at a replication fork. This study also demonstrates the formation of a priming loop during primer synthesis. The mechanism by which leading and lagging strand synthesis is coordinated is discussed.
- 40.Lee J, Chastain PD, 2nd, Griffith JD, Richardson CC. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage t7 replication proteins. J Mol Biol. 2002;316:19–34. doi: 10.1006/jmbi.2001.5325. [DOI] [PubMed] [Google Scholar]
- 41.Kim YT, Richardson CC. Acidic carboxyl-terminal domain of gene 2.5 protein of bacteriophage T7 is essential for protein-protein interactions. J Biol Chem. 1994;269:5270–5278. [PubMed] [Google Scholar]
- 42.Marintcheva B, Hamdan SM, Lee SJ, Richardson CC. Essential residues in the C terminus of the bacteriophage T7 gene 2.5 single-stranded DNA-binding protein. J Biol Chem. 2006;281:25831–25840. doi: 10.1074/jbc.M604601200. [DOI] [PubMed] [Google Scholar]
- 43.Lee SJ, Marintcheva B, Hamdan SM, Richardson CC. The C-terminal residues of bacteriophage T7 gene 4 helicase-primase coordinate helicase and DNA polymerase activities. J Biol Chem. 2006;281:25841–25849. doi: 10.1074/jbc.M604602200. [DOI] [PubMed] [Google Scholar]
- 44. Marintcheva B, Marintchev A, Wagner G, Richardson CC. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc Natl Acad Sci U S A. 2008;105:1855–1860. doi: 10.1073/pnas.0711919105. Chemical cross-linking and nuclear magnetic resonance analysis reveal that the acidic C-terminal of T7 ssDNA-binding protein binds within the DNA binding cleft of the protein. The binding of the C-terminal tail of gp2.5 to its DNA binding cleft, to ssDNA, and to other proteins regulate multiple functions at the replication fork.
- 45.Ghosh S, Hamdan SM, Richardson CC. Two modes of interaction of the single-stranded DNA-binding protein of bacteriophage T7 with the DNA polymerase-thioredoxin complex. J Biol Chem. 2010;285:18103–18112. doi: 10.1074/jbc.M110.107656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh S, Marintcheva B, Takahashi M, Richardson CC. C-terminal phenylalanine of bacteriophage T7 single-stranded DNA-binding protein is essential for strand displacement synthesis by T7 DNA polymerase at a nick in DNA. J Biol Chem. 2009;284:30339–30349. doi: 10.1074/jbc.M109.024059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marintcheva B, Qimron U, Yu Y, Tabor S, Richardson CC. Mutations in the gene 5 DNA polymerase of bacteriophage T7 suppress the dominant lethal phenotype of gene 2.5 ssDNA binding protein lacking the C-terminal phenylalanine. Mol Microbiol. 2009;72:869–880. doi: 10.1111/j.1365-2958.2009.06682.x. [DOI] [PubMed] [Google Scholar]
- 48.Qimron U, Marintcheva B, Tabor S, Richardson CC. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc Natl Acad Sci U S A. 2006;103:19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran NQ, Lee SJ, Richardson CC, Tabor S. A novel nucleotide kinase encoded by gene 1.7 of bacteriophage T7. Mol Microbiol. 2010;77:492–504. doi: 10.1111/j.1365-2958.2010.07221.x. [DOI] [PubMed] [Google Scholar]
- 50.Tran NQ, Rezende LF, Qimron U, Richardson CC, Tabor S. Gene 1.7 of bacteriophage T7 confers sensitivity of phage growth to dideoxythymidine. Proc Natl Acad Sci U S A. 2008;105:9373–9378. doi: 10.1073/pnas.0804164105. [DOI] [PMC free article] [PubMed] [Google Scholar]