Abstract
Human induced pluripotent stem cells (iPSCs) have become an intriguing approach for neurological disease modeling, because neural lineage-specific cell types that retain the donors' complex genetics can be established in vitro. The statistical power of these iPSC-based models, however, is dependent on accurate diagnoses of the somatic cell donors; unfortunately, many neurodegenerative diseases are commonly misdiagnosed in live human subjects. Postmortem histopathological examination of a donor's brain, combined with premortem clinical criteria, is often the most robust approach to correctly classify an individual as a disease-specific case or unaffected control. In this study, we describe iPSCs generated from a skin biopsy collected postmortem during the rapid autopsy of a 75-year-old male, whole body donor, defined as an unaffected neurological control by both clinical and histopathological criteria. These iPSCs were established in a feeder-free system by lentiviral transduction of the Yamanaka factors, Oct3/4, Sox2, Klf4, and c-Myc. Selected iPSC clones expressed both nuclear and surface antigens recognized as pluripotency markers of human embryonic stem cells (hESCs) and were able to differentiate in vitro into neurons and glia. Statistical analysis also demonstrated that fibroblast proliferation was significantly affected by biopsy site, but not donor age (within an elderly cohort). These results provide evidence that autopsy donor-derived fibroblasts can be successfully reprogrammed into iPSCs, and may provide an advantageous approach for generating iPSC-based neurological disease models.
Keywords: induced pluripotent stem cells, genetic disease models, diagnostics, neurodegenerative diseases, postmortem, autopsy, neural differentiation
Introduction
Traditional in vitro disease modeling approaches rely on primary or immortalized somatic cells that are either obtained from a diseased individual, or are genetically modified to exhibit a disease phenotype in vitro [9]. Unfortunately, these models require a physiologically relevant cell line, which may not be easily available, and genetic modification may not recapitulate the etiology of complex diseases. Human induced pluripotent stem cells (iPSCs) can overcome these shortcomings, because genetically identical, tissue-specific cell types that are disease-applicable can be generated in vitro [9, 12, 18, 23]. Donor-specific iPSC models are particularly intriguing for the study of neurological and neurodegenerative diseases, because unlike other tissues of the human body, live brain tissue often cannot be obtained from living subjects without undue risk of cognitive or functional impairment [5].
The statistical power of iPSC-based neurological models is dependent on the diagnostic accuracy of the diseased (case) and unaffected (control) somatic cell donors. For many neurological diseases, premortem clinical criteria do not provide sufficient information for the subject to be given a definite diagnosis, but rather a possible or probable diagnosis [2, 4, 6, 7, 8, 14, 15, 16, 19]. For example, Alzheimer's Disease can be identified through clinical evaluations as the possible or probable cause of dementia; however, the definite diagnosis of this pathology cannot currently be confirmed until clinical criteria are combined with postmortem histopathological observations of the subject's brain [2, 4, 8, 14]. Because clinical phenotypes like dementia can be shared by multiple neuropathies, postmortem brain banking programs have become an exceptional resource for providing neuropathy-associated brain tissue that has been subjected to the most robust diagnostic methods. In fact, much of what is known about many neurological and neurodegenerative diseases has been from analyses (histological, biochemical, molecular, etc.) on autopsy donor-derived brain tissue [2, 6, 7, 8, 12, 15, 16].
Meske et al. have demonstrated that human dermal fibroblasts (HDFs) can be established in cell culture from autopsy donors up to 48 hours postmortem and from individuals up to 99-years-old [13]. To our knowledge, however, there are no published reports of autopsy donor-derived somatic cells being used for human iPSC generation. Because somatic cell senescence has been identified as a potential barrier in iPSC reprogramming, our group sought to investigate whether autopsy donor-derived fibroblasts could be induced to a pluripotent state [1, 3, 10, 20]. In addition, we chose to examine if these iPSCs could be differentiated in vitro into derivatives of the neural lineage, since this approach may be particularly valuable for neurological disease research.
Materials and Methods
Whole Body Donation
Tissue was collected after a postmortem interval (PMI) of approximately three to seven hours during the rapid autopsies of nineteen whole-body donors (ages 72-97). IPSCs were generated from one autopsy donor, a 75-year-old male identified by clinical studies and histopathological examination as being suitable as a “control” subject as he was negative for major neurological and neuropathological conditions (non-demented, without clinical parkinsonism and without diagnostic levels of histopathology for any major neuropathological condition). The subjects were enrolled as whole body donors in the Banner Sun Health Research Institute (BSHRI) Brain and Body Donation Program and had previously signed informed consent approved by the BSHRI Institutional Review Board (IRB) [2]. Fibroblast cell lines were established as previously described by Villegas and McPhaul, with minor modifications (see online supplementary methods) [21].
Statistical Analysis
A One-way Analysis of Variance (ANOVA) and Tukey's HSD Post-hoc test was performed on the cell counts at passage 1 (P:1) from 30 primary fibroblast cell lines established from 3 autopsy donors to determine the effect of biopsy site. A One-way ANOVA was performed on the cell counts from 34 primary fibroblast cell lines (all obtained from the same biopsy site (arm)) established from 18 autopsy donors to determine the effect of age (binned into 4-year groups). Data sets met the parametric assumptions of normality and homogeneity of variances. All statistics were performed using IBM SPSS Statistics, Version 19 software. Graphs were generated using Microsoft Excel 2008.
Lentiviral Transduction and Feeder-Free iPSC Generation/Maintenance
Autopsy donor-derived fibroblast cell line F02AA1 was selected for induced pluripotency experiments. Human iPSCs were generated in a feeder-free culture system by lentiviral transduction of the Yamanaka factors as previously described (see online supplementary methods) [3, 11, 18]. IPSC lines (through P:10) were frozen in cryopreservation media (mFreSrR; Stem Cell Tech.) and stored at -130°C prior to characterization and differentiation experiments, which were performed using iPSC clones 2-13 and 2-21 at passage 8 (P:8).
Embryoid Body Formation and Neural Differentiation
Embryoid Bodies (EBs), with approximately 400 cells per EB, were generated using Agrewell 400 Plates (Stem Cell Tech) according to the manufacturer's instructions. EBs were transferred in suspension to low adherence, non-treated 6-well plates (BD Biosciences) with approximately 500 EBs per well.
Inductive loss of pluripotency (LOP) was performed as previously described with minor modifications [17]. Briefly, EBs were cultured in suspension for 5 days in hESC media without basic fibroblast growth factor (bFGF) (DMEM/F12 (Invitrogen), 20% Knockout Serum Replacement (Invitrogen), 1mM L-Glutamine (Invitrogen), 0.1mM non-essential amino acids (NEAA; Invitrogen), 0.0007% 2-Mercaptoethanol (Sigma Aldrich), 100units/ml penicillin, 100μg/ml streptomycin (Sigma Aldrich), and 5μg/ml Plasmocin prophylactic (Invivogen)), and the media was exchanged every other day. The LOP media was then replaced with NeuroCult NS-A Proliferation media (Stem Cell Tech), supplemented with 20ng/ml recombinant human epidermal growth factor (rhEGF; Stem Cell Tech), 10ng/ml bFGF (Stemgent, Cambridge, MA, USA), 2μg/ml Heparin (Stem Cell Tech), 100units/ml penicillin, 100μg/ml streptomycin (Sigma Aldrich), and 5μg/ml Plasmocin prophylactic (Invivogen, San Diego, CA, USA), and the EBs were cultured in suspension for an additional 5 days. EBs were plated in complete NeuroCult NS-A Proliferation media onto 6-well tissue culture-treated plates (BD Biosciences) pre-coated with 1ml/well hESC-qualified Matrigel matrix (BD Biosciences), diluted 1 aliquot per 10ml DMEM/F12 (Invitrogen). EBs were allowed to collapse onto the matrix, and adherent cells were further cultured in complete NeuroCult NS-A Proliferation media for 14 days. Neural precursors were collected by bulk enzymatic passage with 1mg/ml Collagenase IV (Sigma Aldrich) in DMEM/F12 (Invitrogen) as previously described [17]. Cells were seeded for differentiation experiments at 5×103 cells/cm2 on glass coverslips (VWR, West Chester, PA, USA) pre-coated with 1ml/coverslip of hESC-qualified Matrigel matrix (BD Biosciences), diluted 1:10 as described above.
Cells plated onto glass coverslips were maintained in NeuroCult NS-A Differentiation media supplemented with 100units/ml penicillin, 100μg/ml streptomycin (Sigma Aldrich), and 5μg/ml Plasmocin prophylactic (Invivogen), and the media was exchanged every other day. Cells were allowed to differentiate for 14, 21, 28 or 35 days, at which time the coverslips were fixed in 4% paraformaldehyde (Thermo Fisher Scientific, Waltham, MA, USA) in DPBS (Invitrogen) for 15 minutes at room temperature. The coverslips were then rinsed and stored in DPBS (Invitrogen) at 4°C for later use in immunocytochemistry (ICC) experiments (see below).
Immunocytochemistry and Microscopy
Cells cultured on Matrigel-coated glass coverslips and fixed with 4% paraformaldehyde were used for all ICC assays. Cells were permeabilized for 10 minutes with 0.1% Triton X-100 (Thermo Fisher Scientific) in DPBS (Invitrogen), followed by a 30-minute incubation with blocking solution (2.5% bovine serum albumin (BSA) in DPBS with 0.1% Tween 20 (Sigma)). All primary antibodies were incubated overnight at 4°C and fluorophore-conjugated secondary antibodies were incubated for 2 hours at room temperature. The primary antibodies used for this study were directed against the following antigens: Oct3/4, 1:100 (Stemgent), SSEA4, 1:100 (Stemgent), TRA-1-60, 1:100 (Stemgent), Nanog, 1:100 (Santa Cruz Biotech, Santa Cruz, CA, USA), Neurexin IV (NRXN IV), 1:100 (Santa Cruz Biotech), Neuron-specific Beta III Tubulin (TUBB3), 1:2000 (Abcam, Cambridge, MA, USA), glial fibrillary acidic protein (GFAP), 1:500 (Abcam), and myelin/oligodendrocyte-specific protein (MOG), 1:500 (Millipore). Alexa Fluor-conjugated secondary antibodies (1:2000, excitations 488, 594, and 647; Invitrogen) were used for all experiments. In addition, 0.1ug/ml 4′-6-Diamidino-2-phenylindole (DAPI) was used as a nuclear counterstain. All brightfield, phase contrast and fluorescence images were obtained using the Leica DM IL inverted microscope with the Leica DFC 290 digital camera and Leica Application Suite software (version 2.8.1). All confocal microscopy images were obtained using an Olympus Fluoview FV1000 confocal microscope and the Olympus FV10-ASW 1.7 imaging analysis software.
Protein Extractions and Western Blots
Total soluble protein (TSP) was extracted from autopsy donor-derived fibroblasts (F02AA1, P:4), and two iPSC clones (2-13 and 2-21, P:8) using ice-cold RIPA buffer supplemented with a protease inhibitor cocktail and EDTA as per the manufacturer's instructions (Thermo Fisher Scientific). Protein extracts were concentrated approximately 10-fold with Microcon Centrifugal Filter Devices (30,000 nominal molecular weight limit; Millipore, Billerica, MA, USA), and TSP was quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
Approximately 25μg TSP/lane was separated using 4-12% Bis-Tris precast gels and NuPage electrophoresis system (Invitrogen). Protein was transferred onto Invitrolon PVDF membranes (Invitrogen) that were subsequently incubated in blocking solution consisting of 5% nonfat dry milk (Safeway, Pleasanton, CA, USA) in tris-buffered saline with 0.1% Tween (TBST; Thermo Fisher Scientific). All primary antibodies were incubated overnight at 4°C and horseradish peroxidase (HRP)-conjugated secondary antibodies were incubated for 1 hour at room temperature. The primary antibodies used for this study were directed against the following antigens: Oct3/4, 1:500 (Stemgent), Sox2, 1:150 (Invitrogen), Klf4, 1:100 (Stemgent), c-Myc, 1:100 (Stemgent), and GAPDH, 1:60,000 (Millipore). The HRP-conjugated secondary antibodies used were as follows: Goat anti-rabbit HRP, 1:5,000 (Upstate/Millipore), goat anti-mouse, 1:1000 (Millipore), and rabbit anti-chicken, 1:10,000 (Millipore). Western blots were developed using the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and the chemiluminescence was detected on x-ray film (Kodak, Rochester, NY, USA).
Single Nucleotide Polymorphism (SNP) Genotyping
Genomic DNA was extracted from fibroblast cell line F02AA1 and IPSC clone 2-21 using the Qiagen Blood & Cell Culture DNA Mini Kit as per the manufacturer's instructions (Qiagen, Valencia, CA, USA). Concentration and purity of both DNA extracts were analyzed using a NanoDrop ND-1000 spectrophotometer (Nanodrop, Wilmington, DE, USA) and Quant-iT PicoGreen Kit (Invitrogen). DNA aliquots were prepared at a concentration of 50ng/μl in a total volume of 10μl for use in the Affymetrix Genome-Wide Human SNP Array 6.0, performed as per the manufacturer's instructions (Affymetrix, Santa Clara, CA, USA) (see online supplementary methods). The number of matching and mismatching SNP calls between the two samples were extrapolated in Microsoft Excel 2008 and this was used to calculate the percent concordance. In addition, Affymetrix Genome-Wide Human SNP Array 6.0 results were used for copy number variation (CNVs) analysis. CNVs were initially detected for both the fibroblast and IPSC samples using the free software tool PennCNV (www.openbioinformatics.org/penncnv) [22]. The log R Ratio (LRR) and B Allele Frequency (BAF) plots were manually inspected using the visualize_cnv.pl program available in the PennCNV package, and any identified CNVs that appeared as potential false-positives were systematically removed.
Results and Discussion
The primary aim of this study was to examine if autopsy donor-derived dermal fibroblasts could be reprogrammed to a pluripotent state, for potential use in iPSC-based disease modeling. A visual summary of the various cell types used and generated in this proof-of-principle study are illustrated (Fig.1). IPSC clones from several additional autopsy donors have been generated using the same procedures as described, demonstrating this approach to iPSC generation is reproducible (data not shown).
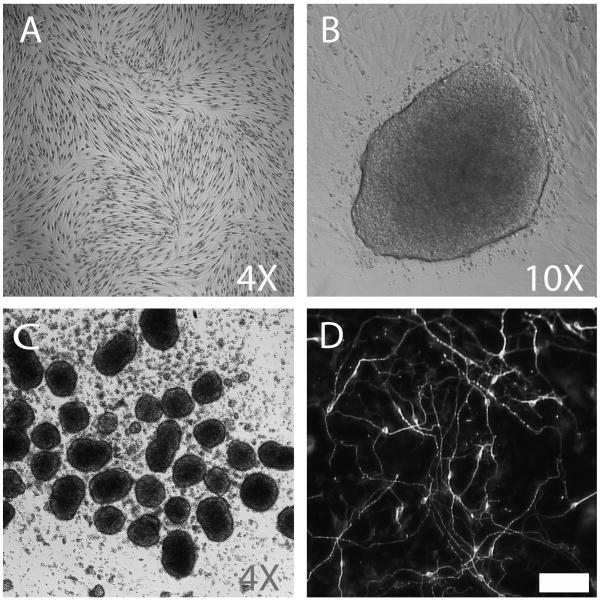
Figure 1.
IPSC induction and neural differentiation from autopsy donor-derived fibroblasts. Brightfield, phase contrast, and immunofluorescence images of (A) autopsy donor-derived dermal fibroblasts (F02AA1; Wright-Giemsa contrast stain), (B) iPSC colony arising from feeder-free conditions 21 days post transduction, (C) EBs generated from iPSC clone 2-13, and (D) neurons after 14 days of in vitro differentiation (neuron-specific beta III tubulin antibody stained). Scalebar = 50μm
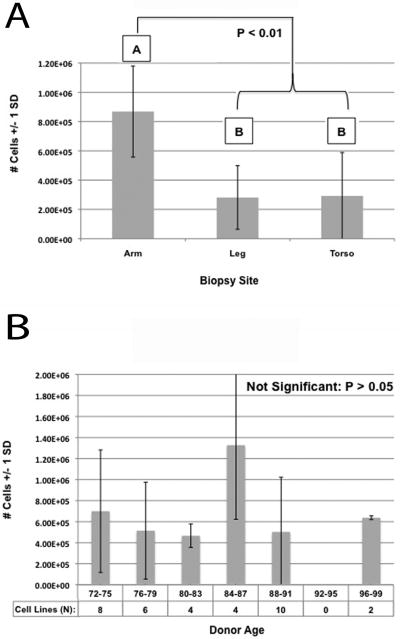
The primary dermal fibroblast cell counts were analyzed to determine if biopsy site or age affected proliferation. Three biopsy sites (arm, leg, and torso) were evaluated, from which all cell lines were established using the same amount of tissue and procedures. A One-Way ANOVA demonstrated a significant difference in fibroblast proliferation between biopsy sites (ANOVA: F= 9.616, v1=2, v2=27, P=0.001). A Tukey's HSD Post-hoc test subsequently revealed the arm biopsy site had statistically greater fibroblast proliferation than either the leg (P=0.003) or torso (P=0.004) sites, while there was no significant difference between the leg and torso (P=0.998) (Fig.2, A). Donor age, within an elderly cohort (ages 72-97), was also analyzed statistically and the One-Way ANOVA showed there was no significant difference in fibroblast proliferation based on donor age (ANOVA: F=1.756, v1=5, v2=28, P=0.155) (Fig.2, B). While these biopsies were collected within a short PMI of three to seven hours, Meske et al. additionally demonstrated that (in a rat model) fibroblast viability/proliferation was negatively affected by longer PMIs [13]. Collectively, these results suggest that researchers aiming to successfully establish fibroblast cell lines from non-optimal biopsy sites or after extended PMIs should collect and use more tissue per cm2 of tissue culture surface.
Figure 2.
Fibroblast proliferation is affected by biopsy site but not autopsy donor age (within an elderly cohort). Number of cells counted (y-axis) in both graphs was recorded 18 days post biopsy (Passage 1). (A) Graphical representation of One-Way ANOVA and Tukey's HSD Post-hoc test results, demonstrating biopsies obtained from the arm result in a significant increase in fibroblast proliferation as compared to biopsies obtained from the leg or torso. (B) Graphical representation of One-Way ANOVA results, demonstrating fibroblast proliferation is not significantly affected by autopsy donor age.
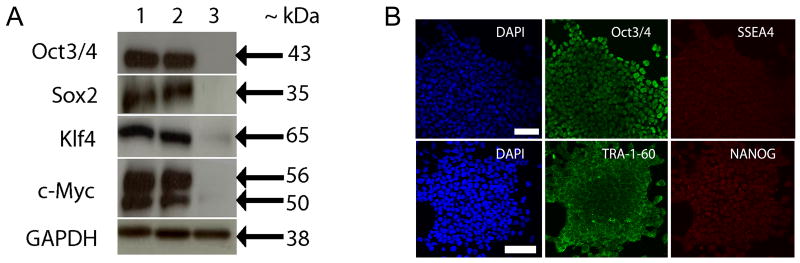
Efficient translation of the Yamanaka factor proteins in the described iPSCs was confirmed by western blot (Fig.3, A). All four factors were strongly expressed in the two iPSC clones evaluated as compared to the autopsy donor-derived fibroblasts. Expression of pluripotency markers was also evaluated by ICC (Fig.3, B). The Yamanaka factor Oct3/4 and the hESC antigens SSEA4, TRA-1-60, and NANOG were all expressed and appropriately localized in the iPSCs.
Figure 3.
Western blot analysis and immunocytochemistry of hESC markers. (A) IPSCs generated from autopsy donor fibroblasts express the Yamanka factor proteins. Lanes: (1) iPSC clone 2-13, (2) iPSC clone 2-21, and (3) autopsy donor-derived dermal fibroblast (F02AA1). (B) Confocal microscopy images of IPSCs shows expression of nuclear and surface pluripotency antigens. Each row of images displays localization of DAPI and two hESC markers within the same field of view. Top panel: 1μm optical section; DAPI, Oct3/4, SSEA4. Bottom panel: 3D reconstruction of 16× 1μm optical sections through the Z-axis; DAPI, TRA-1-60, and NANOG. Scalebars = 50μm
DNA extracted from fibroblast cell line F02AA1 and IPSC clone 2-21 was used for analyzing genotype concordance and CNVs (see online supplementary data). A percent concordance of 99.9625% verified that the IPSC clone had originated from the autopsy donor cell line. In addition, no IPSC-specific CNVs spanned regions greater than 100 kilobases, suggesting that no large chromosomal segments or whole chromosomes had become aneuploid during the reprogramming process.
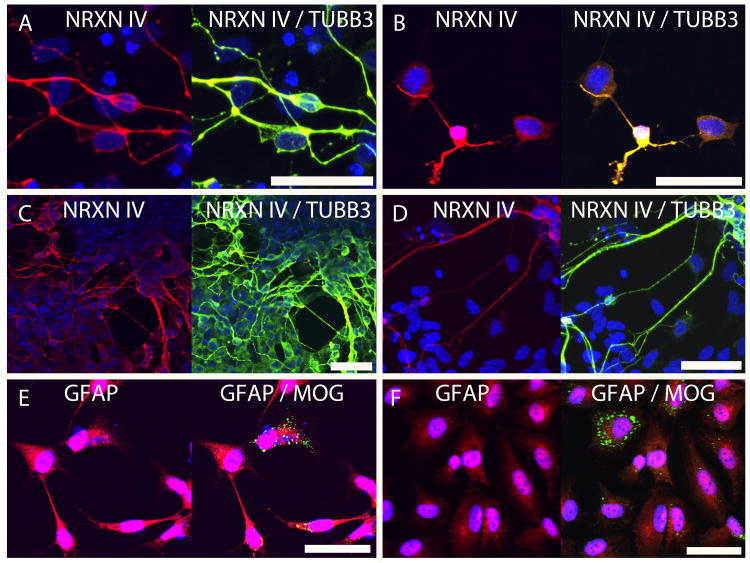
The two iPSC clones (2-13 and 2-21) characterized in this study were differentiated into derivatives of the neural lineage. ICC assays identified neurons after 14 (Fig.1, D; Fig.4, A and B) and 28 (Fig.4, C and D) days of differentiation. TUBB3 and NRXN IV were utilized as terminal neuronal markers; both antigens co-localized specifically to cells exhibiting a neuronal phenotype (Fig.4, A, B, C and D). ICC assays also identified glial subtypes, including astrocytes and oligodendrocytes, after 35 days of differentiation (Fig.4, E and F). GFAP was expressed in the majority of the differentiated, non-neuronal cells, while a small subset of this population expressed MOG (Fig.4, E and F). This data suggests that the majority of the glia were astrocytic in nature, while a small minority was becoming oligodedrocytic.
Figure 4.
IPSCs generated from autopsy donor fibroblasts can be differentiated into neurons and glia. Confocal microscopy images shown are from 1μm optical sections through the Z-axis of cells fixed after 14 (A, B), 28 (C, D) and 35 (E, F) days of differentiation. Left panel: iPSC clone 2-13; Right panel: iPSC clone 2-21. (A, B, C, D) Neurexin IV (NRXN IV) (red), co-localization of Neurexin IV (NRXN IV) and neuron-specific beta III tubulin (TUBB3) (yellow), and the nuclear counterstain DAPI (blue). (E, F) Glial fibrillary acidic protein (GFAP) (red), myelin/oligodendrocyte-specific protein (MOG) (green), and the nuclear counterstain DAPI (blue). Scalebars = 50μm
These results provide evidence that postmortem human tissue can be successfully reprogrammed to a pluripotent state. This approach may be significantly useful for studies investigating diseases and/or drugs that may cause sudden death and for neurodegenerative disease research. The ability to combine both clinical diagnostic criteria and postmortem histopathological observations can greatly increase neurodegenerative diagnostic accuracy; this may subsequently increase the statistical power for donor-specific in vitro disease models. Human iPSC-based disease models generated from postmortem tissue may provide additional confidence for researchers investigating these conditions, and provides an avenue for comparing differentiated in vitro cell types to endogenous adult tissue from the same individual.
Supplementary Material
Highlights.
Human IPSCs provide an avenue for investigating neurological disease mechanisms
Many neurodegenerative diseases are commonly misdiagnosed in live patients
Postmortem histopathological review of a donor brain increases diagnostic accuracy
Postmortem cells can be made into IPSCs that are capable of neural differentiation
Postmortem fibroblast proliferation is effected by biopsy site but not donor age
Acknowledgments
The authors would like to acknowledge the following sources of funding support: The National Institute of Neurological Disorders and Stroke (NINDS, 5U24NS051872-05), The Arizona Alzheimer's Consortium, The National Institute on Aging (P30 AG19610 Arizona Alzheimer's Disease Core Center), The Stardust Foundation, The Marley Foundation, The Arizona Department of Health Services (contract 211002, Arizona Alzheimer's Research Center), The Arizona Biomedical Research Commission (contracts 4001, 0011, and 05-901, Arizona Parkinson's Disease Consortium), The Michael J. Fox Foundation for Parkinson's Research, and the State of Arizona. We are greatly appreciative to the whole body donors and their families, without whom this work would not be possible.
Footnotes
Author Disclosure Statement: The authors have no conflicting financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beach TG, Sue LI, Walker DG, Roher AE, Lue L, Vedders L, Connor DJ, Sabbagh MN, Rogers J. The Sun Health Research Institute Brain Donation Program: description and experience, 1987-2007. Cell Tissue Bank. 2008;9:229–245. doi: 10.1007/s10561-008-9067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 4.Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, Nalls MA, Chen K, Lee W, Chewning K, Villa SE, Meechoovet HB, Gerber JD, Frost D, Benson HL, O'Reilly S, Chibnik LB, Shulman JM, Singleton AB, Craig DW, Van Keuren-Jensen KR, Dunckley T, Bennett DA, De Jager PL, Heward C, Hardy J, Reiman EM, Huentelman MJ. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald S. Brain Biopsy Found Unhelpful for Certain Patients in One Center. Neurology Today. 2010;10:26. [Google Scholar]
- 6.Gelpi E, Heinzl H, Hoftberger R, Unterberger U, Strobel T, Voigtlander T, Drobna E, Jarius C, Lang S, Waldhor T, Bernheimer H, Budka H. Creutzfeldt-Jakob disease in Austria: an autopsy-controlled study. Neuroepidemiology. 2008;30:215–221. doi: 10.1159/000126915. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton RL. The other dementias: the neuropathology of the non-Alzheimer's disease dementias. Rev Neurol. 2003;37:130–139. [PubMed] [Google Scholar]
- 8.Joseph S, Robbins K, Zhang W, Rekaya R. Effects of misdiagnosis in input data on the identification of differential expression genes in incipient Alzheimer patients. In Silico Biol. 2008;8:545–554. [PubMed] [Google Scholar]
- 9.Li Zk, Zhou Q. Cellular models for disease exploring and drug screening, Protein & Cell. 2010;1:355–362. doi: 10.1007/s13238-010-0027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Sumer H, Leung J, Upton K, Dottori M, Pebay A, Verma PJ. Late passage human fibroblasts induced to pluripotency are capable of directed neuronal differentiation. Cell Transplant. 2010;20:193–203. doi: 10.3727/096368910X514305. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, Llanas RA, Thomson JA. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 12.Marchetto MC, Winner B, Gage FH. Pluripotent stem cells in neurodegenerative and neurodevelopmental diseases. Hum Mol Genet. 2010;19:R71–76. doi: 10.1093/hmg/ddq159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meske V, Albert F, Wehser R, Ohm TG. Culture of autopsy-derived fibroblasts as a tool to study systemic alterations in human neurodegenerative disorders such as Alzheimer's disease--methodological investigations. J Neural Transm. 1999;106:537–548. doi: 10.1007/s007020050177. [DOI] [PubMed] [Google Scholar]
- 14.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson PT, Jicha GA, Kryscio RJ, Abner EL, Schmitt FA, Cooper G, Xu LO, Smith CD, Markesbery WR. Low sensitivity in clinical diagnoses of dementia with Lewy bodies. J Neurol. 2010;257:359–366. doi: 10.1007/s00415-009-5324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piguet O, Halliday GM, Reid WG, Casey B, Carman R, Huang Y, Xuereb JH, Hodges JR, Kril JJ. Clinical phenotypes in autopsy-confirmed Pick disease. Neurology. 2011;76:253–259. doi: 10.1212/WNL.0b013e318207b1ce. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz PH, Brick DJ, Stover AE, Loring JF, Muller FJ. Differentiation of neural lineage cells from human pluripotent stem cells. Methods. 2008;45:142–158. doi: 10.1016/j.ymeth.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Tolosa E, Wenning G, Poewe W. The diagnosis of Parkinson's disease. Lancet Neurol. 2006;5:75–86. doi: 10.1016/S1474-4422(05)70285-4. [DOI] [PubMed] [Google Scholar]
- 20.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villegas J, McPhaul M. Current Protocols in Molecular Biology. John Wiley & Sons, Inc.; 2001. Establishment and Culture of Human Skin Fibroblasts. [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.