Abstract
A new computational method to study within-host viral evolution is explored to better understand the evolution and pathogenesis of viruses. Traditional phylogenetic tree methods are better suited to study relationships between contemporaneous species, which appear as leaves of a phylogenetic tree. However, viral sequences are often sampled serially from a single host. Consequently, data may be available at the leaves as well as the internal nodes of a phylogenetic tree. Recombination may further complicate the analysis. Such relationships are not easily expressed by traditional phylogenetic methods. We propose a new algorithm, called MinPD, based on minimum pairwise distances. Our algorithm uses multiple distance matrices and correlation rules to output a MinPD tree or network. We test our algorithm using extensive simulations and apply it to a set of HIV sequence data isolated from one patient over a period of ten years. The proposed visualization of the phylogenetic tree\network further enhances the benefits of our methods.
1. Introduction
The processes involved in intra-host (within a single host or patient) and inter-host evolution are strikingly different for retroviruses such as HIV [12]. In contrast to inter-host evolution, intra-host evolution in HIV is characterized by high rates of evolution, and by strong evidence of positive selection that favors mutations to help the pathogen evade the host immune response. Therefore, intra-host evolution exhibits a strong temporal structure and the positive selection often leads to the extinction of unfavorable lineages. Investigating viral evolution within a single host or patient over a period of time provides a direct and verifiable way to comprehend mutational changes that occur during the replication of a genome over many generations. From the viewpoint of clinical and biomedical research, investigating the intra-host viral evolution through serial sampling of the viral strains over a period of time may lead to a better understanding of the progression of a disease in that patient, or assist in the evaluation of drug therapies or vaccines for a disease. A recent study performed a comprehensive analysis of serially-sampled HIV sequence data from nine patients with data collected over a span of over ten years [19]. Adaptive evolution and the strength of immune selection were investigated in another study with samples from 50 patients [22].
Traditional phylogenetic methods were conceived for the purpose of inferring the history of a set of contemporaneous taxa. In such trees the taxa being analyzed appear at the leaves of the tree. The ancestral sequences are usually unknown. A conflicting situation arises when some of the sequences at the internal nodes are available, such as with serially-sampled viral sequences, but the tree-constructing program interprets all of them as contemporaneous taxa [15]. Ren et al. pointed out that traditional phylogenetic methods do not account for the fact that viral strains can branch, become extinct or revive (after a period of dormancy) between the sampling time periods [13]. Furthermore, the trees resulting from applying the traditional methods are hard to interpret and analyze (see discussion in Section 7). Prior work on phylogeny of non-contemporaneous, serially-sampled sequences includes an algorithm called sUPGMA, a modification of the UPGMA [1] and the work of Ren et al., who modified the neighbor-joining method [13].
An unusually high rate of genetic recombination is yet another factor that sets intra-host evolution of HIV (and other retroviruses) apart from the evolution of other organisms. In this paper we present a distance-based algorithm (called MinPD) to infer evolutionary relationships (including recombination) in serially-sampled sequence data. An important feature of the algorithm is that it does not need the assumption of a molecular clock or an explicit statistical model of evolution. Unlike methods based on maximum likelihood (ML) or maximum parsimony (MP), our method is computationally efficient and can deal with a large number of input sequences. Our method assigns ancestor relationships using minimum pairwise distance without the use of multiple alignments. Ties are broken by resorting to divergence information. Recombinant strains are detected using sequence fragment matrices, correlations and distance rules. The algorithm MinPD was implemented in C. The accuracy of the methods was assessed using extensive experimentation on both simulated data and on real HIV sequence data from the HIV database. The simulated data included sequences at the leaves as well as sequences at internal nodes of a phylogenetic tree. A critical feature of the simulations is that it attempts to mimic the fact that, in reality, only a small random sample of all the viral strains that may be present in a patient is actually sampled. This is achieved by simulating a large number of sequences and discarding a large fraction of them. Another contribution of this work is to show how to incorporate recombination into longitudinal phylogenetic trees without losing any of its essential features and advantages. The resulting phylogenetic networks (see Figure 4 for an example) make it convenient for a biologist to draw useful conclusions. Our work is similar to the work of Ren et al., with the significant added feature that it accounts for recombination.
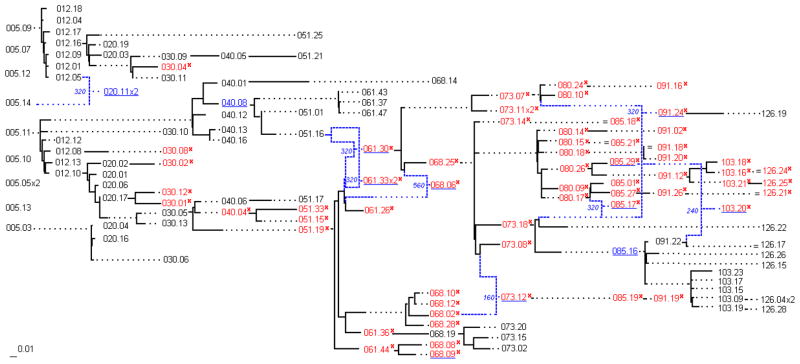
Figure 4.
MinPD Tree of Patient S. Solid lines indicate distances, while dotted lines serve to extend the linking relationships. Each sequence is labeled with the month number and an identification number. Sequences with a mutation predictive of the X4 phenotype are written in red font and also marked with a red “x”. Blue dashed lines are used to link recombinant sequences with their predicted donor sequences. The small numbers in blue next to branch points in the tree are the predicted (approximate) recombination breakpoint positions. Sequences with weaker recombinant signals are underlined in blue, but are not linked to their putative donor sequences. Note that the sequences were divided into 8 and 4 fragments for the recombination analysis.
2. Recombination
An unusually high rate of recombination is one of the evolutionary traits of RNA viruses. During recombination, nucleotide sequences are exchanged among different RNA molecules. Recombination in HIV occurs between two coencapsidated RNA genomes during reverse transcription. During DNA synthesis the reverse transcriptase, which is prone to errors, may switch from one strand to the other, either during the first (−) strand DNA synthesis, or during the second (+) strand DNA synthesis, as part of the mechanism of strand displacement assimilation [3]. If the two ancestral RNA genomes are different, we will call them the donor strains or donor sequences, since the newly synthesized genome will contain fragments of each of them, resulting in the creation of the so-called mosaic genes. Recombination is an important mechanism for producing new genomes with selective growth advantages, by moving functional parts of RNA molecules among different viral strains. It plays a major role in contributing and maintaining genetic diversity in viral populations. Phylogenetic methods that do not account for recombination can make incorrect inferences in the presence of recombination [8, 17, 18]. It is therefore critical to detect recombination. In recent years the development of new tools to model and test for recombination have led to several studies that compare the different methods and assess their accuracy [9, 23]. Methods such as bootscanning detect changes in phylogenetic relationships to detect breakpoints and donors. In bootscanning, the alignment is broken into sequential, overlapping segments (or windows) of 200–500 bases, which are then input to a program for phylogenetic analysis. Bootstrapped phylogenetic trees are built for each segment, and finally the bootstrap value for placing the queried sequence with each of the reference sequences/sequence groups is tabulated and plotted along the genome. High bootstrap values indicate that the reference sequence in that window is a possible donor sequence. Other approaches simply report a recombination rate without identifying breakpoints or the donor strains.
A widely used visual tool for detecting recombination is Stuart Ray’s SimPlot, which uses the bootscanning method [16] and the Maynard Chi-Square method [5] to reveal potential breakpoint positions. This method requires as input in addition to the donor sequences, a reference sequence that is assumed beforehand to have been generated by recombination. An improvement on this method is VisRD, another visual detection method that does not require the reference strain to be identified in advance [20]. The MinPD method, unlike the bootscanning and VisRD methods, identifies recombinant strains, donors, and approximate breakpoint positions, and does not require the intervention of the user. MinPD was explicitly created to study viral quasispecies sampled at different time instances.
3. Evolution of Quasispecies
Viral species have an enormous capacity to adapt to a changing environment, which may change depending on the host’s changing physical condition, immune response, and drug-induced responses. The term quasispecies is applied to closely related genetic variants that differ by small amounts and are affected as a group by natural selection. Virologists use the term to describe the mutant viral strains living within a host. The concepts of quasipecies theory were first introduced by Manfred Eigen with the purpose of describing the molecular evolution of fast-replicating RNA genomes [2]. The evolutionary relationship of these quasispecies over a period of time cannot be revealed using traditional phylogenetic methods as these assume that all species are contemporaneous. Thus new methods are needed to group the genetic variants and to describe their relationships over time.
Drummond and Rodrigo modified the conventional UPGMA method for analyzing serially-sampled sequences [1]. The drawback of their method is that because it is based on the UPGMA method it presupposes that the data has evolved at a constant rate. This work and later improvements of Rambaut et al. (TipDate program) were further constrained by the traditional tree style of handling contemporaneous data where the leaves correspond to the input taxa [11], and thus do not pay tribute to the time-sequential nature of the data, making ancestor-decendant relationships somewhat unclear. Ren et al. proposed a sequential linking algorithm [13, 14], which is computationally inefficient since it is based on the maximum likelihood method. A more efficient method based on the NJ method was proposed by Ogishima et al. in which they were also able to estimate both neutral and selective adaptive evolution patterns [7].
In one of the most comprehensive studies on the evolution of HIV sequences by Shankarappa et al., samples from nine patients were isolated over several time points and studied in relation to the disease progression [19]. The study showed a strong correlation between the emergence of the syncytium-inducing (SI) X4 mutant phenotype and the rapid decline of CD4+ T-cells and a more rapid disease progression. The work of Shankarappa et al. helped to raise a host of questions that are of practical significance with regard to understanding HIV evolution and its relationship to AIDS symptoms.
We devised MinPD as a tree/network-constructing tool to study the evolution of viral quasispecies and to respond to a myriad of questions that may shed light on the progression of the AIDS disease, answering questions such as: (1) Which initial viral strains did the X4 phenotype mutants originate from? (2) Which of the initial strains became extinct and when did this happen? (3) Which strains showed positive selection, proliferating with descendants surviving over extended periods of time? (4) When did most recombinant strains appear? Traditional phylogenetic techniques have severe limitations in addressing such questions.
Data from patient number 2 used in the paper by Shankarappa et al. [19] is also available in the Los Alamos database and has been used as a typical example throughout this paper. Henceforth in this paper, this patient will be referred to as patient S.
4. The MinPD Tree/Network
The MinPD algorithm is based on the concept of minimum pairwise distance. It assumes that an ancestor of any given taxa must have been sampled at one of the previous time points and that the distance to the closest ancestor must be the minimum among all distances to taxa sampled during all prior time points. It utilizes the same criteria to find minimum distance fragments to all other sequences to identify possible recombinant strains. It also assumes that pairwise alignments give less distorted evolutionary distances than do multiple alignments
4.1. The Multiple Alignment Problem
Phylogenetic analysis methods ranging from tree-building methods to recombination detection techniques such as bootscanning, employ (as an initial step) a multiple sequence alignment of all input sequences. Multiple alignments of sequences of different lengths must necessarily add gaps, which often lead to loss of information and gap scoring artifacts, which in turn distort the distance computations. In existing distance-based phylogenetic methods, all distances are computed using this multiple alignment, including the methods that are said to use “pairwise distances.”
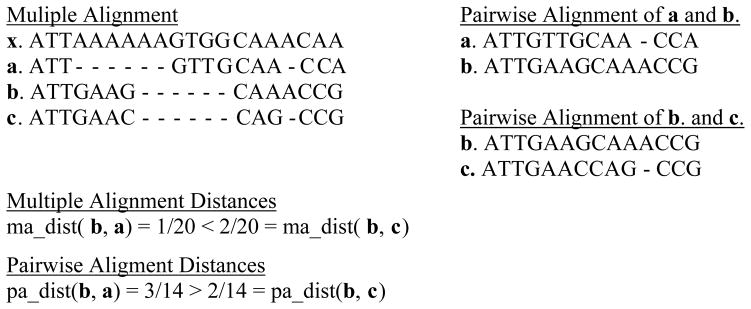
Figure 1 shows a multiple alignment of four sequences and also two pairwise alignments. The gap columns are ignored and do not count as mismatches. However, the pairwise distances in the two alignments are different. What is striking in the example is that the distances between two pairs of sequences exhibited a different order in the multiple alignment as compared to the corresponding distances in the pairwise alignments. It is for the above reasons, that we use pairwise alignments that offer a more accurate distance measure.
Figure 1.
The problem with multiple alignment
4.2. The Algorithm
The inputs to the algorithm are: s, a set of sequences with associated time periods, k, the number of fragments, and t, the threshold for the Pearson Correlation Coefficient. We use the Needleman-Wunsch algorithm to compute an alignment between each pair of sequences. For computing pairwise distances we use the Tamura-Nei Model (TN93) of nucleotide substitution with Gamma-distances [6]. Henceforth, whenever we refer to distance in this text, we mean the TN93 distance, and calculate this distance from a pair of aligned sequences that did not undergo a multiple alignment operation. Finally, we also assume that if the distances indicate two possible candidates for the closest ancestor, then ties are broken using divergence values.
For recombination detection we will assume that there is at most one recombination or crossover point for any recombination between 2 sequences, limiting the number of donor strains to two. The MinPD algorithm is given below.
Algorithm MinPD
-
For each pair of sequences si and sj do
Pairwise align them and compute the distance Dist(si,sj) between them.
Partition si and sj into k fragments and compute the distance vector DistVec(si,sj) of the k distances between the k pairs of aligned fragments. Let its ℓth component be denoted by Dist(si,sj,ℓ), the distance between the ℓth fragments of si and sj.
-
For each sequence si do
if (si passes the test described below for being a recombinant strain) then identify two donor strains and choose them as ancestors of si.
else choose as ancestor of si the sequence at minimum distance from it among sequences sampled at all previous time periods. Break ties using divergence values.
For each set of sequences with the same chosen ancestor, construct a NJ tree and connect the root of the NJ tree to the chosen ancestor.
Note that in Step 2a above, any method can be used to test for recombination or to identify the donor strains. However, below we propose a uniform distance-based method to achieve the same goal. Also, the divergence between two sequences used in step 2b denoted by Div(x,y) is the same as that used in many neighbor joining methods and is given by:
where r(x) = Σj distance(x,sj) is the net total divergence of x to all other sequences, and n is the number of sequences being considered.
MinPD Recombination Test for sequence s
For each of the k fragments of s, select the sequence si whose ith fragment has minimum distance to the ith fragment of s. Put all selected sequences in a list called Candidates. These sequences are candidates for being donors if s is a recombinant strain.
From the list Candidates, let minSeq be the sequence with minimum overall distance to s.
-
For each pair of sequences si and sj from Candidates do
if the Pearson Correlation Coefficient (PCC) between their distance vectors is above a distance threshold, then discard the sequence si or sj with the higher overall distance.
For all sequences in Candidates, discard those that have a fragment with minimum distance in the middle of the sequence, and not at either end.
For each sequence si ≠ minSeq, calculate si_dom = Σ (Dist(minSeq,s,i) − Dist(si,s,i)) in all fragments i where si has the minimum distance to the corresponding fragment in s. If si_dom is below a distance threshold, then discard si.
If exactly two sequences are left undiscarded, then report s as being recombinant with the two sequences as potential donors.
4.3. Fragment and Fragment Distances
The objective of using minimum fragment distances in the MinPD algorithm was to reduce the number of possible candidates for being recombinant donors for a given sequence. For most existing tools that detect recombinant sequences, a good selection of possible donors is critical and improves the chances of getting clear recombination signals. Consider, for example, sequence number 028.415, which was a sequence of length 1000 nucleotides, generated using Treevolve as part of the data sets for our experiments. From the data, we knew beforehand that 028.415 was a result of the recombination of the donor strains 008.384 and 002.97 at breakpoint 469. SimPlot, which uses the bootscanning technique, requires a minimum of 4 and a maximum of 26 sequences to detect recombination. However, as illustrated in the top two bootscanning graphs in Figure 2, SimPlot was able to correctly identify the recombinants when sequence 004.440 was the fourth sequence used, but not when 001.1 was used.
Figure 2.
TOP: SimPlot bootscanning graphs for reference sequence 028.415, but with different “donor” sequences. Upon visual inspection, recombination is clear in the left graph with breakpoint somewhere in the middle of the sequence; in the right graph the recombination signal is lost due to a bad selection of putative donor sequences. BOTTOM: line charts of MinPD distances vectors for reference sequence 028.415 with 4 and 8 fragments respectively.
We tried two sets of experiments, one where each sequence was divided into 4 fragments and the other where each was divided into 8 fragments. It was necessary to fine tune the threshold values used in the algorithm before we were able to get comparable performance in the two sets of experiments. The bottom of Figure 2 shows two graphs, one for the 4 fragment case, and one for the 8 fragment case. Each graph represents the components of the distance vector (with respect to reference sequence 028.415) of the candidate sequences selected in Step 2 of the recombination test for sequence 028.415. Thus only the sequences that have at least one fragment at a minimum distance from the corresponding fragment of 028.415 are represented.
The recombination test described in the MinPD algorithm above was able to successfully identify the recombination donors and the fragment within which the breakpoint may be located. Note that our algorithm works under the assumption that there is at most one recombination breakpoint. This is perhaps justified given that HIV averages about three recombination events for an entire genome and that new strains are produced only if the recombining strains are genotypically distinct on both sides of the breakpoint.
5. Experiments with Simulated Data
To test the MinPD algorithm, two synthetic data collections were generated, the first with recombination, and the second without. The first collection was generated using SeqGen1.2.5., and was enhanced by the twister randomization function of SeqGen 1.2.7.
Each of the 100 data sets in this collection contained 1023 sequences from the leaves and internal nodes of a template tree, out of which an average of 32 were randomly chosen (to simulate sampling from a population) and was input to the MinPD algorithm. The results seen in Table 1 show that more than 90% of the time, MinPD chose the correct closest ancestor (referred to in the table as a Match). A subtree relative (a direct descendant of the correct closest ancestor) was chosen about 9% of the time. All other outcomes were counted as errors. The errors included cases where a grand ancestor (ancestor of actual closest ancestor) was picked, although multiple mutations on the same location during evolution can lead to “backward” substitutions and to a grand ancestor being genetically closer to the queried sequence. The overall error rate was less than 0.5%. Note that picking a subtree relative is not classified as an outright “error” because we consider it as a minor deviation from the correct relationship.
Table 1.
Experiments with non-recombinant sequences
| Runs | Sequence Length | Match | Subtree Relative | Errors | |
|---|---|---|---|---|---|
| 100 | 600n | 90.9% | 8.8% | 0.37% | |
| 100 | 1000n | 90.9% | 9.1% | 0.06% | |
|
| |||||
| Total/Average | 200 | 90.9% | 8.95% | 0.22% | |
The second data collection consisted of 100 data sets each containing about 500 sequences (or slightly more, depending on how many recombination events occur) generated using the software package Treevolve version 1.3. Treevolve was modified to include the Twister randomization function of SeqGen 1.2.7., and to output sequences at the internal nodes. Treevolve evolves a sequence using Hudson’s coalescent method with recombination [4]. As before, to mimic the actual sampling from large populations, an average of 45 of the 500 or more sequences were randomly chosen for input to the MinPD algorithm. The sets of data were simulated under the HKY model of evolution with the alpha parameter of the gamma distribution set to 0.5. A transition/transversion ratio of 4 was chosen and the base frequencies were set to A=0.22, C=0.18, G=0.40, and T=0.2. The results of our experiments on the simulated data are shown in the table below. In order to get realistic data, a mutation rate of 0.5×10−4 and a population growth rate of 0.75×10−4 were selected. The recombination rate of 0.1×10−7 was selected since for the given mutation rate, higher recombination rates gave enormously long lineages. This is because although coalescent events result in a reduction of the number of lineages by one, recombination events cause an increase.
Table 2 shows the results of these experiments. The labels on the columns are explained below. In the presence of recombination events, the ability of the MinPD algorithm to correctly establish phylogenetic relationship among the input sequences is adversely affected. Even on non-recombinant sequences, the percentage of correct predictions dropped from over 90% (Table 1) to under 75% (Non Rec Matches) in Table 2. In each data set, about 3–5% (Rec Count/Total Count) of the strains sampled were recombinant strains. Of these, about 65% (Rec Detected) were correctly detected as being recombinant. The donors were correctly identified in over 50% (Rec Matches) of those cases. In about 15% (Rec Errors) of the cases, the program identified the donors incorrectly. In the remaining 31% (Rec Subtree Relative) of the cases, a subtree relative was determined to be the donor. Of the over 4500 sequences (Total Count) in the 4-fragment (#Frag) run, only 39 non-recombinant sequences were reported as being recombinant sequences by the MinPD program, accounting for a 1% false positive rate (False Pos).
Table 2.
Experiments with recombinant sequences
| # Frag | Thres holds | Runs | Len | Total Count | Non Rec Matches | Non Rec Subtree Relative | Non Rec Errors | Rec Count | Rec Detected | Rec Matches | Rec Subtree Relative | Rec Errors | False Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 0.75 | 100 | 600 | 4540 | 74.4% | 20.9% | 0.7% | 149 | 67.1% | 49% | 37% | 14% | 0.6% |
| 8 | 0.67 | 100 | 600 | 4540 | 73.4% | 20.6% | 0.6% | 149 | 63.3% | 55.8% | 26.3% | 17.9% | 1.9% |
| 4 | 0.9 | 100 | 1000 | 4671 | 72.8% | 21.1% | 0.3% | 212 | 67.9% | 56.9% | 27.1% | 16.0% | 0.8% |
| 8 | 0.8 | 100 | 1000 | 4674 | 74.3% | 19.9% | 0.5% | 199 | 61.3% | 52.5% | 35.3% | 12.3% | 0.8% |
| Total | 400 | 73.7% | 20.6% | 0.5% | 177.3 | 64.9% | 53.6% | 31.4% | 15.0% | 1.0% | |||
The results were somewhat weaker when the sequences were divided into 8 fragments instead of 4, which required additional fine-tuning of the thresholds chosen for the program. Note that when the sequences were divided into 8 fragments, there were more potential candidates for the choice of donors, which complicates the selection of correct donors. The threshold for PCC was set to values between 0.67 to 0.9 and the threshold for si_dom, was set to the average of the distances from si scaled by the quantity n/k, where k is the number of fragments, and n the number of fragments where si has a minimum distance. We conjecture that more fine-tuning can further improve the program’s sensitivity, and that sliding-window methods could improve the specificity.
It would be reasonable to conjecture that detecting recombination signals is harder with shorter sequences, although this was not observed in our experiments with sequences of length 600. It is possible that this is balanced out by the fact that there is a corresponding lower probability of recombination events in smaller sequences.
6. Experiments with Serially-sampled HIV Sequence Data
The final evaluation of the MinPD algorithm was performed by constructing the phylogenetic network for HIV sequence data from patients available from the Los Alamos HIV database. The viral strains were sampled and sequenced for a single patient (patient S) at month numbers 5, 12, 20, 30, 40, 51, 61, 68, 73, 80, 85, 91, 103, and 126. The resulting “longitudinal” phylogenetic network is shown in Figure 4. Each sequence is labeled with the month number and an identification number. There is no reasonable way to evaluate the correctness of the resulting network. Therefore, we focus our discussions on how well it correlates with the emergence of X4 strains (see below), and on how the resulting network makes it convenient to draw a variety of conclusions. It is worthwhile to compare the difficulty of drawing similar conclusions from the ML tree generated for the same data, as shown in Figure 3 below. The longitudinal network shown in Figure 4 is drawn from left to right and requires that sequences sampled at the same time be vertically aligned. This does not mean that all sequences undergo the same amount of evolution from the root sequence. On the contrary every link between a parent and child node consists of straight-line segments. Horizontal thick lines are a measure of the amount of evolutionary changes that take place between the sequences. Horizontal dashed lines are added merely to achieve the vertical alignment of the nodes corresponding to contemporaneous sequences. The only purpose of vertical lines is to ensure the correct connectivity.
Figure 3.
Maximum Likelihood (ML) tree of serially-sampled HIV sequence data from patient S.
All sequences marked with a red “x” have a lysine (K) or arginine (R) at position 320, a mutation that is predictive of the X4 phenotype. With the help of immunological data, it was shown by Shankarappa et al. that patient S’s CD4+ and CD3+ T-cell numbers fell rapidly during the emergence of X4 genotypic strains [19]. The longitudinal network makes it convenient to understand how widespread the X4 genotype is in each sampling period.
Furthermore, it is interesting to note that patient S was prescribed antiretroviral drugs called zidovudine (ZDV) and stavudine (d4T) before the 103 months sampling period, and a few months later was prescribed lamivudine (3TC). The administering of this drug therapy coincides with a decrease in the X4 genotypic strains [19], as is easily observed in the MinPD network in Figure 4.
Before the large-scale emergence of the X4 genotype (up to 51 months), the MinPD network suggests that three groups of genetically similar quasispecies sequences were present in the population. One group became extinct at 51 months, while the other two groups each contributed a sequence, 051.19 and 051.16, that recombined to create strain 061.30, the possible closest ancestor of the large X4 quasispecies that proliferated in the ensuing years.
In the second half of the network corresponding to time periods 61 to 126 months, only two groups of quasispecies, linked by recombinant sequence 073.12, were identified by MinPD, one of the groups becoming extinct probably at the onset of antiretroviral therapy with 091.19 as its last sampled sequence, and the other group formed by descendants from recombinant sequence 061.30, giving rise to a mixed population of X4 and non-X4 genotypic strains. It is also interesting to note that the first X4 mutations that appear at 30 months have a relatively large genetic distance to its ancestor in comparison to the contemporaneous strains, suggesting a higher rate of mutations for those particular strains. It should be noted that the above conclusions are made more convenient by the way the MinPD network is presented.
To make the comparisons more clear, we show the ML tree generated for the same data. We aligned 65 sequences from the first 61 months using ClustalX. Subsequently we did a heuristic search for the ML tree using PAUP (version 4b10) [21]. If the horizontal axis is to be thought of as time, then the ML tree shown in Figure 3 exhibits several anomalies with strain 051.19 (sampled at 51 months) appearing after strain 061.31 (sampled at 61 months), and strain 030.01 appearing after strain 051.51.
Furthermore recombinant data cannot be identified in a traditional phylogenetic tree, but for the fact that it often has very long branches. In the MinPD network, recombinant sequences are linked to their donor ancestors by blue lines and the breakpoint position is added left and next to the recombinant sequence. The recombination results output by MinPD were studied in detail using graph analysis, and only recombination relationships with the strongest signals were added to the network. Sequences with weaker recombinant signals were underlined in blue. A 2002 study of in vitro HIV-1 sequences and recombination site analysis suggested that the C2 env domain was a particularly “hot” region for recombination [10]. The data of patient S does not contain the entire C2 region but includes the regions V3, C3, V4, C4, and V5, all of which were also found to have several recombination sites. Upon inspection of the MinPD network it can be observed that most recombinant sequences and signals were detected at the 61(2) and 68(2) months - these seems to correlate with the X4 emergence. At 85(3) months there is another surge in recombination signals. The recombination signals are markedly stronger for the sequences with the X4 genotype than for those without which corresponds with the higher genetic diversity of those time periods [19].
7. Conclusions
In this paper we have described a new method to study the phylogenetic relationship of serially-sampled quasispecies and to visualize the relationships. We explained our decision to avoid multiple alignments in order to get better distance measures. We presented results of extensive computer simulations in which we mimic random sampling of sequences. We also discussed how to interpret the results of our algorithm in the context of viral disease progression and showed how to incorporate the information in the visualization of the tree. We studied a method to detect recombination in serially-sampled data and presented the results of simulation experiments. Our method is especially helpful in selecting putative recombinant sequences among a large set of sequences. At this point the selected sequences may be analyzed using another tool to find exact breakpoints and detect more than one crossover, but in the cases were the sequence length is short the presence of more than one crossovers is more seldom and our method will return good results. Applying our method on simulated recombinant data returned a 65% success rate and few false positives.
Several issues about the MinPD tree remain to be investigated. Is it possible to improve the recombination detection by a better choice of threshold values and/or distance rules, or by plugging in other recombination methods? The divergence tiebreaker was developed for selecting one common ancestor between two or more possible ancestors with identical distances to the reference sequence. Can a tiebreaker be developed to choose from two or more potential donor sequences?
What similar trends will emerge when we apply the MinPD algorithm to all the nine patient data sets used in the analysis by Shankarappa et al.? The MinPD tree and underlying data invites a more thorough evaluation of the information contained in serially-sampled quasispecies data.
Acknowledgments
P.B. was supported by NIH/NIGMS R25GM61347. G.N. was supported in part by NIH Grant P01 DA15027-01.
Footnotes
This version of the paper is revised from the one that appears in the CSB proceeding. It contains an additional paragraph in section 5.
Contributor Information
Patricia Buendia, Email: pbuen001@cs.fiu.edu.
Giri Narasimhan, Email: giri@cs.fiu.edu.
References
- 1.Drummond A, Rodrigo AG. Reconstructing genealogies of serial samples under the assumption of a molecular clock using serial-sample UPGMA (sUPGMA) Molecular Biology and Evolution. 2000;17:1807–1815. doi: 10.1093/oxfordjournals.molbev.a026281. [DOI] [PubMed] [Google Scholar]
- 2.Eigen M. Viral Quasispecies. Scientific American. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 3.Flint SJ, Enquist LW, Krug RM. Principles of Virology. Washington: ASM Press; 2000. pp. 207–208. [Google Scholar]
- 4.Grassly N, Harvey P, Holmes E. Population dynamics of HIV-1 inferred from gene sequences. Genetics. 1999;151:427–438. doi: 10.1093/genetics/151.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maynard Smith J. Analyzing the Mosaic Structure of Genes. Journal of Molecular Evolution. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 6.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford Univ. Press; 2000. [Google Scholar]
- 7.Ogishima S, Ren F, Tanaka H. Reconstruction and Analysis of Within-host Longitudinal HIV-1 Evolution by a Distance-based Sequential-linking Algorithm. Chem-Bio Informatics Journal. 2001;1(2):73–83. [Google Scholar]
- 8.Posada D, Crandall K. The effect of recombination on the accuracy of phylogeny reconstruction. J Mol Evol. 2002;2002(54):396–402. doi: 10.1007/s00239-001-0034-9. [DOI] [PubMed] [Google Scholar]
- 9.Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A. 2001;98(24):13757–62. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinones-Mateu M, Gao Y, Ball S. In Vitro subtype recombinaints of Human Immunodeficiency Virus Type 1: Comparison to Recent and Circulating In Viva Recombinaint Forms. Journal Of Virology. 2002;76(19):9600–9613. doi: 10.1128/JVI.76.19.9600-9613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rambaut A. Estimating the rate of molecular evolution: Incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395–399. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- 12.Rambaut A, et al. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5(1):52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 13.Ren F, Ogishima S, Tanaka H. Longitudinal phylogenetic tree of within-host viral evolution from noncontemporaneous samples: a distance-based sequential-linking method. Gene. 2003;317(1–2):89–95. doi: 10.1016/s0378-1119(03)00656-5. [DOI] [PubMed] [Google Scholar]
- 14.Ren F, Ogishima S, Tanaka H. A New Algorithm for Analysis of Within-Host HIV-1 Evolution. Pacific Symposium on Biocomputing; 2001; Hawaii. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigo A, Steel M. DIMACS Working Group on Phylogenetic Trees and Rapidly Evolving Diseases. 2004 http://dimacs.rutgers.edu/Workshops/WGPhylogeneticTrees/announcement.html.
- 16.Salminen M, et al. Identification of recombination breakpoints in HIV-1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 17.Schierup M, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–891. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schierup M, Hein J. Recombination And the molecular clock. Mol Biol Evol. 2000;17:1578–1579. doi: 10.1093/oxfordjournals.molbev.a026256. [DOI] [PubMed] [Google Scholar]
- 19.Shankarappa R, et al. Consistent Viral Evolutionary Changes Associated with the Progression of HIV 1 Infection. Journal of Virology. 1999;73(12):10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strimmer K, et al. A novel exploratory method for visual recombination detection. Genome Biology. 2003 doi: 10.1186/gb-2003-4-5-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swofford DL, et al. Phylogenic Inference. In: Mable BK, editor. Molecular Systematics. Sinauer &Associates; Sunderland, MA: 1996. pp. 407–514. [Google Scholar]
- 22.Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Mol Biol Evol. 2003;20(8):1318–25. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
- 23.Wiuf C, Christensen T, Hein J. A simulation study of the reliability of recombination detection methods. Mol Biol Evol. 2001;18:1929–1939. doi: 10.1093/oxfordjournals.molbev.a003733. [DOI] [PubMed] [Google Scholar]