Abstract
Cyanobacteria require efficient protein-quality-control mechanisms to survive under dynamic, often stressful, environmental conditions. It was reported that three serine proteases, HtrA (high temperature requirement A), HhoA (HtrA homologue A) and HhoB (HtrA homologue B), are important for survival of Synechocystis sp. PCC 6803 under high light and temperature stresses and might have redundant physiological functions. In the present paper, we show that all three proteases can degrade unfolded model substrates, but differ with respect to cleavage sites, temperature and pH optima. For recombinant HhoA, and to a lesser extent for HtrA, we observed an interesting shift in the pH optimum from slightly acidic to alkaline in the presence of Mg2+ and Ca2+ ions. All three proteases formed different homo-oligomeric complexes with and without substrate, implying mechanistic differences in comparison with each other and with the well-studied Escherichia coli orthologues DegP (degradation of periplasmic proteins P) and DegS. Deletion of the PDZ domain decreased, but did not abolish, the proteolytic activity of all three proteases, and prevented substrate-induced formation of complexes higher than trimers by HtrA and HhoA. In summary, biochemical characterization of HtrA, HhoA and HhoB lays the foundation for a better understanding of their overlapping, but not completely redundant, stress-resistance functions in Synechocystis sp. PCC 6803.
Keywords: biochemical characterization, oligomerization, PDZ domain, serine protease, substrate specificity, Synechocystis sp. PCC 6803
Abbreviations: Deg, degradation of periplasmic proteins; DTT, dithiothreitol; HtrA, high temperature requirement A; Hho, HtrA homologue; LB, Lysogeny Broth; LB/Amp100, LB minimal medium containing 100 μg·mg−1 ampicillin; ROS, reactive oxygen species
INTRODUCTION
The comparatively self-sufficient lifestyle of photosynthetic organisms comes at a price: a high inherent risk of damage to proteins and pigments by ROS (reactive oxygen species) that are formed as by-products of oxygenic photosynthesis, particularly under unfavourable environmental conditions [1]. In the natural environment, cyanobacteria such as Synechocystis sp. PCC 6803 (hereafter called Synechocystis 6803), which was isolated from a freshwater lake, are exposed to tremendous changes in light intensities and temperatures. Under these conditions, formation of ROS and subsequent damage to proteins is increased [2]. To cope with this problem, Synechocystis 6803 contains a variety of chaperones and proteases that monitor proper folding and function of proteins [3]. Particularly interesting candidates are enzymes of the multifunctional Deg (degradation of periplasmic proteins)/HtrA (high temperature requirement A) family of ATP-independent serine endopeptidases.
Deg/HtrA proteases are defined by the combination of a catalytic domain of the S1B serine proteases, containing His-Asp-Ser as a catalytic triad, with one or more C-terminal PDZ domains [4]. The PDZ domains mediate specific protein–protein interactions by preferentially binding to the C-terminal three or four amino acid residues of the target proteins [5–7]. In Escherichia coli, DegP/HtrA is a major heat-shock protein that acts both as a chaperone and as an unspecific housekeeping protease on denatured proteins [8], whereas its membrane-anchored paralogue DegS/HhoB (HtrA homologue B) acts as a sensor for folding stress in the periplasm and initiates the σE stress response [9–11]. Synechocystis 6803 has three genes encoding Deg/HtrA proteases, named htrA/degP (Cyanobase locus-tag accession number slr1204), hhoA/degQ (Cyanobase locus-tag accession number sll1679) and hhoB/degS (Cyanobase locus-tag accession number sll1427) after their E. coli orthologues [12]. However, phylogenetic analysis showed that the three Deg/HtrA paralogues of Synechocystis 6803 are more closely related to each other than to the E. coli orthologues carrying the same name and vice versa [13]. In order to avoid confusion and prevent assumptions purely based on the name, we will hereafter use the designations HtrA, HhoA and HhoB for the Synechocystis 6803 Deg/HtrA proteases, and DegP, DegQ and DegS for the E. coli paralogues.
Crystallographic studies showed that the basic functional unit of Deg/HtrA proteases is a homotrimer that is stabilized by residues of the protease domains [11,14,15]. The E. coli chaperone/protease DegP further stacks two of the trimers with the catalytic centres facing each other and thus forms a very compact, proteolytically inactive, hexameric complex [14]. DegP is activated through reversible conformational changes triggered by binding of peptides to the first of its two PDZ domains [6,16–19] or by a shift to higher temperature [8]. In solution, activated DegP forms large spherical 12- or 24-mers, composed of four or eight homotrimers, enclosing the substrates [20,21]. On lipid membranes, E. coli DegP/HtrA forms bowl-shaped structures with 4-, 5-, or 6-fold symmetry with a trimer as the basic structural unit [22]. In contrast, the E. coli stress sensor DegS is a trimer that is anchored in the plasma membrane by its N-terminal transmembrane domain, exposing the protease and PDZ domains to the periplasm in a proteolytically inactive conformation [11]. Binding of activating peptides, such as the C-termini of unfolded outer membrane porins, to the PDZ domains triggers conformational changes that activate DegS [5,7,10].
In comparison with the detailed studies on E. coli Deg/HtrA proteases, little is known about the physiological function and biochemical mechanism of Synechocystis 6803 orthologues. Bioinformatic analysis of the primary sequences of HtrA, HhoA and HhoB showed that all three proteins contain only a single C-terminal PDZ domain [13,23,24], predicted secretory signal peptides of 34 and 25 residues for HhoA and HhoB respectively, and an N-terminal transmembrane-spanning domain of HtrA [23]. Biochemical analysis of the recombinant HhoA (rHhoA) suggested that, similar to E. coli DegP [25], this protease also functions as a general housekeeping enzyme in the removal of misfolded proteins.
Earlier studies investigating the physiological function of cyanobacterial Deg/HtrA proteases focused mainly on their proposed role in the selective degradation of the D1 protein from photosystem II, which is particularly susceptible to photo-oxidative damage and quickly turned over under high-intensity light stress (reviewed in [26–28]). Analyses of a Synechocystis 6803 ΔhtrA/hhoA/hhoB triple mutant showed that these proteases are not essential for the degradation of the D1 protein when exposed to high-intensity light [29] or UV-B radiation [30]. Nevertheless, Deg/HtrA proteases play vitally important roles in the resistance to heat and high-intensity-light stresses [3,29,31]. The ΔhtrA/hhoA/hhoB triple mutant accumulated more oxidatively damaged proteins than the did the wild-type, did not grow at high-intensity light or high temperature and showed severe deficits in phototaxis [29]. This study also suggested an astonishing overlap in function of Deg/HtrA proteases, because these severe phenotypes were only observed for the ΔhtrA/hhoA/hhoB triple mutant, but not for any of the double-deletion mutants. This is particularly puzzling, because proteomic analyses found the Synechocystis 6803 Deg/HtrA proteases in different subcellular compartments. HtrA was found in the outer membrane [32], and HhoA in the periplasm [33] and associated with the plasma membrane [34], but no experimental results for the predicted periplasmic localization of HhoB have been reported.
In the present study, we have expressed all three proteases as soluble recombinant enzymes (rHtrA, rHhoA and rHhoB) in active and inactive variants in order to explore similarities and differences in their enzymatic properties. We show that rHtrA, rHhoA and rHhoB can degrade unfolded model substrates, but with different preferences regarding cleavage site, pH and temperature. Furthermore, accumulation of distinct degradation intermediates and formation of different oligomeric complexes indicate diverging molecular mechanisms, both between the three cyanobacterial Deg/HtrA proteases as well as in comparison with the well-studied E. coli orthologues.
EXPERIMENTAL
Strains and culture conditions
A glucose-tolerant wild-type strain of Synechocystis sp. PCC 6803 was grown at a light intensity of 50 μmol of photons·m−2·s−1 at 30°C in liquid BG-11 medium [35] buffered with 10 mM Tes, pH 8.2.
Plasmid construction
Genomic DNA was isolated from Synechocystis 6803 using a kit (Roche Diagnostics). Cloning of recombinant HhoA constructs has been described previously [25]. The sequences encoding N-terminally truncated active HtrA and HhoB protease constructs (Figure 1A) were amplified by PCR from genomic DNA with gene-specific primers (Operon Biotechnologies) and ligated into the pET151-D/TOPO expression vector using the Champion™ directional cloning kit (Invitrogen) using the method in the manufacturer's instructions. The following primers were used: for rHtrA, 5′-CACCTCCATTGCTCCTGCCAATGAG-3′ and 5′-GCATTATTGGGGAGGGGCGCTGG-3′; and for rHhoB, 5′-CACCAGTCTGACTCCTGCCCCGGT-3′ and 5′-G-CATTATTGACCCAAATCTTCGGGA-3′. Plasmids encoding proteolytically inactive variants were obtained by changing the codons for the active-site serine residues to alanine with the QuikChange II point mutagenesis kit (Stratagene). The codon for Ser296 of rHtrA was mutated to alanine (rHtrASA) using the primer pair 5′-CATTAACCCTGGTAACTCTGGTGGTC-3′ and 5′-CACCAGCGTTACCCGGGTTAATGGCC-3′, which also introduce a silent point mutation that generates an additional SmaI restriction site. The codon for Ser258 of rHhoB was mutated to alanine (rHhoBSA) using the primers 5′-GGCAATGCCGGTGGGCCCCTGCTC-3′ and 5′-CAGGG-GCCCACCGGCATTGCCTGG-3′, which also introduces an ApaI restriction site through a silent point mutation. Active and inactive PDZ domain deletion constructs were obtained by the same PCR-based cloning strategy, using the plasmids encoding active or inactive HtrA and HhoB as templates. The following primers were used: for rHtrASAΔPDZ, 5′-CACCTCCATTG-CTCCTGCCAATGAG-3′ and 5′-GCTCATTTGCCCGTAGCA-ATTAACTG-3′; and for rHhoBΔPDZ and rHhoBSAΔPDZ, 5′-CACCAGTCTGACTCCTGCCCCGGT-3′ and 5′-GCTCACA-TTTTGCCCTTGGTAAACAG-3′. All inserted sequences and orientation in the plasmids were confirmed by DNA sequencing (GATC Biotech).
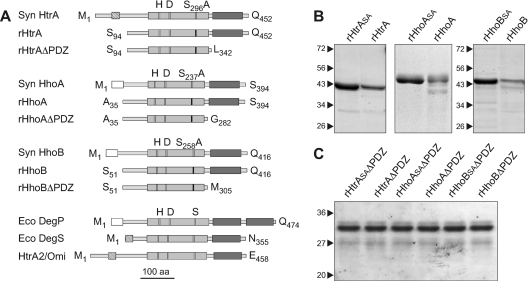
Figure 1. Purification of recombinant active and inactive variants of Synechocystis 6803 Deg/HtrA proteases.
(A) Schematic representations of rHtrA, rHhoA and rHhoB constructs generated for the present study, E. coli DegP and DegS and human HtrA2/Omi. Hatched boxes mark putative transmembrane domains, white boxes show predicted signal peptides, light grey boxes indicate protease domains with the positions of catalytic histidine and and aspartic acid marked in white and the position of a catalytic serine residue mutated to alanine in inactive variants indicated in black, and dark grey boxes mark PDZ domains. Proteins are drawn to scale, with a length of 100 amino acid residues indicated. (B) Coomassie-Blue-stained SDS/PAGE gels showing the elution fractions after Ni2+-affinity chromatography, containing recombinant active rHtrA, rHhoA and rHhoB, and inactive rHtrASA, rHhoASA and rHhoBSA variants of proteases. Molecular masses in kDa are indicated on the left-hand side. (C) Coomassie-Blue-stained SDS/PAGE gels showing the elution fractions after Ni2+-affinity chromatography, containing recombinant active rHtrAΔPDZ, rHhoAΔPDZ and rHhoBΔPDZ, and inactive rHtrASAΔPDZ, rHhoASAΔPDZ and rHhoBSAΔPDZ variants of proteases lacking PDZ domains.
Expression and purification of recombinant protein
Chemically competent E. coli BL21 Star™ (DE3) cells (Invitrogen) transformed with expression plasmids were grown in 50–100 ml of LB/Amp100 [LB (Lysogeny Broth) minimal medium containing 100 μg·mg−1 ampicillin] at 37°C overnight. These pre-cultures were harvested by centrifugation (3000 g, 8 min, 4°C), resuspended and further grown in 1–2 litres of LB/Amp100 at 19°C to a D600 of 0.4–0.6. Expression of recombinant protease constructs was induced by the addition of 0.1 mM IPTG (isopropyl β-D-thiogalactopyranoside). Cultures were further grown overnight at 19°C and then harvested by centrifugation (5000 g, 10 min, 4°C). Cell pellets were resuspended in 10–20 ml of binding buffer (50 mM Hepes/NaOH, pH 8.0, and 300 mM NaCl) and stored at −20°C.
E. coli cells were lysed on ice by 10–15 repeats of 10 s of sonication (Heidolph Instruments) with a 20 s cooling interval and then centrifuged (26000 g, 1 h, 4°C). For protease-activity assays with β-casein and complex-formation assays, recombinant protease constructs were purified from the soluble cell lysate by Ni2+-affinity chromatography with a HisTrap™ column connected to an ÄKTA purifier FPLC system (GE Healthcare) as described previously [25]. For all protease-activity and reconstitution assays, recombinant protease constructs were purified at 5°C from filtered soluble cell lysates (45 μm filter) using His GraviTrap™ affinity columns (GE Healthcare) as described in the manufacturer's instructions. Application of cell lysate in binding buffer was followed by a washing step with 10 ml of binding buffer containing 60 mM imidazole and an elution step with 5 ml of elution buffer (50 mM Hepes/NaOH, pH 8.0, 300 mM NaCl and 500 mM imidazole). The eluate was collected in 1 ml fractions, then diluted 10-fold with distilled water and concentrated using Amicon Ultra centrifugal filter devices with a molecular-mass cut-off of 10 kDa (Amicon) in order to decrease the imidazole concentration and the buffer strength. Protein concentrations were determined by a Bradford protein assay (Bio-Rad Laboratories) with BSA solutions as standards.
Analysis of protease complex formation
Elution fractions containing approximately 200 μg of purified Deg/HtrA protease constructs were incubated either alone or in a 1:3, 1:1 or 3:1 molar ratio with β-casein (Sigma-Aldrich) as a model substrate for 10 min at room temperature (23°C). Complex sizes were estimated by size-exclusion chromatography using a pre-packed Superdex™ 200 column (GE Healthcare). The column was repeatedly calibrated with high- and mid-mass molecular-mass markers (GE Healthcare) as described in the manufacturer's instructions.
Protease-activity assays
For protease-activity assays, purified protease constructs were incubated with an excess of β-casein, BSA or lysozyme in 250 mM Tris/HCl, pH 7.0, supplemented with 20 mM CaCl2 for 2 h at 40°C. Unfolding of lysozyme was induced by incubation with 20 mM DTT (dithiothreitol) for 3 h at 40°C prior to protease-activity assays.
Fluorigenic protease-activity assays were performed using the Enzchek Protease Assay (Invitrogen). For each reaction, 3 μg of purified recombinant protease was incubated with 0.5 μg of BODIPY TR-X casein in a total volume of 200 μl in the presence of 0.1 mM NaN3. To measure pH optima, 10 mM Mes (up to pH 6.5) or Hepes (from pH 7.0) were used to buffer the solutions and the measurements were performed at 30°C. Fluorescence signals measured at a wavelength of 617 nm were followed with an Infinite® 200 microplate reader (Tecan) using 589 nm as λex. For kinetic measurements at different pH values, a fluorescence-signal increase was followed over time, recording six replicates every 10 min, while incubating the sample plate in darkness at 30°C inside the microplate reader. The activity was calculated from the initial rate of fluorescence emission. For determination of the temperature dependence, assays were incubated in 10 mM Mes and at the optimal pH of the three Deg/HtrA proteases (pH 5.5 for rHtrA, and pH 6.5 for rHhoA and rHhoB) at a temperature range from 10°C to 65°C in a PCR cycler before measuring total fluorescence at room temperature after 40 min of incubation.
Sequencing of proteolytic β-casein fragments
To identify β-casein cleavage sites, 200 pmol of purified recombinant proteases (active, or inactive variants as negative controls) were incubated in the presence of 10 mM CaCl2 with 75 μg of β-casein at 40°C for several hours. Samples were taken after 0, 30 and 90 min and 10 h and analysed by SDS/PAGE (see below). Proteolytic fragments of cleaved β-casein were excised from the gel and sent for N- and C-terminal sequencing to the Protein Analysis Centre (PAC, Karolinska Institute, Stockholm, Sweden).
Protein analysis
Proteins were solubilized in LDS (lithium dodecyl sulphate) buffer and separated by SDS/PAGE as described previously [36]. Gels were stained with Coomassie Brilliant Blue R250 [37].
Bioinformatics
Sequence analysis was performed using SMART [38], TargetP [39] and SignalP [40] programs. For engineering of mature soluble protease constructs, the secondary structures of HhoA, HhoB and HtrA were analysed with PsiPred v. 2.5 [41] and compared with published structures of E. coli DegP [14], DegS [11] and human HtrA2/Omi [15] and sequence alignments [4,23].
RESULTS AND DISCUSSION
Engineering and heterologous expression of Deg/HtrA protease constructs of Synechocystis 6803
To investigate the enzymatic properties of three cyanobacterial Deg/HtrA proteases in detail, we expressed rHtrA, rHhoA and rHhoB in their active form as recombinant fusion proteins with a C-terminal His6-tag using the E. coli expression system. For controls, inactive versions of the proteases with mutations in the catalytic site were generated by site-directed point mutagenesis, expressed and purified under similar conditions. On the basis of previous studies of membrane-anchored human HtrA2 [15] and E. coli DegS [10,11], the putative transmembrane domain of HtrA from Synechocystis 6803 was truncated after the residue Gln93 (Figure 1A and Supplementary Figure S1 at http://www.BiochemJ.org/bj/435/bj4350733add.htm). Both active rHtrA, as well as inactive rHtrASA, with Ser296 replaced by alanine, were obtained in sufficient quantity and purity after purification by Ni2+-affinity chromatography (Figure 1B). rHhoA and its proteolytically inactive version rHhoASA, both lacking the predicted signal peptide, were expressed and purified as described previously [25] (Figures 1A and 1B). HhoB contains a predicted N-terminal signal sequence of 25 residues and a long additional N-terminal domain of 56 residues (compared with only 23 residues in HhoA) between the signal peptide and the protease domain. On the basis of our experience with rHhoA, we first tried to express rHhoB lacking only the predicted signal sequence. However, this construct could not be expressed as a soluble protein and its yield was extremely low (results not shown). Therefore we further truncated HhoB after Gln50, placing the introduced His6-tag at approximately the same distance from the protease domain as in rHhoA. A third rHhoB construct was truncated after Gln74, close to the predicted first helix of the protease domain (Supplementary Figure S1). Of these, only the rHhoB construct truncated after Gln50 yielded low amounts of active enzyme (Figure 1B; hereafter rHhoB). Also the inactive version rHhoBSA, with Ser258 replaced by alanine, was obtained with a similar yield and purity (Figure 1B).
To investigate the role of the single PDZ domain in each of the three Synechocystis 6803 Deg/HtrA proteases, we further created PDZ-domain deletion constructs by truncation within the predicted loops connecting the protease and PDZ domains (Figure 1A and Supplementary Figure S1). rHtrA and rHtrASA were truncated after Lys342 to yield rHtrAΔPDZ and rHtrASAΔPDZ respectively. Cloning and expression of rHhoAΔPDZ and its proteolytically inactive variant rHhoASAΔPDZ, both truncated after Gly282, were expressed and purified as described previously [25] (Figures 1A and 1C). Similarly, rHhoB and rHhoBSA were truncated after Met305, resulting in rHhoBΔPDZ and rHhoBSAΔPDZ respectively. All deletion constructs were obtained in good quantity and reasonable purity after Ni2+-affinity chromatography (Figure 1C).
All three Deg/HtrA proteases in Synechocystis 6803 are protein-quality-control factors
We assayed the activity and substrate specificity of the recombinant Deg/HtrA constructs using model substrates that differed with regard to their folding states, such as bovine β-casein, BSA and lysozyme. All three protease constructs with an intact PDZ domain, rHtrA, rHhoA and rHhoB, were able to cleave the naturally unfolded β-casein, but not correctly folded globular BSA or lysozyme (Figure 2A). Reduction of lysozyme with DTT, which induces partial unfolding by breaking disulfide bonds, allowed its cleavage by HhoA and to a lesser extent by rHtrA, but not by HhoB (Figure 2A, lower panel). Under the chosen conditions, equal amounts of rHtrA, rHhoA and rHhoB showed very different activities: little uncleaved β-casein remained in assays with rHtrA, and a prominent degradation fragment appeared; rHhoA completely degraded the excess of β-casein; whereas rHhoB showed only little activity and generated a prominent degradation fragment with a slightly lower molecular mass than intact β-casein (Figure 2A). All substrates tested were not degraded in the presence of inactive protease variants, thus excluding that the substrate degradation by active variants was mediated by impurities in the preparation (Figure 2A).
Figure 2. Proteolytic activity of Synechocystis 6803 Deg/HtrA proteases against substrates with different folding states.
Naturally unfolded β-casein, globular folded BSA, folded lysozyme and partially denatured reduced lysozyme were used as model substrates. (A) Protease constructs with intact PDZ domains. (B) Protease constructs without PDZ domains, e.g. C-terminally truncated after the protease domain.
We consistently assayed a lower activity for rHhoB than for rHtrA and rHhoA. We cannot exclude that this is merely caused by unfortunate engineering of the rHhoB truncation constructs, but the chosen truncation of HhoB at Gln50 should have affected neither the fold of the protease domain nor any other important domain [24]. Instead, this truncation made rHhoB more similar to rHhoA, which is the most active recombinant protease in our hands. Thus the low activity of rHhoB might also reflect, similarly to E. coli DegS [9], a more stringent substrate specificity, a function as a chaperone rather than a protease, a requirement for as yet unidentified cofactors, such as allosteric activators [17–19], or a need for the formation of a hetero-oligomeric complex. The formation of a hetero-hexameric complex was reported for the chloroplast-located Deg5 and Deg8 in Arabidopsis thaliana [42].
Taken together, this provides evidence that all three proteases, HtrA, HhoA and HhoB, might act as protein-quality-control factors degrading denatured and damaged proteins [29]. This further suggests remarkable mechanistic differences to Deg/HtrA proteases in E. coli, where DegP and DegQ with two PDZ domains [17,43] act as quality-control factors, whereas DegS, containing a transmembrane domain and a single PDZ domain, acts as a stress sensor [11,17].
Deletion of the PDZ domain severely impairs proteolytic activity
rHtrAΔPDZ showed only residual activity against β-casein and reduced lysozyme (Figure 2B). In addition, rHhoAΔPDZ degraded both unfolded substrates less efficiently than its variant with the intact PDZ domain, and larger degradation fragments accumulated after proteolysis of reduced lysozyme (Figure 2B). rHhoBΔPDZ showed only minute activity against β-casein and no activity was detected against reduced lysozyme (Figure 2B). As expected, no degradation of substrates tested was observed in the presence of inactive protease variants lacking PDZ domains (Figure 2B). Taken together, these results suggest that the PDZ domains are not absolute requirements for proteolytic activity of all three proteases, but they are important for the regu-lation of proteolytic activity and/or for presenting the substrate to the protease domain.
rHtrA, rHhoA and rHhoB generate distinct β-casein degradation products
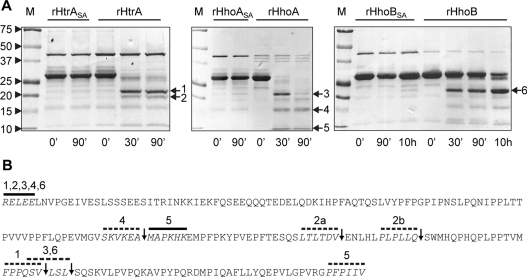
Proteolysis of the model substrate β-casein by recombinant Deg/HtrA proteases with intact PDZ domains was further investigated in more detail, and different enzymatic kinetics and distinct degradation intermediates were observed (Figure 3A). In the presence of rHtrA, only traces of β-casein could be detected after 30 min of incubation, but proteolytic fragments 1 and 2 remained stable and were not further degraded during 90 min of incubation (Figure 3A, left-hand panel). Similarly, most of the β-casein was cleaved by rHhoA within 30 min, and the generated proteolytic fragment 3, but not fragments 4 and 5, disappeared within 90 min (Figure 3A, middle panel). Longer incubation times lead to a complete degradation of all fragments (Figure 2A, [25]). In contrast, a very low proteolytic activity was detected for rHhoB. Significant amounts of β-casein were detected even after 10 h of incubation with this protease and proteolytic fragment 6 accumulated progressively with incubation time (Figure 3A, right-hand panel). This suggests that Synechocystis 6803 Deg/HtrA proteases release their substrates after defined cleavage sites, similar to E. coli DegS [10] and A. thaliana Deg2 [44] and Deg15 [45,46]. This is a remarkable contrast with E. coli DegP, which digests unfolded substrate proteins progressively without accumulation of intermediates [6,21].
Figure 3. Cleavage site specificity of Synechocystis 6803 Deg/HtrA proteases.
(A) Degradation of β-casein by recombinant rHtrA, rHhoA and rHhoB, analysed by Coomassie-Blue-stained SDS/PAGE. M indicates the molecular-mass protein marker; molecular mass in kDa is indicated on the left-hand side. Arrows with numbers highlight bands that were excised from the gel for N- and C-terminal protein sequencing. Proteolytic assays using rHtrA were performed in 50 mM Mes, pH 5.5, and 20 mM CaCl2; assays with rHhoA and rHhoB were carried out in 50 mM Tris/HCl, pH 7.0, and 20 mM CaCl2. Representatives of at least three replicates are shown. (B) Amino acid sequence of mature β-casein. Black bars show residues identified by N-terminal sequencing of fragments excised from gels in (A), broken bars indicate residues identified by C-terminal sequencing and arrows indicate deducted cleavage sites.
Cleavage-site specificity
The cleavage specificity of proteases is defined by the amino acid composition of the substrate around the cleavage site, with non-prime-site residues P3, P2 and P1 preceding the cleavage site and prime-site residues P1′, P2′ and P3′ following the cleavage site [47]. To gain insight into the cleavage-site specificity of rHtrA, rHhoA and rHhoB, we analysed the most abundant proteolytic β-casein fragments generated by each protease by N- and C-terminal sequencing. The majority of the sequenced fragments contained the N-terminus of mature β-casein, whereas cleavage occurred at the C-terminal part of this substrate (Figure 3B and Table 1). Three C-terminal cleavage sites, Val162, Gln141 and Val130, were identified for rHtrA (Figure 3B and Table 1). A common cleavage site at Leu165 was found for rHhoA and rHhoB; however, rHhoA additionally cleaved β-casein at Ala101 generating two proteolytic fragments, the N-terminal 1–101 as well as the C-terminal 102–199 amino acid fragments (Figure 3B and Table 1).
Table 1. Cleavage sites within β-casein for rHtrA, rHhoA and rHhoB assayed by N- and C-terminal sequencing.
The mature secreted version of β-casein (processed at the N-terminal Ala15) was used for the studies of the generated fragments (N1–199 amino acids).
| Generated fragments | Band number in the gel | ||
|---|---|---|---|
| Protease | (amino acids) | Cleavage site | (see Figure 3 in the main paper) |
| rHtrA | N1–162 | FPPQSV↓LSL | 1 |
| N1–130 | LTLTDV↓ENL | 2a | |
| N1–141 | PLPLLQ↓SWM | 2b | |
| rHhoA | N1–165 | QSVLSL↓SQS | 3 |
| N1–101 | SKVKEA↓MAP | 4 | |
| N102–199 | KEA↓MAPKHK | 5 | |
| rHhoB | N1–165 | QSVLSL↓SQS | 6 |
E. coli DegP showed a strong preference for small hydrophobic residues (valine, alanine, threonine and isoleucine) in the P1 position, whereas the P2 and P3 positions are less defined [6]. Our results show a similar trend for rHtrA and rHhoA (Figure 2B). It was further demonstrated that PDZ1, the first of the two PDZ domains of E. coli DegP, recognized the hydrophobic C-terminus of substrates and presented the substrate for cleavage [6]. Since DegP cleavage occurred at a hydrophobic site approximately 12–16 amino acids further towards the N-terminus, the resulting cleavage fragment could again be bound by a PDZ1 domain of the trimer, resulting in processive cleavage of the substrate into peptides of 10–20 residues [6]. Similarly, the PDZ domains of Synechocystis 6803 Deg/HtrA proteases might preferentially bind the hydrophobic residues at the C-terminus of β-casein (–PFPIIV) and present the substrate to the catalytic site for cleavage, explaining the absence of larger C-terminal fragments. However, all three Synechocystis Deg/HtrA proteases are not as processive as E. coli DegP, possibly due to the lack of a second PDZ domain.
Inactive Synechocystis Deg/HtrA proteases form different oligomers
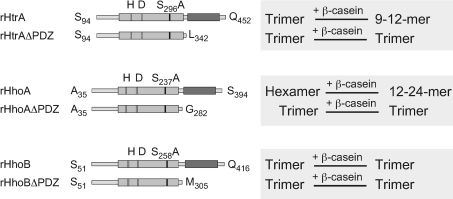
Earlier structural studies have shown that the basic unit of Deg/HtrA proteases is a homotrimer [11,14,15]. Two trimers of E. coli DegP are stacked into a cage-like hexamer by both PDZ domains of the subunits [14,48,49]. Homology modellling predicted all three Synechocystis 6803 Deg/HtrA proteases to form trimers [23]. Surprisingly, we found previously that rHhoA also assembles into a hexamer in solution [25], despite the fact that it only contains one PDZ domain. In the present study, we assayed the oligomerization state of rHtrASA and rHhoBSA by size-exclusion chromatography. rHtrASA eluted with a volume of 13.2 ml, corresponding to an apparent molecular mass of 140 kDa (Figure 4A), which is consistent with trimer formation. Published crystal structures show that Deg/HtrA protease trimers from Homo sapiens and E. coli are disc-shaped and wider than tall if viewed from the side [11,15]. Therefore, such trimers have a larger hydrodynamic radius than globular proteins of the same molecular mass, resulting in a larger-than-expected apparent molecular mass when estimated by the elution volume in size-exclusion chromatography calibrated with globular standard marker proteins. However, the rHtrASA complexes were unstable, and we frequently observed monomers eluting at approximately 14.6 ml (apparent molecular mass of ~60 kDa). In agreement with our previous experiments [25], rHhoASA formed a hexamer eluting with a volume of 11.5 ml (apparent molecular mass 320 kDa) (Figure 4B, [22]). rHhoBSA eluted at 13.1 ml, indicating a trimeric complex with an apparent molecular mass of approximately 160 kDa (Figure 4C). For rHtrASA and rHhoBSA, additional peaks were observed at the void volume of the Superdex™ 200 size-exclusion columns, but very little or no protein was detected in these fractions after SDS/PAGE and Coomassie Blue staining (Figures 4A and 4C, lower panels), indicating that these peaks might represent SDS-insoluble aggregates.
Figure 4. Complex formation by inactive variants of Synechocystis 6803 Deg/HtrA proteases in the absence and the presence of substrate.
Size-exclusion chromatography of (A) rHtrASA (B) rHhoASA and (C) rHhoBSA. Black lines show elution profiles obtained in the absence of substrate, and grey lines show profiles after pre-incubation with β-casein in a 3:1 molar ratio of protease/substrate (A and B) or a 1:3 ratio (C). Lower panels show aliquots of the size-exclusion chromatography fractions separated by SDS/PAGE and visualized by Coomassie-Blue staining. The exclusion volume (V0) of the columns and the elution volumes of selected marker proteins are indicated with black arrowheads. Ve, elution volume.
Substrate binding to rHtrASA and rHhoASA changes their oligomerization states
In the presence of substrates, E. coli DegP massively rearranges and assembles several trimers into large oligomeric complexes, mediated by interactions of both PDZ domains [16,20,21]. To test the effect of substrate binding on the oligomerization state of cyanobacterial Deg/HtrA proteases, we incubated rHtrASA, rHhoASA and rHhoBSA with β-casein as a model substrate prior to size-exclusion chromatography. In the presence of β-casein, the elution diagram of rHtrASA displayed an additional peak at ~10.5 ml, indicating the formation of a complex with an apparent molecular mass of approximately 580 kDa (Figure 4A). β-casein alone elutes at 13.5 ml (results not shown; compare with Figure 4C), but co-elutes with rHtrASA in the 10.5 ml peak, clearly demonstrating that this complex is the result of β-casein binding by rHtrASA (Figure 4A). As for rHtrASA alone, an additional prominent peak is detected at the void volume of the column, but little protein is observed in the sample aliquot of this fraction after SDS/PAGE analysis (Figure 4A). Owing to steric constraints, the formation of such higher-order oligomeric HtrA–substrate complexes appears unlikely under physiological conditions, since HtrA is anchored in the outer membrane [30]. This suggests that HtrA may function similarly to human HtrA2/Omi, which is anchored as an inactive zymogen to the inner mitochondrial membrane [48]. A soluble active form of HtrA2/Omi is released by cleavage of the N-terminal 133 amino acids that contain the transmembrane anchor (Figure 1A). This soluble form translocates from the mitochondria to the cytosol and contributes to apoptosis [50]. A similar release of the cyanobacterial HtrA protease from the outer membrane would also help to explain the overlap in its function with HhoA and HhoB under certain conditions.
Similar to rHtrASA, rHhoASA also changed its complex size dramatically in the presence of β-casein, featuring prominent additional peaks at 8.3 ml and at 9.5 ml (Figure 4B). Both rHhoASA and β-casein co-eluted in the peak at 9.5 ml, indicating the formation of a large rHhoASA–β-casein complex of approximately 670 kDa. Hardly any protein was observed in the fraction containing the early peak at 8.3 ml, which is close to the column void volume and might therefore contain insoluble aggregates. In contrast, the elution profile of rHhoBSA in the presence of β-casein showed no additional peaks and the elution profiles of rHhoBSA and β-casein alone were very similar, indicating that these two proteins did not form a stable complex under the conditions tested (Figure 4C). This observation is consistent with the low activity of rHhoB observed in proteolytic assays (Figure 2A) and may indicate either a problem of the rHhoB constructs that impairs formation of higher-oligomeric complexes or a lack of required cofactors, such as specific peptides [16–19]. It was reported that binding of specific effector peptides to the PDZ1 of E. coli DegP in a resting conformation causes a local rearrangement that is allosterically transmitted to the substrate-binding pocket of the protease domain, and such an activated protease state assembles into larger oligomeric particles [18–21]. In addition, E. coli DegS is activated by the binding of allosteric effectors to the PDZ domain, but, in contrast with DegP, protease activation and substrate proteolysis are separate events [10,11].
Our present studies on cyanobacterial Deg/HtrA proteases indicate that the formation of large oligomeric complexes in the presence of a substrate might be a common feature for this enzyme family (Figure 4). Unfortunately, size-exclusion chromatography is not suitable for a reliable determination of the exact oligomerization state of rHtrASA–β-casein and rHhoASA–β-casein complexes, due to the unknown number of bound β-casein molecules and its unknown effect on the hydrodynamic radius of the complexes. However, in comparison with results published by Krojer et al. [21] and Jiang et al. [20], the formation of 12- or 24-meric complexes for rHhoASA, and 9–12-mers for rHtrASA, is estimated.
Deletion of PDZ domains abolishes formation of oligomers beyond a trimer
We had shown previously [25] that the deletion the PDZ domain of rHhoA prevents formation of a hexamer. Instead, rHhoAΔPDZ assembled into a trimer, which is consistent with the homology model that attributed trimerization to the N-terminus of the protease domain [23]. Similarly, rHtrAΔPDZ and rHhoBΔPDZ were detected as trimers (results not shown). However, the trimers of all three constructs were not very stable and monomers were frequently observed. The equilibrium of trimers and monomers appeared concentration dependent, with monomers prevailing at low protein concentrations. All three PDZ-domain deletion constructs, rHtrAΔPDZ, rHhoAΔPDZ and rHhoBΔPDZ, were not able to assemble into larger complexes in the presence of β-casein (summarized in Figure 5). This is in agreement with reports showing that peptide binding to the PDZ domain of E. coli DegP [16–18] promotes formation of higher-order particles.
Figure 5. Schematic overview of the oligomerization results.
Our present results demonstrated that, in contrast to trimeric rHhoB, trimeric rHtrA and hexameric rHhoA form higher-order oligomers in the presence of β-casein. Deletion of the PDZ domain locks all three proteases in the trimer state.
Mg2+ and Ca2+ increase activity of rHtrA, rHhoA and rHhoB
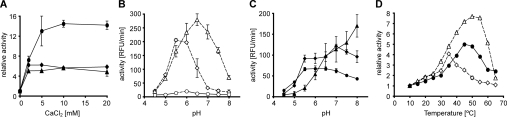
We characterized the proteolytic activity of rHtrA, rHhoA and rHhoB in biochemical terms using BODIPY TR-X-labelled casein as a fluorigenic substrate. The effect of increasing concentrations of divalent ions on the proteolytic activity of the three Deg/HtrA proteases was tested. The values were normalized to the protease activity without the addition of divalent cations to display the actual increase of activity in the presence of divalent ions. Although the activity of rHtrA and rHhoA increased almost 4–6-fold in the presence of 5 mM CaCl2 and remained constant at higher CaCl2 concentrations, the activity of rHhoB increased more gradually up to 14-fold, reaching the maximal level at 10 mM CaCl2 (Figure 6A). Similar results were obtained after the addition of MgCl2 (Supplementary Figure S2A at http://www.BiochemJ.org/bj/435/bj4350733add.htm). The reason for this activation by Mg2+ or Ca2+ is not clear at present, but either (i) divalent cations could interact with the substrate and make it more susceptible for degradation, and/or (ii) binding of divalent cations could affect the structure and thus activate the Deg/HtrA protease. The much stronger effect of Mg2+ or Ca2+ ions on rHhoB activity (Figure 6A) can be explained by the almost negligible activity in the absence of divalent ions (Figure 6B).
Figure 6. Biochemical properties of rHtrA, rHhoA and rHhoB assayed with a fluorigenic casein substrate.
In all panels, data points obtained for rHtrA are represented by diamonds, rHhoA by triangles and rHhoB by circles. Open symbols connected by broken lines indicate the absence of divalent ions, and filled symbols connected by continuous lines indicate the presence of divalent ions. Please note that detector-gain settings may differ between panels. Results are means±S.D. (n=3). (A) Effect of Ca2+ ion concentration on proteolytic activity. Activity was measured by the initial rate of fluorescence generation in 10 mM Mops buffer, pH 6.5, and normalized to the signal observed without CaCl2. (B) Effect of pH on protease activity in the absence of divalent ions. Activity was measured by the initial rate of fluorescence generation. RFU, relative fluorescence units. (C) Effect of pH on protease activity in the presence of 10 mM CaCl2. (D) Effect of temperature on protease activity as measured by the total fluorescence after 40 min, normalized to the fluorescence observed at 10°C.
Overlapping, but distinct, pH and temperature profiles of rHtrA, rHhoA and rHhoB
The pH-preference profiles were measured for all three Deg/HtrA proteases in the absence (Figure 6B) and the presence (Figure 6C) of CaCl2 at a range from pH 4.5 to 8.0. In the absence of CaCl2, rHtrA showed a maximal proteolytic activity between pH 5.5 and 6.0, whereas rHhoA had its optimum at pH 6.5 (Figure 6B). Almost no proteolytic activity was detected for rHhoB (Figure 6B). However, the addition of 10 mM CaCl2 resulted in strong changes in the pH optima, leading to their broadening and a shift towards a basic pH (Figure 6C). For rHtrA, the highest activity was detected at pH 7.0, whereas the activity of rHhoA progressively increased with higher pH (Figure 6C; please note that the fluorescence detector gain was decreased after the addition of divalent cations to avoid saturation). This striking shift in the pH optimum of HhoA in the presence of Ca2+ is in agreement with previous observations [25]. Similar pH-preference profiles were also observed after the addition of MgCl2, ruling out a Ca2+-specific effect (results not shown). The activity of rHhoB in the presence of Ca2+ was constantly high at pH values between 5.5 and 6.5 and decreased slightly at pH 6.5–8.0 (Figure 6C).
In the absence of Ca2+, rHtrA, rHhoA and rHhoB were all most active in slightly acidic conditions. In the presence of 10 mM Ca2+, both rHtrA and rHhoB showed a broader pH optimum, with a maximum between pH 5.5 and 7.0. In contrast, rHhoA showed a striking shift of its pH optimum to basic conditions. To our knowledge, such a strong shift in the pH preference has not been reported for any other protease.
The effect of temperature on the proteolytic activity of Deg/HtrA proteases was assessed by measuring total fluorescence at one fixed time point after 40 min of incubation at the indicated temperature (Figure 6D). To ensure that neither the substrate was limiting at the chosen time point nor saturation of the detector occurred at or near the fluorescence maxima, control experiments were performed in which the fluorescence emission generated by degradation of BODIPY TR-X-labelled casein was measured over time at 30°C (Supplementary Figure S2B). Only low proteolytic activity was recorded for the three active Deg/HtrA proteases at a temperature ranging from 10 to 25°C (Figure 6D). Whereas rHtrA had its temperature optimum between 30°C and 40°C, the activity of rHhoA increased with rising temperature, reaching its maximum at ~50–55°C, and decreased rapidly at higher temperatures, probably due to denaturation of the enzyme (Figure 6D). A similar temperature curve has also been observed previously for rHhoA using resorufin-labelled casein as substrate [25]. The temperature dependency of rHhoB was comparable with rHhoA; however, its optimum was shifted to slightly lower temperatures and the maximal activity was already reached at 45–50°C (Figure 6D).
Implications for the physiological function of HtrA, HhoA and HhoB in Synechocystis 6803
All three Synechocystis 6803 Deg/HtrA proteases were able to act on unfolded protein substrates, suggesting functions as general protein-quality-control proteases consistent with the stress-sensitive phenotype of the triple mutant ΔhtrA/hhoA/hhoB [29]. The overlapping biochemical properties of HtrA, HhoA and HhoB may allow any two of the enzymes to compensate for the lack of the third, which could explain the striking lack of stress-sensitive phenotypes of the double-deletion mutants in comparison with the triple mutant ΔhtrA/hhoA/hhoB [29]. However, our results also suggest that each of the three enzymes could contribute differently to the overall proteolytic potential with varying environmental conditions. For instance, Synechocystis 6803 cells prefer to grow at alkaline conditions [51] and are commonly grown in BG11 media, which contain approximately 0.5 mM of combined Mg2+ and Ca2+ ions. Thus HhoA would be the most active of the three Synechocystis 6803 Deg/HtrA proteases under optimal physiological conditions.
The observed activation of all three proteases by Ca2+ and Mg2+ ions and the slightly acidic pH optimum of HtrA and HhoB may be particularly important for an effective stress response to high-intensity light. Sufficiently high-intensity light saturates photosynthesis rates, causing acidification of the thylakoid lumen and increased formation of ROS [1]. This further leads to oxidative damage of photosynthetic protein complexes and release of Ca2+ from the thylakoid membranes to the lumen [26]. These conditions then would activate any of the three Deg/HtrA proteases. Since no reliable localization studies exist for Deg/HtrA proteases in cyanobacteria, it is not clear whether these enzymes are present in the periplasmic space, in the thylakoid lumen or in both compartments. Unfortunately, purification of the cyanobacterial thylakoid lumen proteome poses massive technical challenges [52] and thus neither a comprehensive proteomic analysis has been reported nor is it possible to probe this compartment for the presence of Deg/HtrA proteases with antibodies. However, three Deg/HtrA proteases have been detected in the thylakoid lumen of higher plants, where no other general proteases were reported [53,54]. Furthermore, out of the reported 16 paralogues of A. thaliana, the three cyanobacterial proteases show the highest similarity to the three thylakoid-lumen-located Deg/HtrA enzymes [24]. Hence it is very likely that at least one of the Synechocystis 6803 Deg/HtrA proteases, such as HhoB, is located in the thylakoid lumen.
The temperature profiles of rHtrA, rHhoA and rHhoB indicate how these enzymes may contribute to high-temperature resistance. Whereas rHtrA activity decreased at temperatures higher than 30°C, which is the preferred growth temperature of Synechocystis 6803, the activity of rHhoA and rHhoB increased to well above lethal temperatures. The soluble enzymes HhoA and HhoB should, therefore, play a more important role in protein quality control at high temperatures than HtrA. Interestingly, htrA is the only gene regulated by the two-component system Hik10 (histidine kinase 10)–Rre3 (response regulator 10) under salt and hyperosmotic stresses, which suggests an additional, more specific, function of HtrA [55].
Conclusions
In summary, the present family-wide biochemical characterization of the Synechocystis 6803 Deg/HtrA proteases suggests complementary physiological functions of the three enzymes and highlights considerable mechanistic differences of HtrA, HhoA and HhoB, as compared with their well-studied E. coli orthologues. We have shown that the three Deg/HtrA proteases of Synechocystis 6803, expressed as soluble recombinant His6-tagged fusion proteins, can act as protein-quality-control proteases. Distinct pH and temperature optima of the three enzymes and different cleavage sites and oligomerization statuses speak in favour of partially overlapping, but not completely redundant, physiological functions.
Online data
AUTHOR CONTRIBUTION
Pitter Huesgen engineered Deg/HtrA constructs, performed oligomerization studies together with Helder Miranda, Manuela Perthold and Holger Schuhmann, analysed the data and wrote the paper. Helder Miranda and XuanTam Lam expressed and purified Deg/HtrA constructs and performed proteolytic assays. Holger Schuhmann performed bioinformatic analysis. Christiane Funk and Iwona Adamska co-conceived the research and edited the manuscript prior to submission.
FUNDING
This work was supported by the Deutsche Forschungsgemeinschaft [grant numbers AD92/8-2 and AD92/8-3 (to I.A.)], the University of Konstanz (to I.A.), the Swedish Energy Agency [grant numbers 2009-000928 (to C.F.)], the Royal Swedish Academy of Sciences [grant number 312-3820-06 (to C.F.)], the Lawsky Foundation (to X.L.) and Umeå University (to C.F.).
References
- 1.Barber J., Andersson B. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem. Sci. 1992;17:61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- 2.Melis A. Photosystem-II damage and repair cycle in chloroplasts: what modulates the rate of photodamage? Trends Plant Sci. 1999;4:130–135. doi: 10.1016/s1360-1385(99)01387-4. [DOI] [PubMed] [Google Scholar]
- 3.Sokolenko A., Pojidaeva E., Zinchenko V., Panichkin V., Glaser V. M., Herrmann R. G., Shestakov S. V. The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 2002;41:291–310. doi: 10.1007/s00294-002-0309-8. [DOI] [PubMed] [Google Scholar]
- 4.Clausen T., Southan C., Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 5.Hasselblatt H., Kurzbauer R., Wilken C., Krojer T., Sawa J., Kurt J., Kirk R., Hasenbein S., Ehrmann M., Clausen T. Regulation of the σE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress. Genes Dev. 2007;21:2659–1670. doi: 10.1101/gad.445307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krojer T., Pangerl K., Kurt J., Sawa J., Stingl C., Mechtler K., Huber R., Ehrmann M., Clausen T. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohn J., Grant R. A., Sauer R. T. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 8.Spiess C., Beil A., Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 9.Alba B. M., Zhong H. J., Pelayo J. C., Gross C. A. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σE activity. Mol. Microbiol. 2001;40:1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- 10.Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 11.Wilken C., Kitzing K., Kurzbauer R., Ehrmann M., Clausen T. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S., et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 13.Huesgen P. F., Schuhmann H., Adamska I. The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol. Plant. 2005;123:413–420. [Google Scholar]
- 14.Krojer T., Garrido-Franco M., Huber R., Ehrmann M., Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 15.Li W., Srinivasula S. M., Chai J., Li P., Wu J. W., Zhang Z., Alnemri E. S., Shi Y. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 2002;9:436–441. doi: 10.1038/nsb795. [DOI] [PubMed] [Google Scholar]
- 16.Krojer T., Sawa J., Huber R., Clausen T. HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat. Struct. Mol. Biol. 2010;17:844–852. doi: 10.1038/nsmb.1840. [DOI] [PubMed] [Google Scholar]
- 17.Meltzer M., Hasenbein S., Mamant N., Merdanovic M., Poepsel S., Hauske P., Kaiser M., Huber R., Krojer T., Clausen T., et al. Structure, function and regulation of the conserved serine proteases DegP and DegS of Escherichia coli. Res. Microbiol. 2009;160:660–666. doi: 10.1016/j.resmic.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Merdanovic M, Mamant N., Meltzer M., Poepsel S., Auckenthaler A., Melgaard R., Hauske P., Nagel-Steger L., Clarke A. R., Kaiser M., et al. Determinants of structural and functional plasticity of a widely conserved protease chaperone complex. Nat. Struct. Mol. Biol. 2010;17:837–844. doi: 10.1038/nsmb.1839. [DOI] [PubMed] [Google Scholar]
- 19.Meltzer M., Hasenbein S., Hauske P., Kucz N., Merdanovic M., Grau S., Beil A., Jones D., Krojer T., Clausen T., et al. Allosteric activation of HtrA protease DegP by stress signals during bacterial protein quality control. Angew. Chem. Int. Ed. 2008;47:1332–1334. doi: 10.1002/anie.200703273. [DOI] [PubMed] [Google Scholar]
- 20.Jiang J., Zhang X., Chen Y., Wu Y., Zhou Z. H., Chang Z., Su S. F. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11939–11944. doi: 10.1073/pnas.0805464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., Clausen T. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 22.Shen Q. T., Bai X. C., Chang L. F., Wu Y., Wang H. W., Sui S. F. Bowl-shaped oligomeric structures on membranes as DegP's new functional forms in protein quality control. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4858–4863. doi: 10.1073/pnas.0811780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen T., Kidron H., Taipaleenmäki H., Salminen T., Mäenpää P. Transcriptional profiles and structural models of the Synechocystis sp. PCC 6803 Deg proteases. Photosynth. Res. 2005;84:57–63. doi: 10.1007/s11120-005-0475-x. [DOI] [PubMed] [Google Scholar]
- 24.Kieselbach T., Funk C. The family of Deg/HtrA proteases: from Escherichia coli to Arabidopsis. Physiol. Plant. 2003;119:337–346. [Google Scholar]
- 25.Huesgen P. F., Scholz P., Adamska I. The serine protease HhoA from Synechocystis sp. strain PCC 6803: substrate specificity and formation of a hexameric complex are regulated by the PDZ domain. J. Bacteriol. 2007;189:6611–6618. doi: 10.1128/JB.00883-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adir N., Zer H., Shochat S., Ohad I. Photoinhibition – a historical perspective. Photosynth. Res. 2003;76:343–370. doi: 10.1023/A:1024969518145. [DOI] [PubMed] [Google Scholar]
- 27.Edelman M., Mattoo A. K. D1-protein dynamics in photosystem II: the lingering enigma. Photosynth. Res. 2008;98:609–620. doi: 10.1007/s11120-008-9342-x. [DOI] [PubMed] [Google Scholar]
- 28.Huesgen P. F., Schuhmann H., Adamska I. Deg/HtrA proteases as components of a network for photosystem II quality control in chloroplasts and cyanobacteria. Res. Microbiol. 2009;160:726–732. doi: 10.1016/j.resmic.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Barker M., de Vries R., Nield J., Komenda J., Nixon P. J. The Deg proteases protect Synechocystis sp. PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J. Biol. Chem. 2006;281:30347–30355. doi: 10.1074/jbc.M601064200. [DOI] [PubMed] [Google Scholar]
- 30.Cheregi O., Sicora C., Kos P. B., Barker M., Nixon P. J., Vass I. The role of the FtsH and Deg proteases in the repair of UV-B radiation-damaged photosystem II in the cyanobacterium Synechocystis PCC 6803. Biochim. Biophys. Acta. 2006;1767:820–828. doi: 10.1016/j.bbabio.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Silva P., Choi Y. J., Hassan H. A., Nixon P. J. Involvement of the HtrA family of proteases in the protection of the cyanobacterium Synechocystis PCC 6803 from light stress and in the repair of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:1461–1470. doi: 10.1098/rstb.2002.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang F., Hedman E., Funk C., Kieselbach T., Schröder W. P., Norling B. Isolation of outer membrane of Synechocystis sp. PCC 6803 and its proteomic characterization. Mol. Cell. Proteomics. 2004;3:586–595. doi: 10.1074/mcp.M300137-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Fulda S., Huang F., Nilsson F., Hagemann M., Norling B. Proteomics of Synechocystis sp. strain PCC 6803. Identification of periplasmic proteins in cells grown at low and high salt concentrations. Eur. J. Biochem. 2000;267:5900–5907. doi: 10.1046/j.1432-1327.2000.01642.x. [DOI] [PubMed] [Google Scholar]
- 34.Huang F., Fulda S., Hagemann M., Norling B. Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics. 2006;6:910–920. doi: 10.1002/pmic.200500114. [DOI] [PubMed] [Google Scholar]
- 35.Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979;111:1–61. [Google Scholar]
- 36.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Letunic I., Copley R. R., Schmidt S., Ciccarelli F. D., Doerks T., Schultz J., Ponting C. P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 41.Bryson K., McGuffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell. 2007;19:1347–1361. doi: 10.1105/tpc.106.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolmar H., Waller P. R., Sauer R. T. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J. Bacteriol. 1996;178:5925–5929. doi: 10.1128/jb.178.20.5925-5929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haussühl K., Andersson B., Adamska I. A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 2001;20:713–722. doi: 10.1093/emboj/20.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helm M., Luck C., Prestele J., Hierl G., Huesgen P. F., Fröhlich F., Arnold G. J., Adamska I., Gorg A., Lottspeich F., et al. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11501–11506. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuhmann H., Huesgen P. F., Gietl C., Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 2008;148:1847–1856. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 48.Iwanczyk J., Damjanovic D., Kooistra J., Leong V., Jomaa A., Ghirlando R., Ortega J. Role of the PDZ domains in Escherichia coli DegP protein. J. Bacteriol. 2007;189:3176–3186. doi: 10.1128/JB.01788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jomaa A., Damjanovic D., Leong V., Ghirlando R., Iwanczyk J., Ortega J. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J. Bacteriol. 2007;189:706–716. doi: 10.1128/JB.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vande Walle L., Lamkanfi M., Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 2008;15:453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- 51.Kurian D., Phadwal K., Mäenpää P. Proteomic characterization of acid stress response in Synechocystis sp. PCC 6803. Proteomics. 2006;6:3614–3624. doi: 10.1002/pmic.200600033. [DOI] [PubMed] [Google Scholar]
- 52.Spence E., Sarcina M., Ray N., Møller S. G., Mullineaux C. W., Robinson C. Membrane-specific targeting of green fluorescent protein by the Tat pathway in the cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 2003;48:1481–1489. doi: 10.1046/j.1365-2958.2003.03519.x. [DOI] [PubMed] [Google Scholar]
- 53.Peltier J. B., Emanuelsson O., Kalume D. E., Ytterberg J., Friso G., Rudella A., Liberles D. A., Soderberg L., Roepstorff P., von Heijne G., et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell. 2002;14:211–236. doi: 10.1105/tpc.010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schubert M., Petersson U. A., Haas B. J., Funk C., Schröder W. P., Kieselbach T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 2002;277:8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- 55.Murata N., Suzuki I. Exploitation of genomic sequences in a systematic analysis to access how cyanobacteria sense environmental stress. J. Exp. Bot. 2006;57:235–247. doi: 10.1093/jxb/erj005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.