Abstract
The acquisition of a mesenchymal phenotype is a critical step in the metastatic progression of epithelial carcinomas. Adherens junctions (AJs) are required for suppressing this epithelial-mesenchymal transition (EMT) but less is known about the role of tight junctions (TJs) in this process. Here, we investigated the functions of blood vessel epicardial substance (BVES, also known as POPDC1 and POP1), an integral membrane protein that regulates TJ formation. BVES was found to be underexpressed in all stages of human colorectal carcinoma (CRC) and in adenomatous polyps, indicating its suppression occurs early in transformation. Similarly, the majority of CRC cell lines tested exhibited decreased BVES expression and promoter DNA hypermethylation, a modification associated with transcriptional silencing. Treatment with a DNA-demethylating agent restored BVES expression in CRC cell lines, indicating that methylation represses BVES expression. Reexpression of BVES in CRC cell lines promoted an epithelial phenotype, featuring decreased proliferation, migration, invasion, and anchorage-independent growth; impaired growth of an orthotopic xenograft; and blocked metastasis. Conversely, interfering with BVES function by expressing a dominant-negative mutant in human corneal epithelial cells induced mesenchymal features. These biological outcomes were associated with changes in AJ and TJ composition and related signaling. Therefore, BVES prevents EMT, and its epigenetic silencing may be an important step in promoting EMT programs during colon carcinogenesis.
Introduction
A hallmark of epithelial cells is the ability to organize through cell-cell adhesion into an epithelium functioning, collectively, as a tissue. Within the epithelium, there is coordinated cell motility, proliferation, and differentiation. The dynamic nature of the epithelium is apparent when there is loss of cell-cell contact, leading to individual epithelial cells assuming a fibroblast-like or mesenchymal morphology. This phenotypic change is termed epithelial-mesenchymal transition (EMT); in many tissues, upon regaining cell-cell contacts, cells undergo reciprocal mesenchymal-to-epithelial transition (MET). Cellular junctional complexes, including tight junctions (TJs) and adherens junctions (AJs), are key regulators of these transitions. For example, the role of the AJ as a modulator of canonical Wnt signaling through sequestration of β-catenin at the cell membrane is well established (1). We postulated that blood vessel epicardial substance (BVES, also known as POPDC1), a junctional associated, 3-pass transmembrane protein, may also regulate intracellular signaling networks and thus affect the balance between mesenchymal and epithelial phenotypes.
BVES was originally isolated from a cDNA screen of the developing heart (2), and initial immunolocalization studies showed BVES at the cell membrane of the proepicardial surface. A subpopulation of cells in this region undergo EMT, which was associated with loss of cell membrane bound BVES (3). Direct evidence for BVES regulating epithelial migration during embryogenesis was reported using a Xenopus developmental model. Injection of antisense morpholinos targeting BVES mRNA into a 2-cell Xenopus embryo led to disorganized migration and disrupted organogenesis (4). These observations suggest that BVES is capable of modulating the transition between epithelial/mesenchymal phenotypes.
The epithelial-to-mesenchymal switch also occurs in pathologic processes such as epithelial tumor progression. For example, colorectal carcinoma (CRC) tumor cells at the invasive front tend to possess mesenchymal traits, such as being hypermigratory, poorly differentiated, hyperproliferative, and incapable of establishing cell-cell contact–mediated growth inhibition (5). Not surprisingly, loss of junctional molecules, such as AJ components, E-cadherin (CDH1), and p120, has been associated with increased tumor invasiveness (6, 7). Conversely, most CRC lines that express E-cadherin have decreased invasiveness (8). Like that of AJs, TJ dysfunction also plays a role in carcinoma, although far less is known about TJ signaling in both normal biology and in malignancy. Claudin-1, -3, -4, and -7 are overexpressed in ovarian, CRC, and gastric cancers (9, 10). However, at least for claudins, this may be tissue specific, as claudin-1 potentially functions as a tumor suppressor in gastric cancer (11). Both AJ and TJ dysfunction contribute to a protumorigenic phenotype, partially by modification of junctional regulation of intracellular signaling pathways (WNT), cytoskeletal networks (Rho/Rac), or via modulation of EMT (12). What role BVES plays in these processes is unknown at this time.
We hypothesized that BVES modulates epithelial-mesenchymal phenotypes by regulating junctional complex formation and associated signaling networks in normal and malignant epithelial cells. We found that BVES expression was reduced in multiple types of solid tumor malignancy, and in CRC BVES levels were decreased in all stages and in adenomas. This occurred via BVES promoter hypermethylation. Manipulating BVES expression in complementary experiments using carcinoma lines and a human corneal epithelial (HCE) cell line resulted in reciprocal changes in epithelial-mesenchymal phenotypes, indicating a role for BVES in regulating EMT processes. BVES influenced multiple signaling pathways potentiating these effects, including TJ-associated RhoA activation and WNT signaling. Lastly, ectopic expression of BVES attenuated CRC tumor growth of orthotopic xenografts and inhibited metastasis. Our data show that BVES coordinately regulates TJ and AJ signaling, impacting the balance between epithelial and mesenchymal phenotypes. Thus, BVES is a potential tumor suppressor in multiple tissues.
Results
BVES expression is reduced in colon adenocarcinoma.
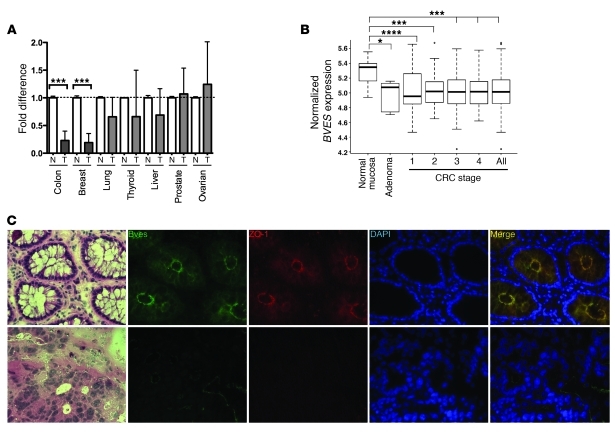
Adherens and tight junctional composition is frequently modified in solid tumor malignancy, with junctional proteins typically involved in maintaining an epithelial phenotype being downregulated (CDH1, Cldn7). We hypothesized that expression of BVES, another TJ-associated protein, would be reduced in carcinoma. Therefore, we screened a multi-tissue cancer panel for BVES mRNA levels (Figure 1A). BVES expression was significantly reduced in colon (6.7×; P = 0.01) and breast carcinoma (7×, P = 0.01) compared with that in respective normal tissues. BVES expression was not significantly reduced in lung, thyroid, and liver cancer, although BVES levels did trend downward. To verify and extend this observation, we analyzed the combined Moffit Cancer Center/Vanderbilt Medical Center colon cancer expression array data set, consisting of 250 tumor samples (see Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI44228DS1) organized by tumor stage, along with adenoma and normal mucosa samples (13, 14). BVES transcript levels were significantly decreased in all stages of CRC compared with those in normal adjacent mucosal specimens (Figure 1B). BVES transcript levels were also reduced in premalignant lesions (adenomas) compared with those in normal mucosa, indicating that BVES loss occurs early in the progression to tumorigenesis. Stratification of clinical treatments and outcomes by BVES transcript levels did not reveal a significant influence of BVES expression levels on either of these clinical variables. Similar expression screening for transcripts of the 2 other Popeye family members showed that popeye domain containing 2 (POPDC2) was not differentially expressed; however, POPDC3, located immediately telomeric to BVES on chromosome 6q21, was markedly downregulated in all stages of CRC (data not shown). These findings indicate that loss of BVES is associated with cancerous transformation.
Figure 1. BVES expression is significantly downregulated, and it is mislocalized in human colon carcinoma.
(A) A human BVES hydrolysis probe was used to perform qPCR on an Origene cDNA array (TissueScan Cancer Survey I). Ct values were normalized to β-actin and then presented as fold reduction from matched normal (N) (n = 3 per tissue type) and tumor (T) tissue (n = 9 per tissue type). ***P < 0.001, 2-tailed unpaired t test. (B) Analysis of the combined Moffit Cancer Center and Vanderbilt Medical Center colon tumor expression array data set (10, normal samples; 6, adenomas; 33, stage I; 76, stage 2; 82, stage 3; and 59, stage 4; for combined total of 250 CRC samples). For whisker plots, the bottom and top of the boxes are the 25th and 75th percentile, respectively (the lower and upper quartiles, respectively), and the band near the middle of the box is the 50th percentile (median). The whiskers extend to the most extreme data points, which are no more than 1.5 times the interquartile range from the box. *P = 0.04, ***P = 0.001, ****P = 0.0001. (C) Representative immunofluorescence images from normal colon or adenocarcinoma (original magnification, ×400) of H&E, BVES (green), ZO-1 (red), DAPI (blue), and merged images.
We noted an inverse relationship between BVES and ZEB1 expression in the CRC data set (P < 0.0001; Supplemental Figure 1). ZEB1 promotes EMT via transcriptional repression of E-cadherin and other cell polarity genes (15). Subsequently, increased ZEB1 expression is associated with a mesenchymal phenotype, while BVES expression promotes an epithelial phenotype. Thus, observing an inverse BVES and ZEB1 relationship reinforces the notion that BVES loss favors mesenchymal transcriptional programs in CRC.
To determine whether BVES protein levels mirrored the changes seen at the mRNA level, we performed immunofluorescence microscopy in matched normal and CRC tissues to localize BVES expression. BVES and ZO-1 colocalized at the apical domain of colonocytes in normal colonic mucosa (Figure 1C, top row). This is similar to a previously reported BVES staining pattern in polarized epithelium (16). In contrast, tumor tissue revealed little to no detectable BVES and ZO-1 staining (Figure 1C, bottom row). We quantified BVES protein levels in tumor and matched normal tissue via immunoblotting coupled with Odyssey systems analysis (see Methods) and observed a 40%–50% reduction in BVES in 70% of the surveyed samples (data not shown). Based on the relative paucity of intratumoral BVES seen in our immunolocalization studies, we suspect that protein levels of BVES seen in the immunoblots of the tumor tissues are inflated due to noncancerous epithelial contamination in tumor lysates.
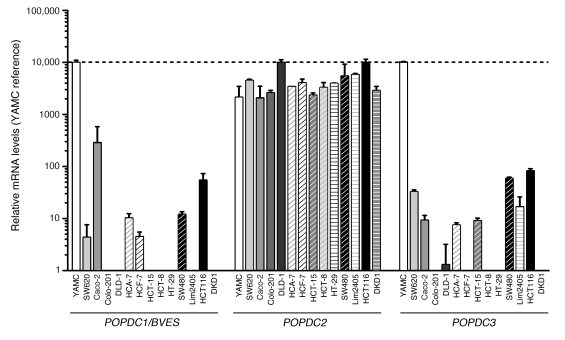
We next surveyed a CRC cell line panel for BVES expression. For comparison, we measured BVES mRNA levels in the YAMC cell line, a noncancerous colonic epithelial cell line. Low to undetectable BVES message levels were present in 12 out of 13 of the carcinoma lines surveyed (Figure 2). The exception was the Caco-2 cell line, which expressed relatively high levels of BVES message and coincidently is also capable of forming a differentiated, polarized epithelium. POPDC3, located immediately telomeric to BVES on chromosome 6q21, had an expression pattern similar to that of BVES. In contrast, POPDC2, the third Popeye family member, had moderate to high levels of expression in all cell lines surveyed. Thus BVES levels in CRC cells paralleled levels seen in CRC clinical samples.
Figure 2. BVES expression is reduced in CRC cell lines.
Popeye family member expression in colorectal cancer cell lines. RT-qPCR for POPDC1 (BVES), POPDC2, and POPDC3 using hydrolysis probes. Relative expression adjusted to the reference gene GAPDH and then standardized to YAMC (non-cancer colon cell line) is shown (mean ± SD).
BVES is silenced via hypermethylation in malignancy.
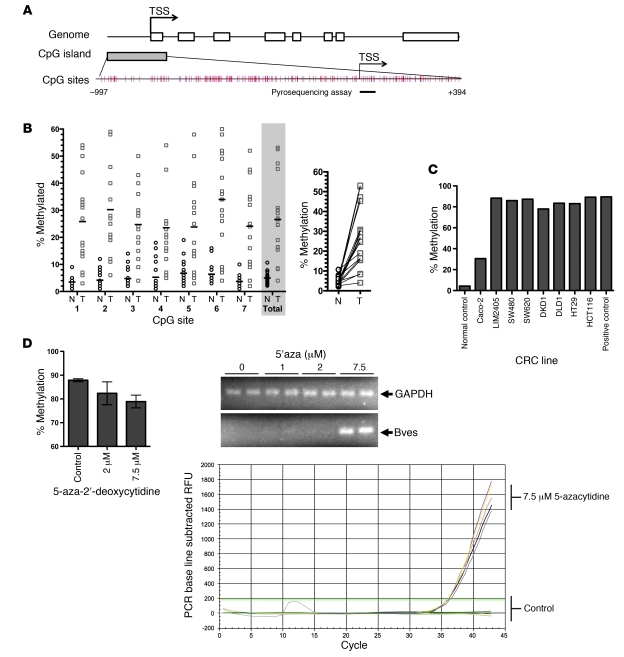
Promoter CpG island hypermethylation is a common feature in diverse malignancies, resulting in transcriptional silencing of many genes. The BVES promoter contains a large CpG island from –997 to +394 from the transcription start site (TSS) (Figure 3A) based on the criteria and algorithm described by D. Takai and P.A. Jones (17). The CpG sites in this region are very dense. We performed quantitative bisulfite pyrosequencing analysis in 18 matched CRC and normal tissues. In normal samples, the BVES promoter demonstrated low methylation levels (average methylation level was 4.9%). In contrast, BVES promoter methylation was significantly increased at each individual CpG site examined and reached an average of 26.6% in cancerous tissues (P < 0.001; Figure 3B, left, and Supplemental Figure 2). A paired sample plot reveals that BVES promoter methylation was increased in every matched pair of normal versus tumor tissues sampled (Figure 3B, right). Analysis of our expression array data indicated that for these 18 samples BVES transcript levels were inversely related to promoter methylation status, consistent with increased methylation silencing gene expression (Supplemental Figure 3). We next determined the methylation status of the BVES promoter in CRC cell lines and found that, with one exception (Caco-2 cells), the BVES promoter was heavily methylated (80%–90%; Figure 3C). This correlates with the general absence of BVES transcripts in CRC cells (see Figure 2). Caco-2 cells demonstrated lower levels of methylation (30%) and exhibit the highest relative levels of BVES mRNA expression among CRC cell lines (Figure 2). Furthermore, of all of the lines surveyed, Caco-2 cells are capable of forming a mature, differentiated epithelium (18).
Figure 3. BVES expression is silenced via promoter hypermethylation.
(A) Schematic representation of the human BVES promoter. The CpG island of BVES extends from –997 to +394 from TSS. Each red tick mark represents 1 CpG site. The arrows indicate the TSS. Pyosequencing assay to determine methylation level (%) was carried out on 7 CpG sites, as underlined by the black bar. TSS, transcriptional start site. (B) BVES promoter methylation status in colon adenocarcinoma compared with that in adjacent normal mucosa samples. Methylation level (%) at each of the 7 CpG dinucleotides was determined (left). For normal versus tumor comparisons, a horizontal bar represents the mean. The average level of the 7 CpG sites is shaded in gray. Individual symbols represent individual samples, and horizontal bars represent the mean. Before/after plot of matched samples, demonstrating 100% concordant increase in BVES methylation in cancers (right) (n = 18). P < 0.001, 2-tailed unpaired t test. N = normal; T = tumor tissue. (C) Pyrosequencing of BVES promoter DNA in the CRC panel. Data are presented as the average methylation percentage of 7 CpG pairs. (D) Pyrosequencing results of LIM2405 cells treated with either vehicle or 5-aza-2′-deoxycytidine for 72 hours (left). Data are presented as the average of triplicate measurements from duplicate experiments. qRT-PCR in LIM2405 cells showed detection of BVES mRNA following treatment with 5-aza-2′-deoxycytidine. RFU, relative fluorescence units. This demonstrates that BVES transcription in both clinical samples and CRC lines is silenced via promoter hypermethylation.
To confirm the role of DNA methylation in the transcriptional regulation of BVES, we treated LIM2405 cells with 7.5 μM 5-aza-2′-deoxycytidine for 72 hours and examined BVES promoter methylation and mRNA expression changes. 5-aza-2′-deoxycytidine treatment leads to DNA demethylation via inhibition of DNA methyl transferase activity. BVES gene expression was restored after 5-aza-2′-deoxycytidine treatment, and this reexpression was accompanied by a decrease in promoter DNA methylation from 94% to 72% (Figure 3D). These results indicate that promoter hypermethylation is one mechanism mediating transcriptional silencing of BVES in colorectal adenocarcinoma. It is possible that DNA methylation represents a common mechanism by which BVES is silenced in multiple solid tumors.
Restoration of BVES induces epithelial features in cancer cells.
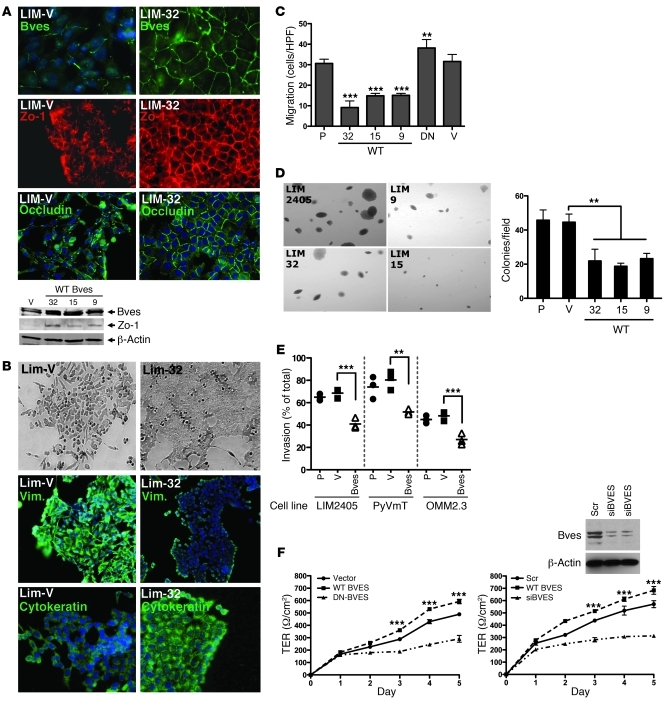
The findings above indicate that loss of BVES expression is associated with a mesenchymal phenotype in CRC cells. This observation leads to the question of whether BVES plays an active role in modulating epithelial-mesenchymal phenotypes or is merely a bystander in the malignant transformation process. We selected the LIM2405 cells to study the effects of restoring BVES expression. The LIM2405 colorectal cancer cell line exhibits prominent mesenchymal features, including anchorage- and cell contact-independent growth, expression of vimentin as the primary intermediate filament, attenuated ability to establish a monolayer with low levels of junctional proteins, and increased motility and invasive properties (19). In addition, LIM2405 cells express little to no detectable BVES. Using stable transfection, we generated a mixed population of LIM2405 cells stably expressing a WT BVES, along with multiple BVES-expressing LIM2405 clones (LIM-32, LIM-19, LIM-15). These stable BVES-expressing cells were evaluated for changes in mesenchymal features.
Immunofluorescent microscopy demonstrated increased BVES expression and concomitant increased ZO-1 levels, both confirmed by immunoblot (Figure 4A). In addition, the stably expressed BVES protein exhibited cell membrane localization coincident with ZO-1 and occludin localization in LIM-32 cells. This is similar to TJ protein expression patterns observed in nonmalignant epithelium (Figure 5B). Morphological restoration of BVES expression in LIM2405 cells induced epithelial features, as these cells grew in flat, adherent sheets and demonstrated reciprocal changes in vimentin and cytokeratin staining (Figure 4B). Pooled or clonal BVES-expressing LIM2405 cells exhibited increased transepithelial resistance (TER), a marker for epithelial polarization, which was reversed by transiently transfecting dominant-negative BVES (DN-BVES), indicating that these effects were consequent to BVES expression (Supplemental Figure 4). Consistent with epithelial characteristics, BVES expression attenuated LIM2405 cell migration by 66% (Figure 4C; P < 0.001) and reduced anchorage-independent growth by 53.5% (P < 0.01; Figure 4D, top left panel) Furthermore, the colonies that did form were notably smaller than either vector control (LIM2405-V) or parental LIM2405 (LIM2405-P) colonies, suggesting that BVES restored anchorage-dependent growth. Reinforcing this observation, BVES increased the doubling time (reduced proliferation rate) of LIM2405 cells and induced a density-dependent accumulation of cells in the G1 fraction (Supplemental Figure 5). In Matrigel invasion assays, BVES expression reduced LIM2405 invasiveness (41% reduction; P < 0.001). The effects of BVES on cellular function were not limited to LIM2405 CRC cells. OMM2.3 (an ocular melanoma line) and PyVmT (a murine breast cancer line) cells, both with low BVES levels, showed decreased invasion when BVES expression was increased (36% and 44%, respectively; P < 0.01 and P < 0.001, respectively) (Figure 4E). Caco-2 cells express BVES and are capable of forming a mature, competent epithelium. In contrast, colon cancer cells, which do not express BVES, appear to be incapable of forming a polarized epithelium. Whether BVES was knocked-down using siRNA (siBVES) or cell membrane localization displaced by transient expression of a DN BVES, there was a significant decrease in TER (Figure 4F) and doubling time (Supplemental Figure 6). These effects are reciprocal to those observed when BVES expression was restored in LIM2405 cells. There were, however, minimal differences in morphology, which is not surprising given that the TER assay requires superconfluent cells. Collectively, these experiments demonstrate that BVES can reverse mesenchymal growth characteristics of carcinoma cells, resulting in restoration of a more epithelial phenotype.
Figure 4. BVES induces an epithelial-like phenotype in carcinoma cells.
(A) Immunofluorescent microscopy for BVES, ZO-1, and occludin in LIM2405-V (LIM-V) or LIM2405 BVES-expressing (LIM-32) cells. DAPI stain (blue) was used to identify nuclei. Western blot analysis of BVES and ZO-1 protein levels in LIM2405-V cells or 3 independent BVES-expressing clones (LIM-32, LIM-15, and LIM-9, referred to as 32, 15, and 9, respectively) (original magnification, ×200). (B) Phase-contrast microscopy and immunofluorescent staining for the vimentin (Vim.) and cytokeratin (original magnification, ×200). (C) Migration assay. LIM2405-P (P), LIM2405-V (V), LIM2405-BVES clones (32,15,9), and LIM2405 cells stably expressing a carboxyterminal deletion BVES construct functioning as a dominant negative (DN) were used. Data is represented as the mean ± SD. ***P < 0.001, **P < 0.01, Student’s t test. (D) Anchorage-independent growth assay (original magnification, ×100). Colonies were tallied on day 14. Colony size and colony number are shown. Data are presented as mean ± SD. HPF, high-powered field. **P < 0.01. (E) Boyden chamber invasion assay was performed using LIM2405, PyVmT (murine breast carcinoma; ref. 53), or OMM2.3 (ocular melanoma) cells. Individual symbols represent individual cells, and horizontal bars represent the median. **P < 0.01, ***P < 0.001, Student’s t test. (F) CACO-2 cells transiently transfected with the indicated constructs. Upon achieving confluence, TER was measured daily (n = 6). ***P < 0.001, Student’s t test.
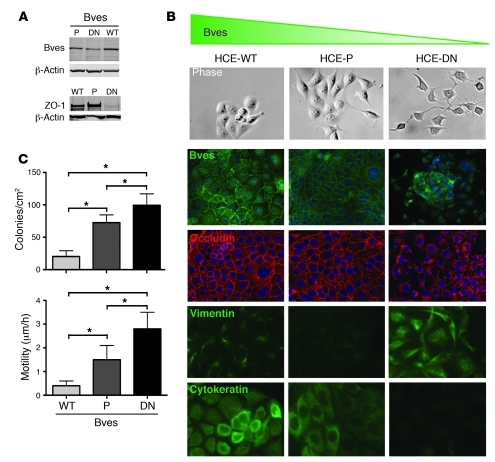
Figure 5. BVES regulates EMT in HCE cells.
(A) Lysates were obtained from parental (untransfected) HCE cells (P), cells stably transfected with WT BVES (WT), or a COOH-truncated BVES (DN) cells functioning as a dominant negative interfering with expression and subcellular localization. Immunoblot for ZO-1, BVES, or β-actin. (B) Representative morphology of the indicated cell lines by phase-contrast microscopy (original magnification, ×400). Immunofluorescent localization of BVES, (original magnification, ×200), occludin (original magnification, ×200), vimentin (original magnification, ×400), and cytokeratin (original magnification, ×400) in confluent cultures. (C) Quantification of both anchorage-independent growth assays (6 replicates were performed) and cellular migration (20 replicates were performed for each cell type). *P < 0.05, Student’s t test. This indicates that BVES can influence epithelial-to-mesenchymal phenotypes.
Loss of BVES induces mesenchymal features in epithelial cells.
Originally, Bader and colleagues associated BVES with EMT based on changes in BVES subcellular distribution during cardiac morphogenesis (2, 3, 20). However, it is not clear whether changes in BVES localization and/or expression actively drive EMT in epithelial cells or are merely bystander events. In our previous studies, transient disruption of either BVES expression or its cell membrane localization attenuated cell-cell contact, resulting in epithelial cells assuming fibroblast morphology, suggesting that transient manipulations of BVES could regulate EMT (21). Collectively, these observations led us to postulate that loss of BVES function could promote EMT. To test this, HCE cells were stably transfected with carboxyterminal truncated DN-BVES, which resulted in abnormal BVES trafficking and had a dominant-negative effect. These cells (HCE-DN cells) were evaluated for epithelial-mesenchymal traits using the accepted criteria (22) of (a) morphologic characteristics, (b) cell membrane localization of junction-associated molecules, (c) markers of epithelial/mesenchymal phenotypes, (d) anchorage-independent growth, and (e) cell motility.
By phase microscopy, HCE-DN cells exhibited a strikingly different morphology in comparison with that of both HCE-P and HCE-WT cells (HCE cells stably overexpressing a WT BVES) (Figure 5B). HCE-DN cells were spindly, with long processes, and had attenuated cell-cell contact. This was reinforced at higher cell density, as HCE-DN cells proliferated excessively, forming irregular, heaped-up foci, exhibiting little or no junctional proteins at the cell membrane. In contrast, HCE-WT and HCE cells proliferated in organized clusters and formed normal-appearing epithelial monolayers (Figure 5B). These monolayers displayed cell membrane adhesion junction proteins outlining cell borders in a classic wire-mesh pattern, consistent with the known TJ association of BVES (16). Both HCE and HCE-WT cells expressed abundant cytokeratin and a paucity of vimentin. In contrast, HCE-DN cells had a reciprocal pattern, expressing predominantly vimentin (Figure 5B) and no cytokeratin. Thus, expression of DN BVES but not WT BVES promotes mesenchymal features in epithelial cells. When we assessed anchorage-independent growth, the parental HCE cells formed 72.5 (SD 12) colonies/cm2. WT BVES blunted colony formation with 20.2 (SD 8.9) colonies/cm2 (P < 0.05), while the DN BVES mutant (HCE-DN) significantly increased colony number with 99.3 (SD 17.8) colonies/cm2 (P < 0.05; Figure 5C). Additionally, HCE-DN cells displayed increased motility in comparison with that of both HCE and HCE-WT cells, as measured by time-lapse phase microscopy (P < 0.05; Figure 5C). Collectively, these observations indicate that BVES negatively regulates EMT programs in HCE cells. These findings corroborate in vivo studies (2, 4, 23) by others, demonstrating the role of BVES in developmental morphogenesis, and suggest that BVES regulates EMT in diverse tissue types.
BVES regulates AJ- and TJ-associated signaling.
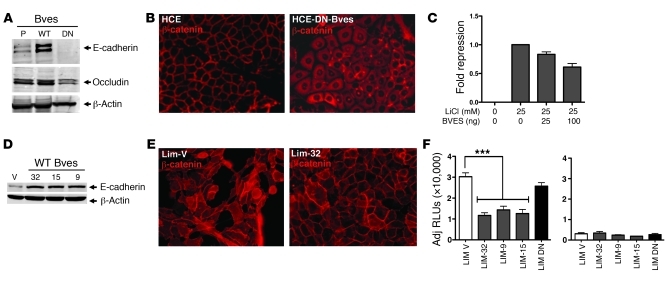
We next determined the influence of BVES on AJ composition and signaling activities in normal and malignant cell lines. We found a direct relationship between BVES expression and E-cadherin expression in HCE-DN BVES and LIM2405 cells expressing BVES (LIM2405, LIM-32, LIM-15, and LIM-9; Figure 6, A and D). Furthermore, β-catenin cellular distribution was perturbed by manipulating BVES, resulting in reciprocal cytoplasmic or membrane-bound localization, respectively, in HCE-DN BVES– and LIM2405-BVES–expressing cells (Figure 6, B and E). Since we noted a marked change in β-catenin localization upon BVES expression, we interrogated the affect of BVES on WNT transcriptional activity using a β-catenin/TCF4 reporter construct. TOPFlash reporter activity was attenuated by BVES expression in the HEK293 WNT reporter line, indicating that BVES-mediated β-catenin redistribution was functionally relevant (Figure 6C). In support of these data, TOPFlash activity was also significantly attenuated in LIM2405-BVES–expressing lines (Figure 6F).
Figure 6. BVES regulates AJ composition and signaling.
(A) Representative immunoblot for E-cadherin and occludin in parental- (P-), DN BVES– (DN-), or WT BVES–expressing (WT-expressing) HCE cells. β-Actin was used to ensure loading consistency. (B) Immunofluorescence shows β-catenin cellular redistribution after BVES knockdown in HCE cells. Representative images are presented (original magnification, ×400). (C) BVES repressed WNT reporter activity in TOPFlash HEK293 (STF293; ref. 46) cells in a dose-dependent fashion. Data shown are from triplicate wells (mean ± SD). (D) Immunoblotting of LIM2405-V (V) or WT BVES (LIM-32, LIM-15, LIM-9) clones for E-cadherin protein. β-Actin was used to ensure consistent loading. (E) β-Catenin immunofluorescent staining in the indicated LIM2405 lines (original magnification, ×400). (F) LIM2405 cell lines were transfected with the SuperTOPFlash TCF4 reporter construct, and luciferase activity was determined 48 hours later. Transfection efficiency was determined via cotransfection with TK-Renilla. Data presented as RLUs/RUs. BVES modulates both TJ and AJ complex composition, affecting AJ-mediated WNT signaling (n = 3). ***P < 0.001, Student’s t test.
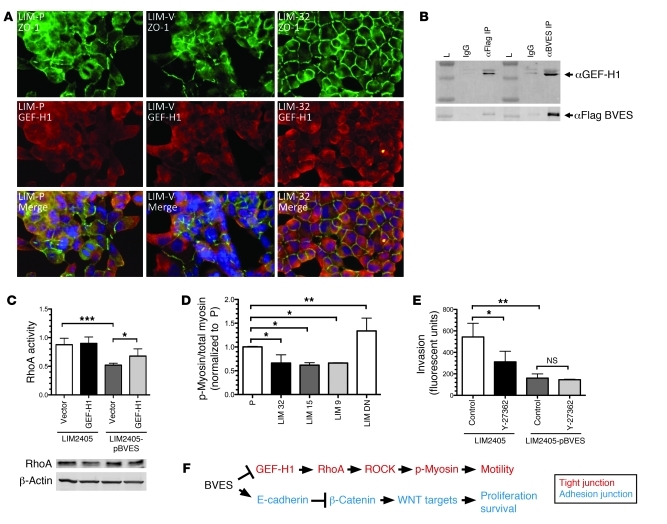
TJs have been reported to directly regulate RhoA activity. As TJs form within an epithelial monolayer, ZO-1 binds GEF-H1, a cytoplasmic RhoA activator, through an adapter protein, cingulin, leading to GEF-H1 membrane sequestration. This reduces GEF-H1 availability, causing decreased levels of activated RhoA (24). We observed increased colocalization of ZO-1 and GEF-H1 at the cell membrane of neighboring cells when LIM2405 cells expressed BVES (LIM-32) (Figure 7A). LIM2405-P and LIM2405-V control cells exhibited only scattered regions of ZO-1/GEF-H1 colocalization. Since BVES/ZO-1 exhibited similar colocalization patterns as ZO-1/GEF-H1 (compare Figure 1C to Figure 7A), we suspected that BVES, ZO-1, and GEF-H1 are capable of (direct or indirect) interactions within a complex. This is supported by immunoprecipitation studies showing GEF-H1 co-immunopurifying with BVES (Figure 7B). Furthermore, increased expression of BVES in pooled BVES LIM2405 cells led to decreased levels of activated RhoA, which is rescued by transient expression of GEF-H1 (Figure 7C). In contrast, increased GEF-H1 expression in LIM2405-P cells had no significant affect on RhoA activation, suggesting that RhoA exists primarily in an activated state in LIM2405 cells. Together, these findings indicate that, in addition to regulating TJ formation, BVES has a modulatory affect on RhoA activation through GEF-H1. Downstream targets and cellular effects of activated RhoA were also investigated. A biochemical end target of activated RhoA is increased myosin light chain (MLC) phosphorylation, which generates cellular contractility needed for motility. With increased BVES expression, we observed decreased levels of phosphorylated MLC (Figure 7D). RhoA exerts these affects on MLC via the intermediary Rho-associated kinase (ROCK). ROCK regulates phosphorylated MLC levels by direct phosphorylation of MLC and deactivation of myosin phosphatase. We next treated BVES-expressing LIM2405 cells with the ROCK inhibitor Y-27362 and used a Boyden chamber assay to quantify invasion. Treatment with Y-27362 resulted in a 50% reduction in LIM2405 invasion. Expression of BVES in LIM2405 cells markedly attenuated invasion, and addition of Y-27362 yielded no additional inhibition, suggesting that BVES acts upstream of ROCK and the majority of the affect of BVES on invasion was via RhoA-mediated signaling. Collectively, these data implicate BVES in regulating TJ and AJ signaling programs and, to our knowledge, identify a new role for BVES in TJ/AJ crosstalk.
Figure 7. BVES regulates TJ-associated RhoA signaling.
(A) Immunofluorescent localization studies for ZO-1 (green) and GEF-H1 (red) in LIM2405-P (LIM-P), LIM2405-V (LIM-V), and LIM-32 clones overexpressing BVES. LIM-32 cells exhibit corresponding cell membrane localization of ZO-1 and GEF-H1 as indicated on the merged image (yellow, cell border). Both LIM2405-P and LIM2405-V exhibit only scattered regions of cell membrane distribution of either ZO-1 or GEF-H1, with little corresponding cell membrane localization (original magnification, ×400). (B) Immunopurification of BVES and GEF-H1. Lysates of cells transiently overexpressing Flag-tagged BVES revealed co-immunopurification of BVES and GEF-H1. Either FLAG or BVES antibodies were used to precipitate proteins. (L= Molelecular Weight Marker). (C) RhoA activity assay measured on LIM2405 and pooled BVES LIM2405 (LIM2405 pBVES) cells after GEF-H1 transfection. Immunoblot of RhoA total protein levels. ***P < 0.005. (D) Ratios of phosphorylated MLC (p-myosin) over total MLC (myosin) were obtained through densitometric analysis of immunoblots (n = 4) of LIM2405 (P), WT BVES (LIM-32, LIM-15, LIM-9) clones, and dominant-negative LIM (DN). WT-BVES clones exhibit a significant decrease in phosphorylated myosin compared the LIM2405-P, while the DN LIM cells exhibited a significant increase in p-myosin. (E) Boyden chamber invasion assay of LIM2405 or pooled BVES LIM2405 cells, comparing effects of ROCK inhibition via treatment with Y-27362 or vehicle (control) (n = 3). *P < 0.05, **P < 0.005, Student’s t test. (F) Schematic of BVES modulating TJ and AJ.
Overexpression of BVES attenuates CRC tumor growth and metastasis in athymic mice.
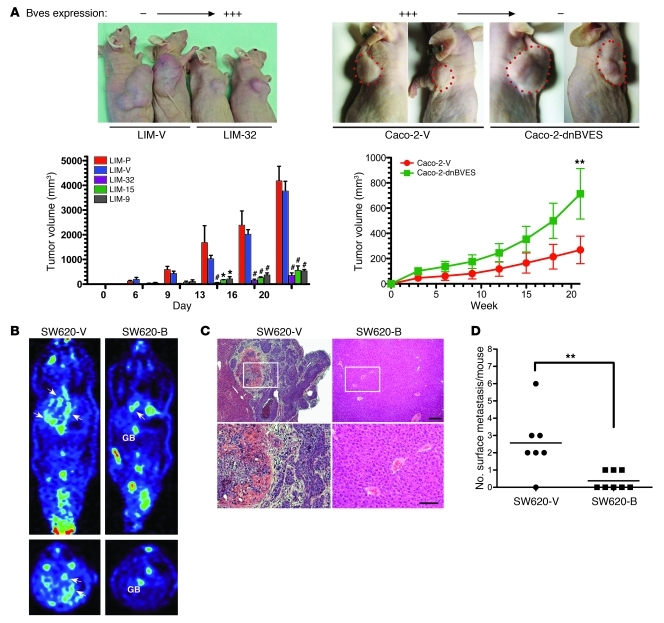
Because BVES restored epithelial features to LIM2405 cells in vitro, we asked whether BVES could attenuate in vivo growth characteristics of LIM2405 cells. Using a xenograft tumor model, LIM2405-P, LIM2405-V, or 3 independent BVES clones (LIM-9, LIM-15, LIM-32) were injected into the dorsal flank of athymic (“nude”) mice. Both the LIM2405-P and LIM2405-V lines formed bulky, large tumors by 20 days (Figure 8A, left) (4.1 cm3 with SD ± 1.0 cm3 and 3.7 cm3 with SD ± 0.96 cm3; no significant difference), with tumors first detectable at day 6. In contrast, all 3 BVES-expressing LIM2405 clones showed an approximate 8-fold reduction in tumor growth, achieving an average tumor volume of 0.484 cm3 with SD ± 0.064 cm3, significantly different to that of either the LIM2405-P or LIM2405-V lines (P < 0.001; Figure 8A, left). Interestingly, we noted modest tumor growth rate acceleration in BVES expressing cells at 20 days. We postulated that BVES was selected against and performed immunofluorescent microscopy, which revealed reduced BVES expression and disorganized intratumoral BVES localization, similar to the BVES staining observed in the LIM2405-P and LIM2405-V control cells (data not shown). Tumors were also evaluated for markers of proliferation (Ki67) and apoptosis (TUNEL) (Supplemental Figure 7). Apoptosis was increased in LIM-32 tumors with unaffected proliferation, indicating that at least a partial effect of introducing BVES expression is restoration of cell death programs. These observations led us to hypothesize that disruption of BVES in a BVES-expressing CRC line could enhance tumor growth. Caco-2 cells express BVES and were chosen for stably transfecting either empty vector (Caco-2-V) or Caco-2 stably expressing DN BVES (Caco-2-dnBves). Stable pooled populations were then injected into the dorsal flank of nude mice, and tumor growth was monitored. Caco-2-dnBves cells produced tumors that were 2.5-times larger than Caco-2-V tumors (714.3 mm3 vs. 269.7 mm3; P < 0.01; Figure 8A and Supplemental Figure 8). The results of these 2 complimentary experiments provide evidence that BVES inversely regulates in vivo tumor growth.
Figure 8. BVES modifies tumor growth and metastasis in athymic mice.
(A) 2.5 × 106 LIM2405-P (n = 3), LIM2405-V (n = 6), Caco-2-V, Caco-2-dnBVES, or LIM2405-BVES–expressing cell lines LIM-9 (n = 6), LIM-32 (n = 3), LIM-15 (n = 3) were implanted in the dorsal flank of 8-week-old athymic nude mice. Quantification of growth rates achieved by measured tumor dimensions at the indicated intervals is presented as tumor volume (volume = [width2 × length] / 2) (average volume ± SEM. *P < 0.05, **P < 0.01, #P < 0.001, 2-way ANOVA . Splenic metastasis assay. SW620-V– or SW620-B–transfected (pooled) cells were injected into the splenic capsule (B) PET imaging (GB = Gallbladder, arrows indicate metastatic foci). (C) Histologic representative livers from indicated cell lines. (D) Metastasis quantification. **P < 0.01, Student’s t test. Small scale bar, 20 mM; large scale bar, 10 mM.
Because our in vitro data indicate that BVES influences EMT, we next determined whether BVES could modify CRC metastasis. SW620 cells lack BVES expression (Figure 2) and readily metastasize in nude mice splenic metastasis assays (9). Subsequently, we generated pooled stable BVES SW620 cells (SW620-B cells) and compared the metastatic potential of this line with that of vector-transfected SW620 cells (SW620-V cells). We noted decreased metastatic foci on microPET imaging at 6 weeks (Figure 8B), correlating with decreased metastatic foci seen at necropsy (Figure 8, C and D). Collectively, these 3 animal experiments indicate that the influence of BVES on cell growth and epithelial-mesenchymal phenotypes is not a cell culture–based phenomena but rather demonstrates that BVES regulates both in vivo CRC tumor growth and metastatic potential in CRC cells.
Discussion
Summary.
Epithelial cells exhibit diverse morphologies ranging from an epithelial phenotype, in which cells organize into epithelia, to a mesenchymal phenotype, with cells functioning mainly as isolated units. Transitions between epithelial-mesenchymal phenotypes are collectively termed either EMT or MET, depending on perspective. We demonstrated reciprocal effects of BVES on epithelial-mesenchymal phenotypes, with loss of BVES inducing EMT in an organized epithelial cell line (HCE cells), while overexpression of BVES in human colon cancer cells, having mesenchymal features, leads to MET, which also significantly attenuated their ability to form tumors as orthotopic implants in nude mice (Figure 8A). Loss of BVES in HCE cells is associated with reduced E-cadherin levels, and both cytoplasmic and nuclear redistribution of β-catenin. Conversely, overexpression of BVES in human colon cancer cells is associated with increased E-cadherin, redistribution of β-catenin to the cell membrane, and decreased TCF4 activity. These results suggest that BVES modulates canonical WNT signaling and that BVES regulates cellular transitions between epithelial-mesenchymal phenotypes and implicates loss of BVES function as contributing to tumorigenesis.
Analysis of a large colorectal cancer expression array data set revealed decreased BVES at all stages of CRC, which further implicates BVES in tumorigenesis. Mechanistically, we determined that BVES promoter methylation was 10-fold higher in cancer samples and also markedly higher in CRC cell lines. Not surprisingly, expression was inversely related to methylation status, and treatment of CRC lines with 5-aza-2′-deoxycytidine restored expression. These findings indicate that BVES promoter methylation suppresses BVES transcription and contributes to epithelial-mesenchymal dysregulation characterized in tumorigenesis.
BVES coordinates junctional signaling.
Junctional complexes, such as AJs and TJs, are classically viewed as mechanical structures linking cells together within an epithelium. The roles of AJs and TJs within a tissue, however, extend beyond their mechanical properties; they also function as signaling complexes allowing intercellular communication. BVES through its regulatory effects on cell-cell interaction modulates both AJ- and TJ-associated signaling. The association of AJs with canonical WNT signaling is well established. Due to the regulatory effect of BVES on E-cadherin levels and subsequent alteration in β-catenin distribution, it was not unexpected that changes in BVES expression were inversely related to TCF4 activity. Our previous studies have established BVES as a regulator of cell-cell adhesion through its modulatory effect in the early assembly and subsequent maturation of adhesion junctions (16). We suspect that it is through this regulatory role that BVES also influences AJ and associated signaling.
BVES also influences TJ-associated signaling by modulating sequestration of proteins at the cell membrane, in a similar fashion to modulation of β-catenin within AJs by AJ-associated proteins. TJs directly sequester GEF-H1, an activator of RhoA, and in the LIM2405 cells, BVES was capable of regulating the sequestration of GEF-H1 at the cell membrane. This, in turn, modulates RhoA activation, resulting in changes to levels of phosphorylated MLC, potentially explaining the effect of BVES on cellular migration (Figure 4C) and invasion (Figure 4E).
These findings identify BVES as a coordinator of TJ and AJ signaling. The existence of cross-talk between TJ and AJ has been demonstrated by others. For example, blocking peptides to the extracellular domain of occludin increased permeability and induce β-catenin expression with activation of WNT target genes (25). Manipulation of claudin-1 expression in SW480 (APC Wildtype, low baseline WNT signaling levels) and SW620 (APC Mutant, high baseline WNT signaling levels) resulted in reciprocal changes in WNT reporter activity, E-cadherin expression, and expression of c-Myc, a WNT target gene (9). We believe that this study is the first report linking BVES with the canonical WNT pathway and that it adds BVES to the short list of TJ transmembrane proteins capable of influencing WNT signaling, thus reinforcing the notion that there is cross-talk between TJ and AJ signaling.
BVES and EMT.
EMT is a biologic phenomenon wherein cells contained in a well-organized epithelium, in response to environmental or genetic cues, break cell-cell contact and initiate programs resulting in adoption of mesenchymal or fibroblast morphology. This transition is reversible and is associated with a variety of physiologic and pathologic processes ranging from developmental to wound healing and tumor progression (22, 26, 27). A classification scheme has recently been proposed classifying EMT into 3 types based on biologic effects (22, 26) Type 1 EMT occurs during embryonic development. Type 2 EMT is associated with wound healing, tissue regeneration, and organ fibrosis. Type 3 EMT occurs during metastatic progression. BVES appears to contribute to all 3 of these types of EMT. This study provides direct evidence that BVES regulates EMT/MET in a reversible manner in normal and cancer epithelial cells. In vivo, it is likely that BVES also plays a role in regulating all 3 types of EMT. The majority of the evidence for BVES as participating in type 1 EMT is provided by Bader and colleagues. In their original studies isolating BVES from the embryonic chick heart, they described the dynamic spatiotemporal localization pattern of BVES in a subset of proepicardial cells that participate in the coronary artery morphogenesis (3, 20). Recently, in cell culture models, BVES has been shown to regulate cell contacts by modulating levels of TJ and AJ proteins (16, 21). Furthermore, corneal epithelial cells expressing mutant BVES took on a mesenchymal phenotype with inability to form a monolayer (21). These observations indirectly point to BVES as a regulator of EMT. Later, using morpholinos to knockdown BVES levels, Ripley and Bader provided direct evidence of BVES regulating organogenesis in multiple systems (4). Brand and colleagues provided evidence for BVES participating in type 2 EMT in their report of delayed skeletal muscle wounding in Bves–/– animals (28). Finally, this report provides evidence for BVES modulation of type 3 EMT by linking loss of BVES with tumor progression and demonstrating that loss of BVES in CRC samples is secondary to hypermethylation of the BVES promoter. Together these observations point to BVES as a regulator of EMT/MET in diverse physiologic and pathologic settings.
BVES and epithelial malignancy.
TJ proteins are common targets for transcriptional silencing in malignancy. Both claudin-3 and -7 are hypermethylated in esophageal squamous cell carcinoma (29), with the absence of claudin-7 associated with loss of E-cadherin and increased invasiveness (30). Claudin-6 (31), -7 (32), and occludin (33) are hypermethylated in breast cancer, and Feng et al., using a candidate gene approach and MethyLight assays, found that BVES was hypermethylated in non–small cell lung cancer (34); both POPDC3 and BVES were hypermethylated and underexpressed in gastric carcinoma (35). Presumably, these epigenetic events translate into skewing toward a mesenchymal phenotype ultimately realizing a survival advantage. In addition to epigenetic regulation of TJ protein expression, altered TJ function is found in lung (36), hepatocellular, breast (37), and colon cancer (38). Identifying BVES as a target for silencing and demonstrating the functional significance of this event further expands the array of targets for pharmacologic exploitation in cancer therapy.
There are multiple mechanisms whereby selective pressures result in gene inactivation in cancer. Our data suggest that promoter hypermethylation is a common mechanism of BVES inactivation in CRC cases. Similar to BVES, the major mechanism of E-cadherin inactivation in carcinoma is via promoter hypermethylation (39). Other mechanisms, such as LOH, mutations, and transcriptional regulation, have also been reported in regulating E-cadherin (40). BVES is located on 6q21 where deletions or LOH of this region have been widely reported in breast (41), ovarian (42), and prostate (43) carcinoma. We speculate that additional mechanisms inactivating BVES in carcinoma exist, and we are in the process of sequencing the normal to high BVES-expressing CRC samples to determine whether BVES inactivating mutations are present.
Given the role of BVES in EMT, one might suspect that loss of BVES would occur late in tumorigenesis. Our data indicate otherwise, as BVES was silenced in all stages of CRC and even early tumorigenesis with decreased expression in adenomas. This is consistent with reports that CpG island methylation in carcinoma occurs early during tumorigenesis and typically is present in normal appearing mucosa (44). Promoter hypermethylation is thought to be involved in progression from adenoma to carcinoma via silencing of tumor suppressor genes (45). BVES hypermethylation at a premalignant stage may be secondary to nonspecific methylation patterns affecting many genes and a selective advantage may only be realized late in tumorigenesis during the process of EMT. Alternatively, loss of BVES expression may affect cellular growth pathways (i.e., WNT/Zonab) and thus exert a selective advantage during premalignant tumor expansion. Nevertheless, loss of BVES expression early in tumorigenesis may represent an additional epigenetic marker that could be combined with other markers, such as loss of E-cadherin, p16, hMLH1, and VHL, in predicting CRC progression and/or risk.
In summary, BVES, the prototypic member of the POPDC gene family, is underexpressed in both colon carcinoma and adenoma via promoter hypermethylation. BVES repression appears to have functional consequences, as reciprocal manipulations of BVES expression in normal and malignant cells result in EMT/MET changes, which in malignancy resulted in conversion to a nonmalignant phenotype. Mechanistically, some of these protumorigenic effects may be secondary to its influence on AJ and TJ composition and related signaling. BVES transcriptional suppression occurs during early tumorigenesis, raising the possibility that BVES could be a therapeutic or preventative target in CRC and perhaps other solid tumor malignancies. Collectively, these observations suggest that BVES functions as a tumor suppressor whose functions are selected against in the course of malignant progression.
Methods
Cell lines.
HCE, murine breast cancer (PvYmT), and ocular melanoma (OMM2.3) cell lines were cultured in DMEM. Colorectal cancer lines (LIM2405 and SW620) were cultured in 10% FBS RPMI 1640, and Caco-2bbe CRC cells were cultured in DMEM. These lines were transfected with WT chick BVES, a carboxyterminal truncated form of BVES, or vector according to established methods (16, 21). The WT chick and carboxyterminal truncated BVES have a FLAG tag at the C terminal. G418 selection was used to obtain a mixed population of stably transfected cells, and transgene expression was verified. In addition, to obtain pooled stable lines, individual stable clones for HCE and LIM2405 cell lines were generated after G418 selection. Approximately, 55 and 25 stable isolates were obtained, respectively, for LIM2405 and HCE cells using cloning rings. Stable clonal isolates were screened for FLAG expression by immunofluorescent localization, yielding 36 clonal LIM2405 lines and 20 clonal HCE lines. SW-480, DKD-1, DLD-1 cells were maintained in RPMI 1640 and HT29 and HCT116 in (McCoy’s 5A Medium modified). All media were supplemented with 10% FBS. These lines were either obtained from ATCC or as a gift from Robert Whitehead, Vanderbilt University, Nashville, Tennessee, USA.
Western blotting.
Protein extraction and Western blotting were performed as previously described (16). Membranes were probed with antibodies against ZO-1, occludin, E-cadherin, BVES, β-actin, and GEF-H1. GEF-H1 antibodies were provided as gifts by M.S. Balda and K. Matter (Institute Ophthalmology, University College London, London, United Kingdom). Alexa Fluor 680– or IRDye 800–conjugated secondary antibodies were applied, and blots were scanned and analyzed using the Odyssey Infrared Imaging System (Li-Cor Biosciences).
Immunofluorescence.
Immunofluorescence staining of cultured HCE and LIM2405 cells as well as CRC tissue sections obtained from the Vanderbilt Human Tissue Acquisition Core was performed by first fixing in 70% methanol, permeabilizing in PBS with 0.25% Triton X-100, and blocking with PBS containing 2% BSA for 1 hour at room temperature. Primary antibodies were diluted in 1% BSA and incubated overnight at 4°C. The cells and sections were washed with PBS, and secondary antibodies were added for 3 hours at room temperature. After PBS and water washes, slides were mounted in fluorescence mounting media and captured using a Nikon fluorescence microscope at the indicated magnification.
Anchorage-independent growth assay.
Soft agar assay was carried out by allowing cells to proliferate within 0.5% agarose at a density of 1,000 cells per ml. After 14 days, the CellTracker Red Vital Dye (Invitrogen) was added according to the manufacturer’s directions to stain for viable cell colonies. Viable colonies consisting of more than 20 cells were counted using a fluorescent microscope. A total of 6 wells were counted for each cell type. HCE cells were generated via SV40 transformation of primary corneal epithelial cells and can form colonies in anchorage-independent growth assays (46).
WNT reporter assays.
HEK293 cells stably transfected with the SuperTOPFlash TCF4 reporter construct (STF293) (47) were transiently transfected with 25–250 ng of BVES and 1 ng/well TK-Renilla and supplemented with pcDNA4 to keep total transfection mass at 1 μg. Cell lines were trypsinized and transferred to 12-well plates at 2 × 105 cells per well in quadruplicate. Twenty-four hours after plating, WNT activity was stimulated using the GSK-3β inhibitor LiCl at 25 mM. Cells were harvested 24 hours later, and both Luciferase and Renilla activity was assessed after following the manufacturers protocol (Dual-Luciferase Reporter Assay System, Promega) on a BD Monolight 2010 Luminometer. Adjusted RLUs were determined (Luciferase/Renilla) and presented as fold change from LiCl-induced activity. The indicated LIM2405 clone was transfected with 300 ng Super (8×) TOPFlash or FOPFlash (48) and 250 ng of pCMV5-secreted alkaline phosphatase (SEAP) plasmids. Luciferase activity was measured and normalized to SEAP activity and is presented as adjusted RLUs. Triplicate samples were measured, and the experiment was repeated 3 times yielding similar results.
Tumor xenograft studies.
Six- to eight-week-old athymic mice (Foxn1 nu/nu mice) were purchased from Harlan Sprague Dawley. LIM2405, Caco-2bbe, and related sublines were grown on plastic culture dishes according to standard culture techniques (49). 2.5 × 106 cells in 200 μl of 1× PBS were injected into the dorsal flank. Results are reported as mean tumor volume (volume = [width2 × length]/2). Five-μm sections of formalin-fixed tumors were prepared and stained with H&E and αBrdU (50), and in situ TUNEL staining (ApopTag, catalog no. S7100; Chemicon) was performed using standard methods and per manufacturers protocols.
Splenic metastasis model.
To asses the impact of overexpression of BVES on metastasis in vivo, male athymic/nude mice (7 weeks old; n = 16) were injected in the spleen with 5 × 106 SW620-V cells or SW620-BVES cells. The spleen was removed after tumor cell injection, and the splenic vein was cauterized. Small-animal microPET imaging was used to screen for nonpalpable lesions in the liver, using 100–150 μCi of 18F-deoxyglucose injected i.p. to detect metabolically active foci in the abdomen. When the tumors were visible by microPET (6 weeks), the mice were sacrificed. The number of metastatic tumor foci on the surface of the livers and identified on microscopic examination of bread-loafed livers was documented. All in vivo procedures were carried out in accordance with protocols approved by the Vanderbilt Institutional Animal Care and Use Committee.
Time-lapse motility assay.
Cell motility was assessed by phase-contrast time-lapse microscopy of individual cells beginning 24 hours after plating and captured hourly. The total distance migrated by individual cells was obtained from these sequential time-lapsed images, and the motility rate was calculated and presented graphically.
Invasion assay.
WT and BVES stably transfected OMM2.3, PMT, and LIM2405 cell lines were evaluated for relative invasive capacities using the Trevigen BME Cell Invasion Kit (Trevigen) according to the manufacturer’s instructions. Cells were serum-starved overnight and plated the following day in serum-free media at 50,000 cells per well into the upper chamber of a 96-well microplate well fitted with 8-μm pore size cell culture inserts precoated with Matrigel basement membrane extract. The lower chamber was prefilled with complete media without antibiotics containing 10% FBS as a chemoattractant. After 24 hours, cell transmigrating inserts were detached in the lower chamber using Accutase (Innovative Cell Technologies) containing 1 μM calcein-AM fluorescent cell labeling dye and incubated at 37°C/5% CO2 for 30 minutes. Microplates were assayed for fluorescence at 485 nm excitation/520 nm emission. Relative fluorescence units were converted to absolute cell number using a standard curve, and cell invasion was quantified as the ratio of invaded cells to the starting cell number.
Promoter methylation analysis.
Pyrosequencing was used to quantify methylation of selected CpG sites in the BVES promoter. We designed a pyrosequencing assay that covers 7 CpG sites in the BVES CpG island (as shown in Figure 3A). Tumor and matched normal adjacent mucosa were homogenized and digested overnight with proteinase K. Clinical sample DNA was provided by Christina Bailey (Vanderbilt University) and quantitated using a Nanodrop instrument (Thermo Scientific); 100–500 ng DNA per sample was bisulfite modified, using a Zymo EZ Methylation Direct Kit (Zymo Research). Modified DNA (40 ng per reaction) was amplified by PCR, using 0.2 μM of each primer, 2 units of hot start Taq DNA polymerase, and 0.2 mM of each dNTP per reaction. Primer sequences used are as follows: Bves, forward, Biotin-TGGGAGTTGAGGTTATAGAGATTTT, and reverse, ACCAAACTCACCAAAAAACTTACC. Cycling programs were 95°C for 15 minutes, then 51 cycles of 95°C for 30 seconds, 51°C for 30 seconds, and 72°C for 30 seconds, followed by a 5-minute incubation at 72°C. PCR products were examined after gel electrophoresis in 1.5% agarose in order to confirm that a single band was obtained. In preparation for pyrosequencing, the biotinylated strand of each PCR product was isolated from 15 μl of each PCR product using a Vacuum Prep Tool (Qiagen), according to the manufacturer’s protocol. Pyrosequencing reactions were performed in a PyroMark MD Pyrosequencing instrument (Qiagen) in the presence of 500 nM sequencing primer (AAACTCACCAAAAAACTT), following the manufacturer’s recommendations. The positive control for methylation was methylated HeLa DNA (New England Biolabs); the negative control was normal human blood DNA (Promega). Positive and negative controls were run with each experiment. Pyrosequencing assays contained an internal control for incomplete bisulfite modification.
RNA expression analysis.
RNA was isolated from LIM2405, HCT-15, HCA-7, OVCAR-3, CACO-2bbe, HT-29, DLD-1, SW480, Colo201, HCT-8, and HCT-7 cells using the RNAEasy Mini Kit (Qiagen). One μg of total RNA was used as template for reverse transcription using the iScript cDNA Synthesis Kit (Bio-Rad). qRT-PCR was performed using human BVES/PopDC1/Pop1 (Hs00362584_m1; Applied Biosystems), PopDC2 (Hs00223273_m1; Applied Biosystems), and PopDC3 (Hs01592415_m1; Applied Biosystems) TaqMan probes according to the manufacturers protocol on a MyIQ (Bio-Rad). Relative expression was standardized to GAPDH and calculated using the comparative cycle threshold method (2–ΔΔCt) (51). Reaction products were run out on a 2% Tris-acetate EDTA agarose gel. BVES TaqMan qRT-PCR was performed on a multicancer tissue cDNA array (TissueScan Cancer Survey I, OriGene) according to the manufacturers protocol.
Microarray experiments — human tissues and microarray platform.
Representative sections of fresh tissue specimens were flash frozen in liquid nitrogen and stored at –80°C until RNA isolation. Quality assessment slides were obtained to verify the diagnosis of cancer or normal adjacent mucosa. Stage was assessed using American Joint Commission on Cancer guidelines for both cohorts of tumor samples. RNA for human tissue was purified using the RNeasy Kit (Qiagen). Samples were hybridized to the Human Genome U133 Plus 2.0 GeneChip Expression Affymetrix array. The expression array data has been deposited, and the GEO accession number is GSE17538. Complete minimum information about a microarray experiment–compliant data sets for analysis are available ( http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17538).
Statistics.
For all presented experiments involving comparison among more than 2 groups, multiple comparison 1-way ANOVA was used. For experiments in which 2 groups were compared, either a 2-tailed Student’s t test or a Mann-Whitney U test was used. Values were considered significant if P was less than 0.05. Where error bars are presented, they represent ± SEM, unless otherwise stated. Microarray data were normalized with the Robust MultiChip Averaging algorithm (52) as implemented in the Bioconductor package Affy. For pair-wise group comparisons, the t test in the Limma package (53) in Bioconductor was used to identify differentially expressed probe sets between the 2 groups under comparison (e.g., normal adjacent specimens versus stage I cancers). The implementation of t test in Limma uses an empirical Bayes method to moderate the standard errors of the estimated log-fold changes; this results in a more stable inference, especially for experiments with a small number of arrays.
Study approval.
The protocols and procedures for this study were approved by the Institutional Review Boards at University of Alabama-Birmingham Medical Center, Vanderbilt Medical Center, Veterans Administration Hospital (Nashville, Tennessee, USA), and H. Lee Moffitt Cancer Center, and informed consent was obtained from each subject per institutional protocol.
Supplementary Material
Acknowledgments
We thank Richard Peek, Keith Wilson, William Grady, Scott Hiebert, Raymond DuBois, Maria S. Balda, and Karl Matter for discussions helpful in conducting the experiments presented in this manuscript. We also thank Amber Burch for reviewing the manuscript, Christina Bailey for providing DNA from clinical samples for the methylation analysis, and Natasha Deane for assistance with the splenic metastasis model. This work was supported by NIH grant K08 DK080221-01 (to C.S. Williams). This work was also funded by DDRC Pilot award P30 DK058404 (to C.S. Williams and M.S. Chang) and GI SPORE CDA award 5P50CA095103 (to C.S. Williams). This work was supported by the following grants from the NIH: CA069457 (to R.D. Beauchamp); TL1 RR024978 (to J.J. Smith); CA106183 (to J.J. Smith); T32 GM07347 (to J.J. Smith); CA112215 (to T.J. Yeatman); CA068485 (to R.D. Beauchamp); CA077839 (to R.D. Beauchamp); National Eye Institute (EY017185) (to M.S. Chang), The Leslie and Judy Smith Discovery Grant (to M.S. Chang), and P30 EY008126 (Core Grant in Vision Research). Other sources of funding were as follows: Society of University Surgeons-Ethicon Scholarship Award (to J.J. Smith), the Knights Templar Eye Foundation (to A. Jayagopal), and a Research to Prevent Blindness Robert E. McCormick Scholar Award and an unrestricted research grant to the Vanderbilt Eye Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(10):4056–4069. doi:10.1172/JCI44228.
References
- 1.Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61(4):514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4<514::AID-JCB4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 2.Reese DE, Bader DM. Cloning and expression of hbves, a novel and highly conserved mRNA expressed in the developing and adult heart and skeletal muscle in the human. Mamm Genome. 1999;10(9):913–915. doi: 10.1007/s003359901113. [DOI] [PubMed] [Google Scholar]
- 3.Reese D, Zavaljevski M, Streiff N, Bader D. bves: A novel gene expressed during coronary blood vessel development. Dev Biol. 1999;209(1):159–171. doi: 10.1006/dbio.1999.9246. [DOI] [PubMed] [Google Scholar]
- 4.Ripley AN, Osler ME, Wright CV, Bader D. Xbves is a regulator of epithelial movement during early Xenopus laevis development. Proc Natl Acad Sci U S A. 2006;103(3):614–619. doi: 10.1073/pnas.0506095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 6.Thoreson MA, Reynolds AB. Altered expression of the catenin p120 in human cancer: implications for tumor progression. Differentiation. 2002;70(9–10):583–589. doi: 10.1046/j.1432-0436.2002.700911.x. [DOI] [PubMed] [Google Scholar]
- 7.Yap AS. The morphogenetic role of cadherin cell adhesion molecules in human cancer: a thematic review. Cancer Invest. 1998;16(4):252–261. doi: 10.3109/07357909809039774. [DOI] [PubMed] [Google Scholar]
- 8.Van Aken J, Cuvelier CA, De Wever N, Roels J, Gao Y, Mareel MM. Immunohistochemical analysis of E-cadherin expression in human colorectal tumours. Pathol Res Pract. 1993;189(9):975–978. doi: 10.1016/S0344-0338(11)80667-9. [DOI] [PubMed] [Google Scholar]
- 9.Dhawan P, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115(7):1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang TL, et al. Claudin-1 has tumor suppressive activity and is a direct target of RUNX3 in gastric epithelial cells. Gastroenterology. 2010;138(1):255–265. doi: 10.1053/j.gastro.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 12.Debruyne D, Oliveira MJ, Bracke M, Mareel M, Leroy A. Colon cancer cells: pro-invasive signalling. Int J Biochem Cell Biol. 2006;38(8):1231–1236. doi: 10.1016/j.biocel.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Smith JJ, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam KT, et al. Loss of Rab25 promotes the development of intestinal neoplasia in mice and is associated with human colorectal adenocarcinomas. . J Clin Invest. 2010;120(3):840–849. doi: 10.1172/JCI40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aigner K, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26(49):6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osler M, Chang M, Bader D. Bves modulates epithelial integrity through an interaction at the tight junction. J Cell Sci. 2005;118(pt 20):4667–4678. doi: 10.1242/jcs.02588. [DOI] [PubMed] [Google Scholar]
- 17.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rousset M, et al. Enterocytic differentiation and glucose utilization in the human colon tumor cell line Caco-2: modulation by forskolin. J Cell Physiol. 1985;123(3):377–385. doi: 10.1002/jcp.1041230313. [DOI] [PubMed] [Google Scholar]
- 19.Whitehead RH, Zhang HH, Hayward IP. Retention of tissue-specific phenotype in a panel of colon carcinoma cell lines: relationship to clinical correlates. Immunol Cell Biol. 1992;70(pt 4):227–236. doi: 10.1038/icb.1992.30. [DOI] [PubMed] [Google Scholar]
- 20.Wada AM, Reese DE, Bader DM. Bves: prototype of a new class of cell adhesion molecules expressed during coronary artery development. Development. 2001;128(11):2085–2093. doi: 10.1242/dev.128.11.2085. [DOI] [PubMed] [Google Scholar]
- 21.Kawaguchi M, Hager HA, Wada A, Koyama T, Chang MS, Bader DM. Identification of a novel intracellular interaction domain essential for Bves function. PLoS ONE. 2008;3(5):e2261. doi: 10.1371/journal.pone.0002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasavada TK, DiAngelo JR, Duncan MK. Developmental expression of Pop1/Bves. J Histochem Cytochem. 2004;52(3):371–377. doi: 10.1177/002215540405200308. [DOI] [PubMed] [Google Scholar]
- 24.Balda MS, Matter K. Tight junctions and the regulation of gene expression. Biochim Biophys Acta. 2009;1788(4):761–767. doi: 10.1016/j.bbamem.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Vietor I, Bader T, Paiha K, Huber LA. Perturbation of the tight junction permeability barrier by occludin loop peptides activates beta-catenin/TCF/LEF-mediated transcription. EMBO Rep. 2001;2(4):306–312. doi: 10.1093/embo-reports/kve066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119(6):1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Andree B, Fleige A, Arnold HH, Brand T. Mouse Pop1 is required for muscle regeneration in adult skeletal muscle. Mol Cell Biol. 2002;22(5):1504–1512. doi: 10.1128/MCB.22.5.1504-1512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth MJ, et al. p16, MGMT, RARbeta2, CLDN3, CRBP and MT1G gene methylation in esophageal squamous cell carcinoma and its precursor lesions. Oncol Rep. 2006;15(6):1591–1597. [PubMed] [Google Scholar]
- 30.Lioni M, et al. Dysregulation of claudin-7 leads to loss of E-cadherin expression and the increased invasion of esophageal squamous cell carcinoma cells. Am J Pathol. 2007;170(2):709–721. doi: 10.2353/ajpath.2007.060343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osanai M, Murata M, Chiba H, Kojima T, Sawada N. Epigenetic silencing of claudin-6 promotes anchorage-independent growth of breast carcinoma cells. Cancer Sci. 2007;98(10):1557–1562. doi: 10.1111/j.1349-7006.2007.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kominsky SL, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22(13):2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 33.Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N. Epigenetic silencing of occludin promotes tumorigenic and metastatic properties of cancer cells via modulations of unique sets of apoptosis-associated genes. Cancer Res. 2006;66(18):9125–9133. doi: 10.1158/0008-5472.CAN-06-1864. [DOI] [PubMed] [Google Scholar]
- 34.Feng Q, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17(3):645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M, et al. Frequent silencing of popeye domain-containing genes, BVES and POPDC3, is associated with promoter hypermethylation in gastric cancer. Carcinogenesis. 2010;31(9):1685–1693. doi: 10.1093/carcin/bgq144. [DOI] [PubMed] [Google Scholar]
- 36.Moldvay J, Jäckel M, Páska C, Soltész I, Schaff Z, Kiss A. Distinct claudin expression profile in histologic subtypes of lung cancer. Lung Cancer. 2007;57(2):159–167. doi: 10.1016/j.lungcan.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 37.Sauer T, Pedersen MK, Ebeltoft K, Naess O. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology. 2005;16(4):193–198. doi: 10.1111/j.1365-2303.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 38.Gröne J, et al. Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of claudin-1, -8 and -12. Int J Colorectal Dis. 2007;22(6):651–659. doi: 10.1007/s00384-006-0197-3. [DOI] [PubMed] [Google Scholar]
- 39.Tamura G, et al. E-Cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst. 2000;92(7):569–573. doi: 10.1093/jnci/92.7.569. [DOI] [PubMed] [Google Scholar]
- 40.Guilford P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392(6674):402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 41.Utada Y, et al. Mapping of target regions of allelic loss in primary breast cancers to 1-cM intervals on genomic contigs at 6q21 and 6q25.3. Jpn J Cancer Res. 2000;91(3):293–300. doi: 10.1111/j.1349-7006.2000.tb00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen LL, et al. Allelic imbalance in selected chromosomal regions in ovarian cancer. Cancer Genet Cytogenet. 2002;139(1):1–8. doi: 10.1016/S0165-4608(02)00620-9. [DOI] [PubMed] [Google Scholar]
- 43.Hyytinen E-R, et al. Defining the region(s) of deletion at 6q16-q22 in human prostate cancer. Genes Chromosomes Cancer. 2002;34(3):306–312. doi: 10.1002/gcc.10065. [DOI] [PubMed] [Google Scholar]
- 44.Issa JP. Cancer prevention: epigenetics steps up to the plate. Cancer Prev Res (Phila Pa). 2008;1(4):219–222. doi: 10.1158/1940-6207.CAPR-08-0029. [DOI] [PubMed] [Google Scholar]
- 45.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159(3):1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Araki-Sasaki K, et al. An SV40-immortalized human corneal epithelial cell line and its characterization. Invest Ophthalmol Vis Sci. 1995;36(3):614–621. [PubMed] [Google Scholar]
- 47.Xu Q, et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116(6):883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 48.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13(8):680–685. doi: 10.1016/S0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 49.Williams CS, Tsujii M, Reese J, Dey SK, DuBois RN. Host cyclooxygenase-2 modulates carcinoma growth. J Clin Invest. 2000;105(11):1589–1594. doi: 10.1172/JCI9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez J, et al. Deletion of Mtgr1 sensitizes the colonic epithelium to dextran sodium sulfate-induced colitis. Gastroenterology. 2006;131(2):579–588. doi: 10.1053/j.gastro.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 53.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.