Abstract
Aging is a major risk factor for the progression of neurodegenerative diseases, including Huntington disease (HD). Reduced neuronal IGF1 or Irs2 signaling have been shown to extend life span in mice. To determine whether Irs2 signaling modulates neurodegeneration in HD, we genetically modulated Irs2 concentrations in the R6/2 mouse model of HD. Increasing Irs2 levels in the brains of R6/2 mice significantly reduced life span and increased neuronal oxidative stress and mitochondrial dysfunction. In contrast, reducing Irs2 levels throughout the body (except in β cells, where Irs2 expression is needed to prevent diabetes onset; R6/2•Irs2+/–•Irs2βtg mice) improved motor performance and extended life span. The slower progression of HD-like symptoms was associated with increased nuclear localization of the transcription factor FoxO1 and increased expression of FoxO1-dependent genes that promote autophagy, mitochondrial function, and resistance to oxidative stress. Mitochondrial function improved and the number of autophagosomes increased in R6/2•Irs2+/–•Irs2βtg mice, whereas aggregate formation and oxidative stress decreased. Thus, our study suggests that Irs2 signaling can modulate HD progression. Since we found the expression of Irs2 to be normal in grade II HD patients, our results suggest that decreasing IRS2 signaling could be part of a therapeutic approach to slow the progression of HD.
Introduction
Understanding the molecular mechanisms that coordinate nutrient homeostasis and life span is clinically important and scientifically challenging (1). In contrast to the lengthy and poorly defined degenerative processes associated with mortality during normal aging, Huntington disease (HD) is characterized by progressive neuronal dysfunction in selected brain regions (2). HD is caused by an expansion of unstable CAG repeats in exon 1 of the huntingtin gene (HTT) that encodes an elongated polyglutamine repeat (polyQ-HTT) (3). Several mouse models have been developed to investigate the pathophysiology of HD, including the transgenic expression of full-length polyQ-HTT, or R6/2 mice, which express a transgene that encodes exon 1 of the human mutant HTT (4–6). R6/2 mice are used widely to investigate neurodegeneration of HD, as they develop a progressive and fatal neurological disease resembling human HD (6). Although all tissues are potentially exposed to the consequences of the mutant HTT protein, striking neuropathological changes develop within the neostriatum: gross atrophy of the caudate nucleus and putamen is accompanied by marked neuronal loss and astrogliosis (7). Other tissues in R6/2 mice, including pancreatic β cells, can degenerate owing to mutant HTT, which can lead to glucose intolerance and diabetes (8).
The function of HTT in normal cell biology is unknown (2); however, mutant polyQ-HTT disrupts many cellular processes: autophagy, energy metabolism, gene transcription, clathrin-dependent endocytosis, intraneuronal trafficking, and postsynaptic signaling (9). In particular, mutant HTT represses the expression of PPARγ, coactivator 1 α (Pparγc1α), a transcriptional coactivator that regulates several metabolic processes, including mitochondrial biogenesis and respiration (10, 11). Recent evidence reveals early defects in mitochondrial function and CNS energy metabolism, which might initiate oxidative stress before the onset of behavioral and neuronal pathology (9, 12). Thus, protection against oxidative stress might slow the progression of HD.
A few common interventions consistently extend animal life span and attenuate neurodegeneration, including calorie restriction or reduced insulin or IGF signaling (13). Reduced insulin-like signaling extends the life span of C. elegans, Drosophila, and mice (14–16). Moreover, calorie restriction extends the survival of polyQ-HTT mutant mice, which suggests that it can protect against neurodegeneration (17). Calorie restriction increases mitochondrial biogenesis and protects against the effects of polyQ-HTT upon mitochondrial gene deletion, enzyme abnormalities, and oxidative damage (18–20). Calorie restriction and reduced insulin/IGF signaling might be linked, because fewer calories reduce the intensity and duration of insulin secretion needed to maintain peripheral glucose homeostasis, and reduced insulin signaling can enhance the expression of antioxidant enzymes and proteins that prevent cellular and DNA damage (21, 22).
In mice, insulin and IGF1 signaling have important effects in the central nervous system, including the regulation of energy homeostasis, neuronal survival, learning and memory, and animal life span (23). Insulin and IGF1 stimulate tyrosine phosphorylation of the IRS proteins (mainly Irs1 and Irs2), which activate the PI3K→AKT and mTOR signaling cascades in peripheral and central tissues (21). In mice, less IGF1 receptor (Igf1r) in the brain extends life span (24). Moreover, reduced Irs2 signaling in the brain can increase the life span of mice maintained on a high-energy diet by nearly 6 months: long-lived brain-specific heterozygous Irs2 knockout (Irs2+/–) mice are slightly overweight and hyperinsulinemic, but are more active and display greater systemic glucose oxidation (25). Thus, less brain Irs2 signaling can extend life span regardless of peripheral insulin resistance or the circulating insulin needed to prevent hyperglycemia.
The role of insulin/IGF→IRS2 signaling in the progression of neurodegeneration and life span is controversial (26, 27). Cell-based studies showed that activation of the IGF→AKT pathway protects neurons from polyQ-mediated toxicity (28, 29). However, the inactivation and nuclear exclusion of forkhead box O (FoxO) transcription factors by AKT signaling reduces the expression of genes that protect against oxidative stress and promote macroautophagy, which might exacerbate the accumulation and deleterious effects of polyQ-HTT (30–33). Consistent with this observation, autophagy-mediated clearance of polyQ-HTT triggered by Irs2 includes a PI3K-independent mechanism (34). Regardless, less neuronal Irs2 signaling attenuates the progression of neurodegeneration through the IGF1 signaling pathway in aggregate-prone mouse models of Alzheimer disease (35–37). In this report, we established the relation between Irs2 signaling and mitochondrial dysfunction during the progression of behavioral and neuronal pathology in R6/2 mice.
Results
Effect of Irs2 signaling on R6/2 mouse life span.
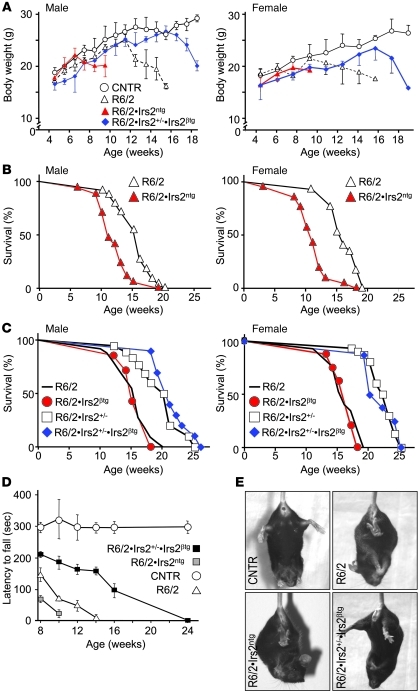
Many components of the insulin/IGF signaling cascade, including Irs2, are expressed in the striatum of patients with grade II Huntington disease (HD; Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI46305DS1). To investigate whether neuronal Irs2 modulates the progression of HD, we intercrossed transgenic B6CBA-Tg(HDexon1)62Gpb/3J mice (a strain of R6/2 mice with a transgene composed of the human HTT gene promoter and exon 1 with 120 ± 5 CAG repeats) with mice that express transgenic Irs2 in the CNS (Irs2ntg mice) to generate R6/2•Irs2ntg mice (Supplemental Figure 2A). R6/2 mice display a neurological phenotype that mimics many of the features of HD (5). The Irs2ntg mice grew to a normal body weight and displayed normal blood glucose concentrations (Supplemental Figure 2, B and C). Compared with control and R6/2 mice, Irs2 concentration and AKT phosphorylation increased significantly in brain extracts from random-fed Irs2ntg mice and R6/2•Irs2ntg mice (Supplemental Figure 2, D–F). Conversely, the mRNA concentration of the R6/2 transgene was indistinguishable in the extracts from R6/2 and R6/2•Irs2ntg mice (Supplemental Figure 2E). The body weights of male and female R6/2 mice increased normally until 12 weeks of age, then declined almost 40%, until the mice died between 15 and 16 weeks of age (Figure 1, A and B, and Supplemental Table 1). In contrast, the body weights of R6/2•Irs2ntg mice increased normally until 7 weeks, but afterward declined rapidly, and half the mice died between 11 and 12 weeks of age (Figure 1, A and B, and Supplemental Table 1). Based on Cox regression, neuronal Irs2 in R6/2•Irs2ntg mice significantly increased the risk of death 3.6-fold compared with R6/2 mice; sex of the mice was not a significant covariate (Supplemental Table 2).
Figure 1. Effect of Irs2 signaling on life span and motor function.
(A) Average body weight of male and female control (CNTR; B6CBAF1/3J without the R6/2 transgene), R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg mice (n = 20 per group). (B) Kaplan-Meier plots showing survival probability for R6/2 and R6/2•Irs2ntg male and female mice. (C) Kaplan-Meier plots showing survival probability for R6/2, R6/2•Irs2βtg, R6/2•Irs2+/–, and R6/2•Irs2+/–•Irs2βtg male and female mice. See Supplemental Tables 1 and 2 for details on statistical analysis. (D) Motor performance was measured biweekly using an accelerating Rotarod apparatus, and latency to fall was recorded for control, R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg male mice. Average time on the rod for each study group is shown (n = 20). (E) Representative images of limb clasping of experimental mice.
Next, we intercrossed R6/2 mice with Irs2+/– mice to generate R6/2•Irs2+/– mice. Male and female Irs2+/– mice express half the normal concentration of Irs2 throughout the body and live about 20% longer than controls (25). Consistent with the inverse relationship between Irs2 and life span, male and female R6/2•Irs2+/– mice displayed a longer life span than did R6/2 mice, even though the mRNA concentration of the human HTT transgene was unchanged (Supplemental Figure 2E). Most of the female R6/2•Irs2+/– mice survived to a mean age of 22.4 ± 0.4 weeks (Figure 1C) and never displayed hyperglycemia (blood glucose >200 mg/dl); however, 3 of the female and all of the male R6/2•Irs2+/– mice developed hyperglycemia between 4 and 8 weeks of age and survived to a mean age of 17.1 ± 0.5 and 19 ± 1 weeks, respectively (Supplemental Figure 3A and Supplemental Table 1).
Since mutant HTT can have a negative effect upon β cell function, and β cells without Irs2 fail sooner in males than in females (38, 39), we reasoned that Irs2 heterozygosity promoted β cell failure that contributed to the reduced life span of male R6/2•Irs2+/– mice. Based on our previous work showing that Irs2 promotes β cell growth, function, and survival (40), we intercrossed the R6/2•Irs2+/– mice with transgenic mice overexpressing Irs2 in pancreatic β cells (Irs2βtg mice). As expected, male R6/2•Irs2+/–•Irs2βtg mice displayed normal blood glucose (Supplemental Figure 3B). Moreover, the mean life span of male and female R6/2•Irs2+/–•Irs2βtg mice was indistinguishable and equal to that of the nondiabetic R6/2•Irs2+/– female mice (Figure 1C and Supplemental Table 1). Transgenic expression of Irs2 in β cells alone did not change the mean life span of R6/2•Irs2βtg compared with R6/2 mice (Figure 1C and Supplemental Table 1). Based on Cox regression using glucose, sex, and genotype as covariates, 50% less Irs2 expression throughout the body (except in β cells, where it was needed to prevent diabetes) significantly reduced the risk of death of R6/2 mice 13-fold; however, hyperglycemia (glucose >200 mg/dl) was a significant covariate that increased the risk of death about 3.4-fold (Supplemental Table 2). To avoid the complications of hyperglycemia in R6/2•Irs2+/– mice, we conducted subsequent experiments with male R6/2•Irs2+/–•Irs2βtg mice, which never developed high circulating glucose.
Effect of Irs2 signaling on motor function of R6/2 mice.
Motor function deteriorates as HD progresses, which we evaluated in mice by stability on the Rotarod, hind limb clasping, and paw print intensity (6). At all ages tested, control mice remained on the rod for about 300 seconds (Figure 1D). In contrast, 8-week-old R6/2 mice fell off the rod after 150 seconds, and fell sooner with increasing age. R6/2•Irs2ntg mice were less capable on the rod than were R6/2 mice, consistent with negative effects of more neuronal Irs2. Conversely, R6/2•Irs2+/–•Irs2βtg mice remained on the rod longer than R6/2 mice, but not as long as the controls (Figure 1D). Compared with R6/2 mice at 11 weeks of age, R6/2•Irs2+/–•Irs2βtg mice displayed fewer tremors and less frequent episodes of hind limb clasping while held in vertical suspension, whereas R6/2•Irs2ntg mice displayed more tremor and hind limb clasping (Figure 1E and Supplemental Figure 4, A and B).
Next, abnormal movement characteristic of HD was investigated using hind paw print patterns (41). The 11-week-old R6/2 and R6/2•Irs2ntg mice displayed weak and disperse paw prints, characteristic of HD, whereas paw print intensity and dispersion of R6/2•Irs2+/–•Irs2βtg mice were normal (Supplemental Figure 4, C–E). Consistent with these results, the fore limb grip of R6/2 and R6/2•Irs2ntg mice was significantly weaker than those of control and R6/2•Irs2+/–•Irs2βtg mice (Supplemental Figure 4F). Thus, reduced expression of Irs2 reduced the progression of motor abnormalities in R6/2 mice.
Effect of Irs2 signaling on brain neuropathology.
Serial brain sections were examined to assess astrogliosis (GFAP immunostaining), degenerated neuron accumulation (fluoro-jade immunostaining), and polyQ-HTT aggregate formation (5, 6). GFAP-positive astrocytes increased significantly in cortex and hippocampus sections from 11-week-old R6/2 and R6/2•Irs2ntg mice compared with controls, whereas the number of GFAP-positive astrocytes in R6/2•Irs2+/–•Irs2βtg sections was indistinguishable from the control (Figure 2, A and B). Punctate fluoro-jade immunostaining, which was entirely absent in control sections, was strongly detected in sections from 11-week-old R6/2 and R6/2•Irs2ntg mice, but was reduced significantly in R6/2•Irs2+/–•Irs2βtg sections (Supplemental Figure 5, A and B). Thus, relative to impaired motor function in short-lived R6/2•Irs2ntg mice, the neuropathological sequela did not worsen compared with R6/2 mice.
Figure 2. Effect of Irs2 signaling on brain neuropathology.
(A) Sections of the cortex and dentate gyrus (DG) from 11-week-old age-matched control, R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg mice immunostained with GFAP antibody. Scale bars: 100 μm (cortex); 20 μm (cortex, insets); 50 μm (DG). (B) GFAP signal, quantified from A. (C) Cortex from 6- and 11-week-old R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg mice stained with anti-Htt antibody (red). Nuclei are stained with DAPI (blue). Representative images of cortical neurons are shown. Scale bars: 50 μm. (D–G) Quantification of inclusion body frequency (D and E) and size (F and G) in cortex (D and F) and striatum (E and G). (H) Western blots of brain extracts prepared from 11-week-old control, R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg mice solubilized in Triton or SDS, resolved by SDS-PAGE, and probed with Htt antibody. Tubulin was used for normalization. (I) SDS-insoluble Htt from H, normalized to tubulin and expressed as a percentage of R6/2 (n = 4). *P < 0.05; **P < 0.01.
Htt antibody never detected inclusions in control mice (data not shown), whereas approximately the same number of inclusions, characteristic of HD, were detected in the neocortex and striatum of 6-week-old R6/2 and R6/2•Irs2ntg mice, and these increased equally by 11 weeks of age (Figure 2, C–E). However, compared with R6/2 mice, the frequency and size of the inclusions progressed more slowly in R6/2•Irs2+/–•Irs2βtg mice (Figure 2, D–G, and Supplemental Figure 5C). Consistent with these results, brain extracts from R6/2•Irs2+/–•Irs2βtg mice contained significantly less detergent-insoluble aggregated Htt than did R6/2 and R6/2•Irs2ntg mice (Figure 2, H and I). These findings showed that decreased expression of Irs2 reduced the progression of neuropathology in R6/2•Irs2+/–•Irs2βtg mice.
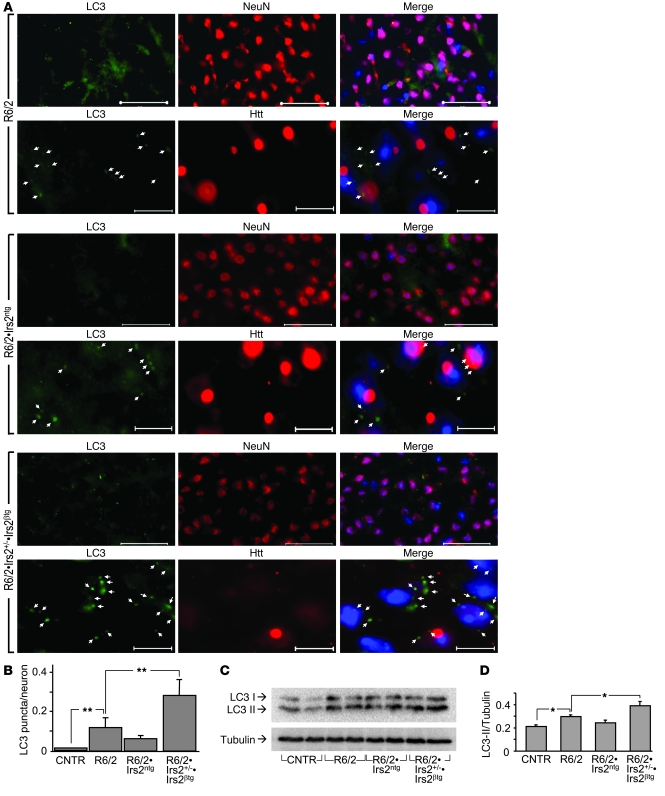
Macroautophagy might regulate the clearance of HTT aggregates in HD (42). Macroautophagy sequesters damaged organelles and unused long-lived proteins — including soluble and aggregate forms of mutant HTT — into double-membrane structures (autophagosomes) for degradation (42, 43). The expression of light chain type 3 (LC3; responsible for assembly of autophagosomes) and autophagy-related gene–6 (Beclin; involved in the recruitment of membranes to form autophagosomes) increased significantly in R6/2•Irs2+/–•Irs2βtg mice (Supplemental Figure 6). We used LC3 immunostaining to assess the assembly of autophagosomes in brain sections (44). LC3 immunostaining was barely detected in control samples, but increased substantially in neurons of R6/2 and R6/2•Irs2ntg mice (Figure 3A). Compared with control and R6/2 mice, the number of neurons stained by LC3 antibody increased significantly in R6/2•Irs2+/–•Irs2βtg mice (Figure 3, A and B). Moreover, the level of LC3-II, a marker of autophagosome induction, also increased significantly in R6/2•Irs2+/–•Irs2βtg mice (Figure 3, C and D). These findings show that increased macroautophagy might be responsible for the reduced HTT aggregates and slower HD progression in R6/2•Irs2+/–•Irs2βtg mice.
Figure 3. Effect of Irs2 signaling on autophagy in brains of R6/2 mice.
(A) Cortex sections from 11-week-old R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg mice stained with LC3 antibody (green). For each genotype, anti-NeuN antibody (as a neuronal marker) and anti-Htt antibody are also shown (both red). Nuclei were stained with DAPI (blue). Representative images of cortical neurons are shown. Arrows indicate LC3 puncta. Scale bars: 50 μm (top); 10 μm (bottom). (B) Quantification analysis of LC3 puncta per neuron in 10 sections per mouse (n = 5 per genotype). (C) Representative Western blot of LC3 of soluble fractions of total brain lysates. (D) LC3-II, normalized to tubulin (n = 4). *P < 0.05; **P < 0.01.
Effect of Irs2 signaling on FoxO1 activity in brains of R6/2 mice.
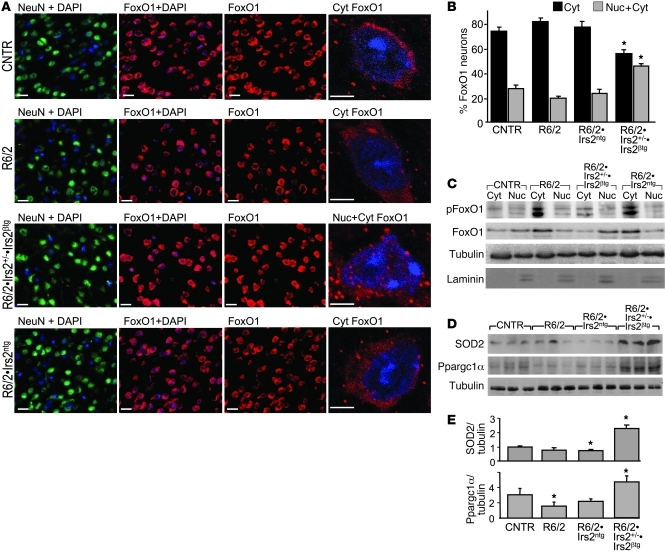
FoxO transcription factors induce the expression of genes that promote autophagy (LC3 and Beclin), mitochondrial function (Ppargc1a), and resistance to oxidative stress (Sod2), which can protect cells and tissues against physiologic stress (45). Insulin/IGF1 activates the PI3K→AKT signaling cascade, which includes the phosphorylation and nuclear exclusion of FoxO1 and FoxO3 (46). We immunostained striatum sections with specific FoxO1 antibodies to distinguish neurons without nuclear FoxO1 from those with nuclear FoxO1 (47). We identified neurons in these sections by immunostaining with NeuN and verified the location of the nucleus by DAPI staining (Figure 4A). Based on manual scoring, 30% of the neurons in control sections displayed nuclear FoxO1 staining, and 70% of the neurons displayed no evidence of nuclear FoxO1 staining (Figure 4, A and B). This distribution was not changed significantly in R6/2 and R6/2•Irs2ntg mice; however, R6/2•Irs2+/–•Irs2βtg sections displayed significantly more neurons with nuclear FoxO1 (Figure 4, A and B). The fractionation of brain homogenates and immunoblotting confirmed that phosphorylated FoxO1 was largely cytoplasmic in R6/2 and R6/2•Irs2ntg mice, whereas FoxO1 was weakly phosphorylated and most strongly detected in the nuclear (laminin-containing) fractions from R6/2•Irs2+/–•Irs2βtg brains (Figure 4C). Compared with control mice, levels of SOD2 and Ppargc1α (products of FoxO1-regulated genes) decreased in brain extracts of R6/2 and R6/2ntg mice, but significantly increased in brains of R6/2•Irs2+/–•Irs2βtg mice (Figure 4, D and E). Thus, reduced Irs2 signaling attenuated phosphorylation and cytoplasmic retention of FoxO1, which can increase the expression of genes in R6/2 mice that promote mitochondrial function and attenuate oxidative stress (11).
Figure 4. Effect of Irs2 signaling on FoxO1 activity in brains of R6/2 mice.
(A) FoxO1 localization (red) in brain sections of 11-week-old control, R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg mice. Sections were also stained with neuronal marker NeuN (green); nuclei were stained with DAPI (blue). A representative confocal cell image of FoxO1 localization is shown at right. Cyt, cytoplasmic; Nuc, nuclear. Scale bars: 20 μm; 5 μm (far right). (B) FoxO1-positive neurons were sorted into nuclear/cytoplasmic or cytoplasmic localization. (C) Western blot analysis of phospho-FoxO1S256 and total FoxO1 in cytoplasmic and nuclear fractions. Tubulin was used for normalization; laminin was used to confirm purity of the nuclear fraction. (D) Western blot of SOD2 and Ppargc1α expression in brains of 11-week-old mice. (E) Quantification of Ppargc1α and SOD2 expression, normalized to tubulin (n = 6). *P < 0.05 vs. control.
Effect of Irs2 signaling on oxidative energy balance in R6/2 mice.
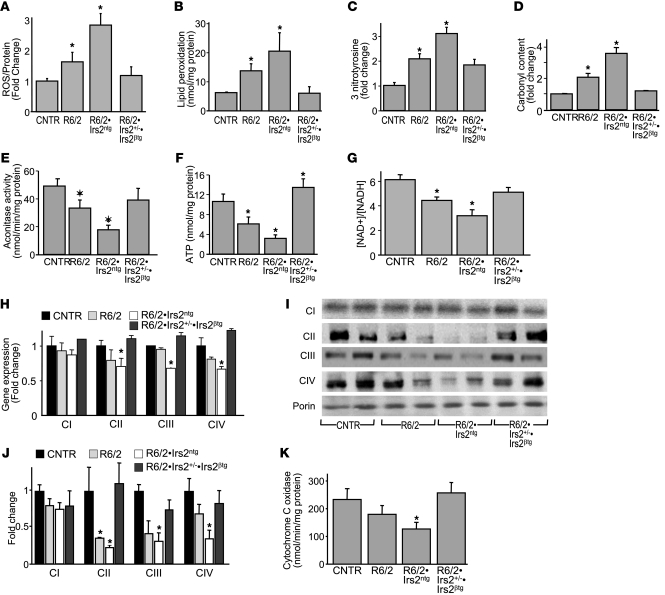
Previous work in livers of mice lacking both Irs1 and Irs2 suggests that nuclear FoxO1 attenuates oxidative stress, at least in part, by promoting mitochondria biogenesis that is modulated by the capacity of the electron transport chain (ETC) to control the [NAD+]/[NADH] ratio (48). Energy deficit is a well-known characteristic of HD, and oxidative damage was reported in early-stage HD progression in R6/2 mice, suggesting that mitochondrial dysfunction might promote neurodegeneration (9). Before evidence of overt neuronal dysfunction, full-length mutant polyQ-HTT can promote mitochondrial fission by interacting with dynamin-related protein–1 (12, 49). Thus, we next determined whether Irs2 modulates the concentration of ROS in the brains of R6/2 mice. Compared with 11-week-old control mice, ROS was slightly but significantly increased in age-matched R6/2 mice and increased 3-fold in R6/2•Irs2ntg mice; however, ROS in R6/2•Irs2+/–•Irs2βtg mice was indistinguishable from the control (Figure 5A). Increased ROS was detected in 5-week-old R6/2•Irs2ntg mice when the ROS concentration in age-matched R6/2 mice equaled that in controls (data not shown).
Figure 5. Effect of Irs2 signaling on oxidative energy balance in R6/2 mice.
(A–D) ROS concentration (A), lipid peroxidation (B), 3-nitrotyrosine levels (C), and carbonyl content (D) in brains of 11-week-old R6/2, R6/2•Irs2ntg, and R6/2•Irs2+/–•Irs2βtg mice (n = 6). (E–G) Mitochondrial aconitase activity (E), ATP levels (F), and [NAD+]/[NADH] ratio (G) of mitochondria from 11-week-old mice. (H) RT-PCR analysis of genes encoding mitochondrial proteins involved in oxidation phosphorylation (ETC). CI, complex I (mt-Nd6); CII, complex II (Sdha); CIII, complex III (Uqcrc1); CIV, complex IV (mt-Co1). (I) Expression pattern of protein markers of complexes I–IV (as in H). (J) Density quantification of bands in I, normalized to porin. (K) Cytochrome C oxidation rate (n = 3). *P < 0.05 vs. control.
ROS promotes cellular damage owing to lipid peroxidation, nitrosylation, and carbonylation (50, 51). Consistent with these previous results, the concentration of lipid peroxides, protein carbonyls, and 3-nitrotyrosine increased significantly in R6/2 mice (Figure 5, B–D). In contrast, these markers of oxidative damage increased further in R6/2•Irs2ntg brain extracts, but decreased to control levels in R6/2•Irs2+/–•Irs2βtg samples. Mitochondrial aconitase activity is highly sensitive to oxidative damage and can reflect the progression of cellular and tissue damage (52, 53). Compared with that in 11-week-old control mice, aconitase activity in the striata decreased 2-fold in R6/2 mice and 4-fold in age-matched R6/2•Irs2ntg mice, but was indistinguishable from control in R6/2•Irs2+/–•Irs2βtg mice (Figure 5E). Thus, less Irs2 signaling consistently reduced the oxidative stress induced by mutant polyQ-HTT in R6/2 mice.
Mitochondria are the principal source of ROS in cells (9). In HD, mitochondrial dysfunction is associated with ATP depletion and increased ROS (54). Compared with that in control brains, ATP concentration decreased 2- and 4-fold in R6/2 and R6/2•Irs2ntg brains, respectively, but increased significantly in R6/2•Irs2+/–•Irs2βtg brains (Figure 5F). Moreover, the [NAD+]/[NADH] ratio in the brains of R6/2 and R6/2•Irs2ntg mice decreased 1.5- and 2-fold, respectively, compared with the normal ratio in control and R6/2•Irs2+/–•Irs2βtg brains (Figure 5G). Thus, Irs2 signaling can augment the deleterious effects of mutant HTT upon mitochondrial function in R6/2 mice, whereas less Irs2 signaling can attenuate these damaging effects.
Next, we investigated the integrity of the ETC in brain extracts from control and R6/2 mice. The ETC is composed of a series of sequentially acting electron carriers that consist of integral membrane proteins with prosthetic groups (55). Complexes I and II pass electrons from NADH and FADH2 to complexes III and IV to generate the electrochemical proton gradient that drives ATP generation by complex V (55). Previous studies reveal deficiency of complexes II–IV in the caudate and putamen of symptomatic HD patients, and the reduction of complex II might precede the death of HD neurons (56). Consistent with the relative progression of mitochondrial dysfunction in R6/2 and R6/2•Irs2ntg mice, the expression and protein concentration of succinate dehydrogenase complex, subunit A, complex II (Sdha), ubiquinol-cytochrome c reductase core protein I, complex III (Uqcrc1), and cytochrome c oxidase subunit 1, complex IV (mt-Co1), were reduced in R6/2 and R6/2•Irs2ntg mice compared with controls. Importantly, these components were restored to normal concentrations in age-matched R6/2•Irs2+/–•Irs2βtg mice (Figure 5, H–J), consistent with improved mitochondrial function. The effect of Irs2 on the terminal step in the ETC was confirmed by progressive loss of cytochrome c oxidation rate in R6/2 and R6/2•Irs2ntg brain that was restored to control levels in R6/2•Irs2+/–•Irs2βtg mice (Figure 5K). Thus, less Irs2→PI3K→AKT signaling could attenuate the progression of HD in R6/2 mice by promoting mitochondrial function and diminishing oxidative stress associated with polyQ-HTT.
Discussion
Our results show that increased IRS2 signaling in the CNS (i.e., in Irs2ntg mice) exacerbates the deleterious consequences associated with mutant HD exon 1 in the brains of male and female R6/2 mice. Mice of the R6/2 strain used herein died between 15 and 16 weeks of age; however, 2-fold more Irs2 in the CNS decreased the life span of R6/2•Irs2ntg mice significantly, to 11–12 weeks. Among age-matched mice, R6/2•Irs2ntg mice were least capable during behavior tests and displayed the most severe oxidative stress and mitochondrial dysfunction. In contrast, R6/2•Irs2+/–•Irs2βtg mice — in which glucose intolerance owing to decreased insulin concentrations is prevented by transgenic β cell–specific Irs2 — showed delayed progression of all mutant HD-induced defects investigated, including behavioral and neuronal pathology, oxidative stress, and mitochondrial dysfunction. Based on these results and previous reports (25, 35, 36), we conclude provisionally that strategies to reduce neuronal Irs2 signaling have the potential to slow the progression of neurodegeneration, whether induced acutely by mutant HTT, chronically by Alzheimer disease, or by natural aging.
PolyQ-HTT causes pathological sequela in R6/2 mice that develop progressively until death — including mitochondrial dysfunction and oxidative stress, accumulation of protein aggregates in the CNS, neuroinflammation, and abnormal behavior and movement (6, 53, 57, 58). However, only mitochondrial dysfunction and oxidative stress increased significantly in the CNS of R6/2•Irs2ntg mice, revealing a major consequence of excess IRS2 signaling on acute neurodegeneration and early death of these HD mice. Irs2 signaling in the CNS can modulate the life span of normal mice, as partial or complete deletion of brain Irs2 increases life span by 20% (25). Our previous work shows that less neuronal Irs2 signaling preserves a youthful diurnal transition between glucose and fat oxidation ordinarily lost in old wild-type mice (25). Thus, excess Irs2 signaling might have negative effects on energy homeostasis during normal aging. Whether central IRS2 signaling exacerbates neuronal mitochondrial damage and oxidative stress during normal aging might have broad consequences for the progression of debilitating disease in an aging population.
Consistent with the inverse relationship between Irs2 expression and life span established in ordinary mice, less Irs2 signaling also significantly extended the life span of R6/2 mice. Whereas the molecular effects of Irs2 upon neuronal function are difficult to establish in ordinary mice, less Irs2 signaling was shown to have many protective effects in R6/2 mice. Mitochondrial function deteriorated more slowly in the R6/2•Irs2+/–•Irs2βtg brain, as the negative effects of PolyQ-HTT on ATP concentration and [NAD+]/[NADH] ratio were largely normalized in age-matched mice. Moreover, nuclear FoxO1 increased in the R6/2•Irs2+/–•Irs2βtg brain, which led to higher expression of Ppargc1α and SOD2, both of which have strong effects on energy homeostasis and prevention of oxidative stress: Ppargc1α promotes mitochondrial biogenesis and function needed for normal oxidative phosphorylation, and SOD2 reduces ROS damage (59–61).
The presence of polyQ-HTT aggregates in the striatum is one of the hallmarks of HD (62, 63). Aggregate accumulation is thought to reflect cellular damage, as cells that form aggregates earlier tend to die earlier (64). PolyQ-HTT aggregates might impair mitochondrial oxidative phosphorylation and contribute to free radical damage (53); however, aggregate formation could be a protective mechanism that sequesters damaged toxic monomers and small oligomers (65, 66). The accumulation of protein aggregates during HD progression might be caused by the instability of polyQ-HTT exacerbated by the effects of progressive oxidative stress. Although more Irs2 signaling exacerbated oxidative stress in R6/2•Irs2ntg mice, more Irs2 did not increase the aggregate load; however, comparable amounts of insoluble polyQ-HTT in R6/2 and R6/2•Irs2ntg mice does not rule out differences in the levels of toxic species in these mice.
The accumulation of fewer inclusions in the R6/2•Irs2+/–•Irs2βtg brain might be related to the improved redox status (67); however, we cannot exclude other modes of protection, such as increased autophagy to facilitate the clearance of protein aggregates. Increased nuclear FoxO-mediated gene expression owing to less Irs2 signaling can increase the expression of genes that mediated autophagy, including LC3 and Beclin. Compared with R6/2 or R6/2•Irs2ntg mice, more autophagosomes accumulated in the striatum of R6/2•Irs2+/–•Irs2βtg mice, consistent with fewer Htt aggregates in these samples.
Recent findings show that mutant polyQ-HTT causes mitochondrial fragmentation before aggregates accumulate in a mouse model of HD (12). Furthermore, 8-week-old R6/2 mice display overt lipid peroxidation in parallel with impaired motor abilities (9). Similar to this, the alterations in brain and muscle metabolism in patients with presymptomatic stages of HD suggest energy deficit to be an early phenomenon in the cascade of events leading to HD pathogenesis (68, 69). In agreement with these observations, the most vulnerable target of oxidative stress, mitochondrial aconitase, decreased and ATP synthesis fell in short-lived R6/2•Irs2ntg mice. Although we did not detect mitochondrial fragmentation in our EM studies (data not shown), the respiratory chain complexes II and III were progressively damaged in R6/2 and R6/2•Irs2ntg mice, but protected in R6/2•Irs2+/–•Irs2βtg mice. Studies of symptomatic HD patients showed marked deficiency of complexes II and III and lesser deficiency of complex IV in the caudate or putamen (53, 70, 71). Thus, reduced mitochondrial capacity caused by the physiological stress of polyQ-HTT can cause stress-induced cellular death pathways and organ failure (72). Less Irs2 signaling can attenuate this destructive cascade.
We do not believe there is a physical interaction between Irs2 protein and HTT; instead, our results suggest that the interface between their functions occurs at the mitochondrial level. For example, mutant HTT abnormally interacts with the mitochondrial fission GTPase dynamin-related protein–1 (12), whereas less Irs2 signaling promotes mitochondrial oxidative phosphorylation capacity. Moreover, the protective effects of calorie restriction could be affected by transcription factors with redox sensitivities, such as Sirt1 (73). Sirt1 activation is neuroprotective in several models of HD (74, 75). In a HD C. elegans model, Sir2/SIRT1 deacetylate DAF16, which induces genes that promote oxidative stress resistance (76). Taken together, these data suggest that the point of interaction between Irs2 signaling and polyQ-HTT is at the level of transcription regulation.
Published reports construct a complicated relation between IGF1 signaling and progression of neurodegeneration. Recently, it was suggested that IGF1 expression is under the direct control of HTT, since IGF1 levels correlated with either wild-type or mutant HTT levels (77). However, the data collected on centenarians show that modulations in serum IGF1 does not necessarily correlate with IGF1R signaling (78). Reduced neuronal IGF1 might predict HD progression (77), whereas more plasma GH and IGF1 levels might predict a faster progression of cognitive decline in HD (79). In contrast, cell-based experiments suggest that more IGF1→PI3K/Akt signaling can inhibit mutant HTT–induced cell death and formation of intranuclear inclusions of polyQ-HTT in HD (28, 29). Regardless, heterozygous deletion of the IGF1R increases longevity in mice (80). Moreover, a recent study showed that a protection mechanism in an AD animal model involves effects of reduced IGF1 signaling on Aβ toxicity (37), pointing to the possibility that manipulation of the IGF signaling pathway can be a promising target for AD therapy.
In mammals, Irs2 signaling plays an important role in maintaining peripheral nutrient homeostasis and pancreatic β cell function. Deletion of Irs2 in the liver causes glucose intolerance, and deletion in pancreatic β cells causes diabetes (25, 81, 82). Thus, more Irs2 protected β cell function in R6/2 mice. However, insulin→Irs2 signaling displays tissue-specific effects, as less Irs2 signaling in the brain extends life span (25), consistent with the role of insulin/IGF1 signaling in the brain during aging. In support of this notion, several recent studies demonstrated that less Irs2 signaling also ameliorates the pathologies associated with AD (35, 36). Although the expression levels of Irs2 and other insulin signaling components were comparable in normal and grade II HD patients, we cannot rule out that their expression or activation might change with the pathologic grade. While we do not suggest that IRS2 signaling defects are the cause of HD, our murine studies nevertheless suggest that Irs2 signaling might modulate HD progression, which is consistent with our previous results on the healthy life span of old mice.
Taken together, our findings demonstrated that reduced Irs2 protected against oxidative stress, mitochondrial dysfunction, and neuropathology and delayed lethality in a mouse model of HD. Since Irs2 and other insulin signaling components are expressed in the human striatum, our findings in mice might be relevant to the progression of human HD. Since calorie restriction also delays HD progression in mice (17), we conclude provisionally that brain Irs2 signaling might integrate the effects of peripheral nutrient homeostasis and insulin signaling with life span in HD. This is an important conclusion, as work in the field moves forward to establish strategies to prevent neurodegeneration in general, and HD progression in particular. Although is it not obvious how to modulate Irs2 signaling in the brain, it is important to appreciate that increased neuronal Irs2 signaling is not a desirable goal in the intact mammal battling neurodegeneration.
Methods
Animals.
Male transgenic B6CBA-Tg (HDexon1)62Gpb/3J mice (Jackson Laboratories), a strain of R6/2 mice founders with a transgenic line of the human HD gene carrying approximately 120 ± 5 CAG repeat expansions, were bred with females from their background strain (B6CBAF1/3J). The length of the CAG repeats was monitored (Laragen) throughout the study. Brain Irs2 transgenic female mice expressing recombinant Irs2 under the control of the neural enolase promoter (Irs2ntg) were crossed to R6/2 male mice to generate R6/2•Irs2ntg mice. In addition, Irs2+/– mice (39) were crossed to Irs2βtg mice (40) to generate female Irs2+/–•Irs2βtg mice. Female Irs2+/–•Irs2βtg mice were further crossed to R6/2 male mice to generate R6/2•Irs2+/–•Irs2βtg mice.
Generation of Irs2ntg mice.
The FLAG tag sequence was added to the 3′ end of mouse Irs2 by PCR, and the tagged cDNA was then inserted directly after the rat neuronal enolase promoter using the SalI/HindIII sites. A linearized DNA fragment containing the Nse-Irs2 transgene was excised and microinjected into fertilized eggs of C57BL/6 mice according to standard techniques by the Beth Israel Deaconess Transgenic Mouse Facility. Germline transmission was confirmed by Southern blotting. Founders were bred with C57BL/6 mice and maintained on this pure background. Routine genotyping was executed by PCR using primers derived from an internal Irs2 sequence.
Survival analysis.
Mice were undisturbed except for routine cage maintenance until their date of death (the day when they no longer had a heartbeat). Although we did not determine the median life span of B6CBAF1/3J control mice, all control mice were alive at 18 months of age, when our observations were terminated (data not shown). Mice used for invasive experiments were maintained in the same facilities but not included in the life span study. To establish whether gender and date of birth influenced life span, we used semiparametric (Cox) regression (COXREG, version 18; SPSS) to control for these covariates (83). Categorical independent variables — representing genotype, sex, and blood glucose concentration — were replaced with a set of indicator variables, representing the presence or absence of category membership, and entered into the model as a single step. Backward stepwise removal of the covariates produced similar results; and the log-minus-log plots confirmed that the baseline hazard functions were proportional across groups of categorical variables. Based on the Cox regression, glucose was a significant confounder, whereas sex was not significant. Thus, Kaplan-Meier plots were used to estimate median and mean life span of the life span data stratified for blood glucose (version 15; SPSS).
Motor performance and body weight assessment.
The Rotarod apparatus (Columbus Instruments) was used to assess motor performance. Mice were evaluated weekly from 4 weeks of age. Mice were tested in 3 5-minute trials with an accelerating speed (0–40 rpm in 5 minutes) separated by a 30-minute interval. The time until a fall from the rod was recorded. Mice remaining on the rod for more than 300 s were removed, and their time was scored as 300 s. Body weights were obtained once a week at the same day and time.
Histological evaluation.
Experimental mice were anesthetized and perfused with 4% PFA. The brains were postfixed at least overnight in the same fixative solution plus 20% sucrose and cryoprotected as previously described (6). Immunofluorescent detection of mutant HTT, GFAP, LC3, and FoxO1 was performed as previously described (6, 11, 47). Sections were incubated overnight in mouse anti-HTT recognizing the N-terminal portion of human and murine HTT (Chemicon), rabbit anti-GFAP (Chemicon), rabbit anti-LC3 (Abgent), or rabbit anti-FoxO1 (Cell Signaling). Fluoro-jade B staining was performed according to the manufacturer’s protocol (Millipore). All fluorescent images were acquired with an Imager Z1 microscope (Zeiss) equipped with an AxioCam Mrm (CCD camera) and a LSM 510 (confocal). The images were quantified using Imaris (versions 6.4 and 7.0; Bitplane), using the function spot to count nuclei and surface to measure the area or the volume of the different objects.
Pyridine nucleotide measurements.
We analyzed the pyridine nucleotides (NAD+ and NADH) in brains with EnzyChrom assay kits according to the manufacturer’s instructions (BioAssay Systems).
Nitrotyrosine formation.
Detection and quantitation of 3-nitrotyrosine in protein samples was carried out with OxiSelect Nitrotyrosine Elisa kit (Cell Biolabs).
Protein carbonyls content.
The quantity of protein carbonyls in a brain protein sample was determined by derivatizing with dinitrophenylhydrazine (DNP) and measuring bound DNP according to the manufacturer’s instruction (Enzo Life Sciences).
Lipid peroxidation products.
The lipid peroxidation products level was evaluated using the Bioxytech LPO-586 kit (OxisResearch). The kit uses a chromatogenic reagent that reacts with the lipid peroxidation products malondialdehyde and 4-hydroxyalkenales.
ROS measurements.
ROS concentration in brain extracts was carried out using the cell-permeable dye 2′,7′-dichlorofluorescein diacetate (H2DCFDA). Brain tissues were snap-frozen in liquid nitrogen followed by quick thawing in buffered medium (5 mmol/l HEPES in PBS) to improve probe diffusion. After rapid thawing, medium was discarded. Samples were processed as previously described (48).
ATP levels.
Detection of ATP levels was carried out by StayBrite ATP Assay kit, according to the manufacturer’s instructions (BioVision).
Cytochrome c oxidase activity.
Cytochrome c oxidase activity in mitochondrial samples was determined using Cytochrome c Oxidase Assay Kit according to the manufacturer’s instruction (Sigma-Aldrich).
Aconitase activity.
Mitochondrial aconitase activity was measured using Aconitase Assay Kit according to the manufacturer’s instructions (Cayman Chemical).
Immunoprecipitation and Western blot.
Immunoprecipitation and immunoblotting were performed as described previously (40). Antibodies were as follows: rabbit anti-FoxO1, pThr24-FoxO1, Akt, IR, and tubulin (Cell Signaling Technology); rabbit anti-Pparg1α, Irs2, and Irs1 (Millipore); SOD2 (Santa Cruz Biotechnology); and mouse anti-Sdha, Uqcrc1, mt-Co1, and Nd6 (Abcam). Images were analyzed and quantified with Kodak MI software 4000MM Pro (version 5.02; Carestream Health Inc.). Mitochondria isolation and nuclear extracts of the studied brains were carried with Mitochondria isolation kit and NE-PER Nuclear and cytoplasmic extraction reagents kit, respectively, according to the manufacturer’s instruction (Thermo Scientific). Experiments with human samples were performed with postmortem grade II HD brains. Age-matched control brains were used in each experiment.
Real-time PCR.
Total RNA was extracted from brain with TRIzol (Gibco; Invitrogen), and reverse transcribed using iscript cDNA kit. cDNA was analyzed using SYBR Green real-time PCR (BioRad Laboratories Inc.). Each reaction was carried out in triplicate as described previously (40). The primers used for the R6/2 transgene were as follows: forward, CCCTGGAAAAGCTGATGAAG; reverse, GCTGTTGTTGCTGCTGTTGT.
Metabolic analysis.
Blood insulin was determined from tail bleeds using a Rat Insulin ELISA kit (Crystal Chem Inc.). Blood glucose levels were measured on random-fed or overnight-fasted animals in mouse-tail blood using Glucometer Elite (Bayer), and serum samples were collected for the insulin measurements. Intraperitoneal glucose tolerance test (d-glucose; 2 g/kg) was performed on mice fasted overnight for 16 hours as previously described (84). Blood glucose was measured 0, 15, 30, 60, and 120 minutes after insulin injection.
Gait analysis.
Animals’ hind paws were painted with nontoxic paint, and the animals were allowed to walk across an 84-cm uncovered corridor lined with paper. The amount of dispersion of the footprint reflected the degree of tremor and abnormal movements. A decrease in the dispersion of the footprints indicated that mice had less tremor and fewer abnormal movements. The intensity of the paw print represented the abnormal posture of the paws as well. The mean intensity of 4 steps from individual mice in either group was measured. Paw prints were scanned and analyzed using Kodak MI software 4000MM Pro (Carestream Health).
Limb movements.
Limb movements, in the form of clasping, were recorded at 11 weeks of age. Clasping phenotype was measured by suspending each mouse by its tail for 30 s and scoring as previously described (5): 0, no clasp; 1, mild clasp (only fore or hind limbs press into the stomach); 2, severe clasp (both fore and hind limbs touch and press into the stomach).
Grip strength analysis.
Grip strength analysis was performed as previously described (57). Briefly, a 100-g spring balance was supported horizontally in a box. One end was permanently fixed; the other was attached to a horizontal metal wire that was free to move, whose ends protruded through slots on the side of the box. To measure grip strength, a mouse was swung gently by the tail so that its fore limbs contacted the wire. The mouse instinctively gripped the wire and was pulled horizontally backward, exerting a tension on the spring balance. When the tension became too great, the mouse released the wire. The scale recorded the maximum load pulled. Each mouse was used for 5 consecutive tests, and the best 3 scores were used for statistical analysis.
Statistics.
All results were analyzed by generalized linear regression (version 18; SPSS), using the Bonferroni correction to adjust the observed significance level when multiple contrasts were tested, and expressed as mean ± SD. A P value less than 0.05 was considered statistically significant.
Study approval.
Animals involved in this study were approved by the IACUC of Children’s Hospital Boston. Human postmortem brain tissues were from obtained from the Harvard Brain Tissue Resource Center; IRB approval for these tissues was P-0001158 (approved for the use of discarded human material).
Supplementary Material
Acknowledgments
This project was supported by NIH grants DK38712, DK55326 (to M.F. White), and R01NS051303 (to D. Krainc) and by the Ellison foundation (to M.F. White and M. Sadagurski).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(10):4070–4081. doi:10.1172/JCI46305.
References
- 1.Wisse BE, Kim F, Schwartz MW. Physiology. An integrative view of obesity. Science. 2007;318(5852):928–929. doi: 10.1126/science.1148032. [DOI] [PubMed] [Google Scholar]
- 2.Stack EC, Ferrante RJ. Huntington’s disease: progress and potential in the field. Expert Opin Investig Drugs. 2007;16(12):1933–1953. doi: 10.1517/13543784.16.12.1933. [DOI] [PubMed] [Google Scholar]
- 3.Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 4.Phillips W, Michell A, Pruess H, Barker RA. Animal models of neurode-generative diseases. Methods Mol Biol. 2009;549:137–155. doi: 10.1007/978-1-60327-931-4_10. [DOI] [PubMed] [Google Scholar]
- 5.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 6.Stack EC, et al. Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington’s disease transgenic mice. J Comp Neurol. 2005;490(4):354–370. doi: 10.1002/cne.20680. [DOI] [PubMed] [Google Scholar]
- 7.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44(6):559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Bjorkqvist M, et al. The R6/2 transgenic mouse model of Huntington’s disease develops diabetes due to deficient beta-cell mass and exocytosis. Hum Mol Genet. 2005;14(5):565–574. doi: 10.1093/hmg/ddi053. [DOI] [PubMed] [Google Scholar]
- 9.Browne SE, Beal MF. Oxidative damage in Huntington’s disease patho-genesis. Antioxid Redox Signal. 2006;8(11–12):2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 10.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurode-generation. Cell Metab. 2006;4(5):349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Tran-scriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127(1):59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Song W, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17(3):377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes KA, Reynolds RM. Evolutionary and mechanistic theories of aging. Annu Rev Entomol. 2005;50:421–445. doi: 10.1146/annurev.ento.50.071803.130409. [DOI] [PubMed] [Google Scholar]
- 14.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120(4):449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice. Nature. 2001;414(6862):412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- 16.Partridge L, Gems D, Withers DJ. Sex and death: what is the connection? Cell. 2005;120(4):461–472. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 17.Duan W, Guo Z, Jiang H, Ware M, Li XJ, Mattson MP. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc Natl Acad Sci U S A. 2003;100(5):2911–2916. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton ML, et al. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98(18):10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKiernan SH, Tuen VC, Baldwin K, Wanagat J, Djamali A, Aiken JM. Adult-onset calorie restriction delays the accumulation of mitochondrial enzyme abnormalities in aging rat kidney tubular epithelial cells. Am J Physiol Renal Physiol. 2007;292(6):F1751–F1760. doi: 10.1152/ajprenal.00307.2006. [DOI] [PubMed] [Google Scholar]
- 20.Lee CM, Aspnes LE, Chung SS, Weindruch R, Aiken JM. Influences of caloric restriction on age-associated skeletal muscle fiber characteristics and mitochondrial changes in rats and mice. Ann N Y Acad Sci. 1998;854:182–191. doi: 10.1111/j.1749-6632.1998.tb09901.x. [DOI] [PubMed] [Google Scholar]
- 21.Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu Rev Physiol. 2008;70:191–212. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- 22.Narasimhan SD, Yen K, Tissenbaum HA. Converging pathways in lifespan regulation. Curr Biol. 2009;19(15):R657–R666. doi: 10.1016/j.cub.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freude S, Schilbach K, Schubert M. The role of IGF-1 receptor and insulin receptor signaling for the pathogenesis of Alzheimer’s disease: from model organisms to human disease. Curr Alzheimer Res. 2009;6(3):213–223. doi: 10.2174/156720509788486527. [DOI] [PubMed] [Google Scholar]
- 24.Kappeler L, et al. Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol. 2008;6(10):e254. doi: 10.1371/journal.pbio.0060254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317(5836):369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 26.Selman C, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2007;22(3):807–1. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 27.Taguchi A, White MF. Response to comment on “Brain IRS2 signaling co-ordinates life span and nutrient homeostasis.”. Science. 2008;320(5879):1012. doi: 10.1126/science.1152366. [DOI] [PubMed] [Google Scholar]
- 28.Colin E, et al. Akt is altered in an animal model of Huntington’s disease and in patients. Eur J Neurosci. 2005;21(6):1478–1488. doi: 10.1111/j.1460-9568.2005.03985.x. [DOI] [PubMed] [Google Scholar]
- 29.Humbert S, et al. The IGF-1/Akt pathway is neuroprotective in Huntington’s disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2(6):831–837. doi: 10.1016/S1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 30.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301(5638):1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 31.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17(9):422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Brault JJ, Schild A, Goldberg AL. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy. 2008;4(3):378–380. doi: 10.4161/auto.5633. [DOI] [PubMed] [Google Scholar]
- 33.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172(5):719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Killick R, et al. Deletion of Irs2 reduces amyloid deposition and rescues behavioural deficits in APP transgenic mice. Biochem Biophys Res Commun. 2009;386(1):257–262. doi: 10.1016/j.bbrc.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freude S, et al. Neuronal IGF-1 resistance reduces Abeta accumulation and protects against premature death in a model of Alzheimer’s disease. FASEB J. 2009;23(10):3315–3324. doi: 10.1096/fj.09-132043. [DOI] [PubMed] [Google Scholar]
- 37.Cohen E, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139(6):1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurlbert MS, Zhou W, Wasmeier C, Kaddis FG, Hutton JC, Freed CR. Mice transgenic for an expanded CAG repeat in the Huntington’s disease gene develop diabetes. Diabetes. 1999;48(3):649–651. doi: 10.2337/diabetes.48.3.649. [DOI] [PubMed] [Google Scholar]
- 39.Withers DJ, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391(6670):900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 40.Hennige AM, et al. Upregulation of insulin receptor substrate-2 in pancreatic beta cells prevents diabetes. J Clin Invest. 2003;112(10):1521–1532. doi: 10.1172/JCI18581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karpuj MV, et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med. 2002;8(2):143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 42.Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6(2):e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol. 2002;17(3):897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- 44.Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem. 2009;284(4):2363–2373. doi: 10.1074/jbc.M806088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117(4):421–426. doi: 10.1016/S0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 46.Cheng Z, Tseng Y, White MF. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol Metab. 2010;21(10):589–598. doi: 10.1016/j.tem.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitamura T, et al. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat Med. 2006;12(5):534–540. doi: 10.1038/nm1392. [DOI] [PubMed] [Google Scholar]
- 48.Cheng Z, et al. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15(11):1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schon EA, Manfredi G. Neuronal degeneration and mitochondrial dysfunction. J Clin Invest. 2003;111(3):303–312. doi: 10.1172/JCI17741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stentz FB, Umpierrez GE, Cuervo R, Kitabchi AE. Proinflammatory cytokines, markers of cardiovascular risks, oxidative stress, and lipid peroxidation in patients with hyperglycemic crises. Diabetes. 2004;53(8):2079–2086. doi: 10.2337/diabetes.53.8.2079. [DOI] [PubMed] [Google Scholar]
- 51.Berlett B, Stadtman E. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1999;272(33):20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 52.Yan LJ, Levine RL, Sohal RS. Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci U S A. 1997;94(21):11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabrizi SJ, et al. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann Neurol. 2000;47(1):80–86. doi: 10.1002/1531-8249(200001)47:1<80::AID-ANA13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 54.Milakovic T, Johnson GV. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J Biol Chem. 2005;280(35):30773–30782. doi: 10.1074/jbc.M504749200. [DOI] [PubMed] [Google Scholar]
- 55.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 56.Benchoua A, et al. Involvement of mitochondrial complex II defects in neuronal death produced by N-terminus fragment of mutated huntingtin. Mol Biol Cell. 2006;17(4):1652–1663. doi: 10.1091/mbc.E05-07-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hockly E, Woodman B, Mahal A, Lewis CM, Bates G. Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res Bull. 2003;61(5):469–479. doi: 10.1016/S0361-9230(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 58.Sathasivam K, et al. Identical oligomeric and fibrillar structures captured from the brains of R6/2 and knock-in mouse models of Huntington’s disease. Hum Mol Genet. 2010;19(1):65–78. doi: 10.1093/hmg/ddp467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 60.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444(7117):337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esposito L, et al. Reduction in mitochondrial superoxide dismutase modulates Alzheimer’s disease-like pathology and accelerates the onset of behavioral changes in human amyloid precursor protein transgenic mice. J Neurosci. 2006;26(19):5167–5179. doi: 10.1523/JNEUROSCI.0482-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57(5):369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 63.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 64.Gong B, Lim MC, Wanderer J, Wyttenbach A, Morton AJ. Time-lapse analysis of aggregate formation in an inducible PC12 cell model of Huntington’s disease reveals time-dependent aggregate formation that transiently delays cell death. Brain Res Bull. 2008;75(1):146–157. doi: 10.1016/j.brainresbull.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Schaffar G, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15(1):95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 66.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95(1):55–66. doi: 10.1016/S0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 67.Wyttenbach A, Sauvageot O, Carmichael J, az-Latoud C, Arrigo AP, Rubinsztein DC. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum Mol Genet. 2002;11(9):1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- 68.Aziz NA, van der Marck MA, Pijl H, Olde Rikkert MG, Bloem BR, Roos RA. Weight loss in neurodegenerative disorders. J Neurol. 2008;255(12):1872–1880. doi: 10.1007/s00415-009-0062-8. [DOI] [PubMed] [Google Scholar]
- 69.Goodman AO, et al. The metabolic profile of early Huntington’s disease--a combined human and transgenic mouse study. Exp Neurol. 2008;210(2):691–698. doi: 10.1016/j.expneurol.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 70.Stahl WL, Swanson PD. Biochemical abnormalities in Huntington’s chorea brains. Neurology. 1974;24(9):813–819. doi: 10.1212/wnl.24.9.813. [DOI] [PubMed] [Google Scholar]
- 71.Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapira AH. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39(3):385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 72.Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J Clin Invest. 2011;121(2):493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merry BJ. Oxidative stress and mitochondrial function with aging--the effects of calorie restriction. Aging Cell. 2004;3(1):7–12. doi: 10.1046/j.1474-9728.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 74.Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37(4):349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 75.Kumar P, Padi SS, Naidu PS, Kumar A. Effect of resveratrol on 3-nitropropionic acid-induced biochemical and behavioural changes: possible neuroprotective mechanisms. Behav Pharmacol. 2006;17(5–6):485–492. doi: 10.1097/00008877-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 76.Catoire H, et al. Sirtuin inhibition protects from the polyalanine muscular dystrophy protein PABPN1. Hum Mol Genet. 2008;17(14):2108–2117. doi: 10.1093/hmg/ddn109. [DOI] [PubMed] [Google Scholar]
- 77.Pouladi MA, et al. Full-length huntingtin levels modulate body weight by influencing insulin-like growth factor 1 expression. Hum Mol Genet. 2010;19(8):1528–1538. doi: 10.1093/hmg/ddq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suh Y, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105(9):3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saleh N, et al. High insulinlike growth factor I is associated with cognitive decline in Huntington disease. Neurology. 2010;75(1):57–63. doi: 10.1212/WNL.0b013e3181e62076. [DOI] [PubMed] [Google Scholar]
- 80.Holzenberger M. The role of insulin-like signalling in the regulation of ageing. Horm Res. 2004;62 suppl 1:89–92. doi: 10.1159/000080765. [DOI] [PubMed] [Google Scholar]
- 81.Lin X, et al. Dysregulation of insulin receptor substrate 2 in beta cells and brain causes obesity and diabetes. J Clin Invest. 2004;114(7):908–916. doi: 10.1172/JCI22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong X, Park S, Lin X, Copps K, Yi X, White MF. Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth. J Clin Invest. 2006;116(1):101–114. doi: 10.1172/JCI25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Allison PD.Survival Analysis Using SAS: A Practical Guide . Cary, North Carolina, USA: SAS Press; 1995. [Google Scholar]
- 84.Dong XC, et al. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8(1):65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.