Abstract
The specialized epithelial cell of the kidney, the podocyte, has a complex actin-based cytoskeleton. Dynamic regulation of this cytoskeleton is required for efficient barrier function of the kidney. Podocytes are a useful cell type to study the control of the actin cytoskeleton in vivo, because disruption of components of the cytoskeleton results in podocyte damage, cell loss, and a prototypic injury response called focal segmental glomerulosclerosis (FSGS). Searching for actin regulatory proteins that are expressed in podocytes, we identified a RhoA-activated Rac1 GTPase-activating protein (Rac1-GAP), Arhgap24, that was upregulated in podocytes as they differentiated, both in vitro and in vivo. Increased levels of active Rac1 and Cdc42 were measured in Arhgap24 knockdown experiments, which influenced podocyte cell shape and membrane dynamics. Consistent with a role for Arhgap24 in normal podocyte functioning in vivo, sequencing of the ARHGAP24 gene in patients with FSGS identified a mutation that impaired its Rac1-GAP activity and was associated with disease in a family with FSGS. Thus, Arhgap24 contributes to the careful balancing of RhoA and Rac1 signaling in podocytes, the disruption of which may lead to kidney disease.
Introduction
The kidneys filter plasma and reabsorb solutes and nutrients to maintain the appropriate extracellular environment. The proximal component of the nephron, the glomerulus, is the primary filtration barrier that prevents the loss of serum proteins into the primary filtrate. The glomerular filtration barrier consists of fenestrated endothelial cells, a thick glomerular basement membrane, and specialized epithelial cells (podocytes) arranged in series. Genetic or acquired diseases of the filtration barrier, especially of the basement membrane or the podocyte, result in the loss of serum proteins into the urine, progressive damage to the glomerulus, and loss in renal function (1).
The podocyte has a complex cellular architecture composed of a parachute-like cell body that attaches to the basement membrane through primary foot processes that in turn arborize into smaller actin-based secondary foot processes (2). In proteinuric kidney diseases, such as focal segmental glomerulosclerosis (FSGS) and minimal change disease, podocytes rearrange their actin cytoskeleton and retract or efface their foot processes (3). While it is unclear how this change results in the leakage of serum proteins into the urine, podocyte foot process effacement appears to be a key step in the breakdown of the filtration barrier. Studies to understand the molecular basis of podocyte foot process effacement in vitro have shown that, in response to stress, podocytes switch from a RhoA-dependent stationary state to a Cdc42- and Rac1-dependent migratory state (4, 5). These studies suggest the intriguing possibility that altered membrane dynamics and increased cell motility are the mechanisms underlying foot process effacement in vivo.
Since podocyte membrane reorganization is a common feature of proteinuric kidney diseases, we sought to understand the regulation of membrane dynamics of these cells. Using an in vitro model of podocyte differentiation, we found that podocytes reduced their membrane ruffling activity as they were differentiated. We found that decreased membrane ruffling in differentiated podocytes was dependent on the presence of the GTPase-activating protein (GAP), Rho-GAP 24 (Arhgap24). Previous work from Stossel and colleagues has shown that Arhgap24 (also known as Filamin A–binding RhoGAP [FilGAP]) is a GAP for Rac1 and that it suppresses lamellipodia formation and cell spreading downstream of RhoA signaling (6). Their work showed that the highest level of Arhgap24 transcript was present in the kidney. Here we show that Arhgap24 was highly expressed in podocytes of the kidney and was upregulated as these cells differentiate in vivo. The ARHGAP24 gene is highly conserved, implying an important role for the gene product. When we sequenced the DNA from patients with FSGS, we identified a loss-of-function mutation in the ARHGAP24 gene in a kindred with familial kidney disease. Taken together, these results suggest that Arhgap24 controls the RhoA-Rac1 signaling balance in podocytes that appear to be dysregulated in proteinuric kidney diseases, such as FSGS.
Results
Differentiated podocytes reduce ruffling of their cell membranes.
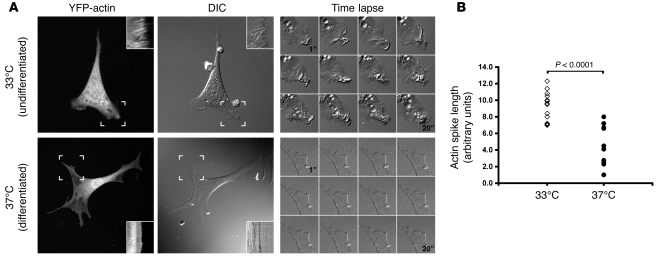
Podocytes can be propagated in vitro by conditional expression of a temperature-sensitive SV40 large T antigen at the permissive temperature of 33°C. Shifting the cells to the nonpermissive temperature (37°C) induces destabilization of the large T antigen, growth arrest, and morphologic changes that mimic podocyte differentiation in vivo (7). During development, podocytes reorganize their cell membranes from broad sheets into sieve-like foot processes (2). Conversely, in proteinuric diseases, podocytes lose this membrane complexity and exhibit effacement of their foot processes (3). Therefore, we hypothesized that the membrane dynamics of podocytes would be regulated in this in vitro model of cell differentiation. When undifferentiated podocytes were cultured at the permissive temperature, they exhibited highly ruffled plasma membranes (Figure 1A). In contrast, the plasma membranes of the differentiated podocytes had a very smooth, flat appearance. In order to quantitate these membrane dynamics, we performed time-lapse imaging of live podocytes transduced with yellow fluorescent protein–actin (YFP-actin). While undifferentiated podocytes rapidly ruffled their membranes, differentiated podocytes had reduced membrane motility, correlating with the reduced frequency of ruffled cell membranes (Supplemental Videos 1 and 2; supplemental material available online with this article; doi: 10.1172/JCI46458DS1). This difference is also apparent by kymographic analysis (Figure 1A, insets, and Figure 1B).
Figure 1. Differentiated podocytes show reduced membrane ruffling.
(A) YFP-actin–transduced podocytes were cultured at the permissive temperature (33°C) or differentiated for 7 to 14 days at the nonpermissive temperature (37°C). Kymographs (insets) obtained at the sites of active ruffling (hatched boxes) show prominent ruffling at 33°C that is reduced at 37°C (also see Supplemental Videos 1 and 2). DIC, differential interference contrast. Original magnification, ×400; approximately ×1,000 (insets and time-lapse panels). (B) Quantification of actin spike lengths in kymographs shows that differentiated podocytes (n = 11) have a significant decrease in ruffling activity compared with that of undifferentiated podocytes (n = 15). Individual symbols represent data from individual podocytes. P = 3.29 × 10–6 by ANOVA with post-test correction.
Podocytes upregulate Arhgap24 when they differentiate.
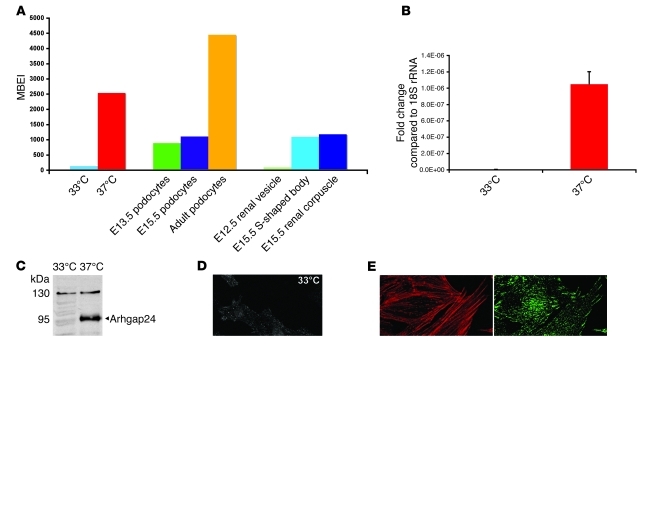
Next, we evaluated the expression of the small G proteins and their regulators that might control membrane motility in this in vitro model of podocyte differentiation. We isolated RNA from undifferentiated and differentiated mouse podocytes and performed gene expression analysis using microarrays. Our cultured podocytes expressed genes for several known podocyte-specific proteins, such as Wilms tumor protein 1 (Wt1), podocin (Nphs2), CD2-associated protein (Cd2ap), podocalyxin (Podxl), synaptopodin (Synpo), α-actinin-4 (Actn4), and to a lower extent nephrin (Nphs1). We focused our analysis on the expression patterns of known regulators of the actin cytoskeleton and membrane dynamics, such as GAPs and guanine nucleotide exchange factors (GEFs) for Rac1, RhoA, and Cdc42 (a list of these genes is available as a supplemental data table in the Gene Expression Omnibus (GEO) database, series GSE23856; http://www.ncbi.nlm.nih.gov/gds/). Of these known GAPs and GEFs, the message for Arhgap24 was highly upregulated when podocytes were differentiated in vitro. Arhgap24 was an intriguing gene, because previous work from Stossel and colleagues had shown high transcript levels in the kidney (6). However, the cell expressing Arhgap24 within the kidney was unknown. We found that RNA from in vivo–isolated podocytes also had a high level of Arhgap24 transcript (8). Next, we examined Arhgap24 gene expression in publicly available microarray data (the GenitoUrinary Molecular Anatomy Project; www.gudmap.org) generated from microdissected glomeruli isolated at various stages of kidney development. The Arhgap24 message was upregulated as glomeruli progressed from the E12.5 renal vesicle stage to the E15.5 S-shaped body to the almost mature E15.5 renal corpuscle (Figure 2A).
Figure 2. Arhgap24 transcript and Arhgap24 protein are specifically expressed in podocytes.
(A) MBEI values for microarray data for in vitro–cultured podocytes (33°C, 37°C), ex vivo–isolated podocytes (E13.5, E15.5, and adult), and laser-capture-microdissected glomeruli (E12.5 renal vesicle, E15.5 S-shaped body, and E15.5 renal corpuscle) show an increase in the Arhgap24 transcript with differentiation in vitro and in vivo. (B) By quantitative RT-PCR, differentiated podocytes have higher Arhgap24 mRNA levels compared with those of undifferentiated podocytes, after normalization to 18S rRNA. (C) Immunoblotting shows that differentiated podocytes have higher levels of Arhgap24 protein than undifferentiated podocytes. (D) Confocal imaging of cultured podocytes also shows an increase in levels of Arhgap24 in differentiated podocytes, concentrated in punctate structures at the base of the cell. (E) Arhgap24 colocalizes with the focal adhesion marker vinculin at the tips of actin stress fibers. Original magnification, ×600 (D and E).
Next, we confirmed the microarray results by quantitative RT-PCR. Arhgap24 transcript levels increased approximately 70 fold when podocytes were differentiated (Figure 2B). To confirm upregulation at the protein level, we generated Arhgap24-specific antiserum in Armenian hamsters and rabbits. Preabsorption of these antisera with Arhgap24 antigen abolished specific signal (Supplemental Figure 1). When podocytes were differentiated in vitro, they upregulated a 95-kDa band corresponding to the Arhgap24 protein (Figure 2C). The lower band seen in the undifferentiated podocytes (at approximately 50 kDa) did not correspond to the predicted molecular weight of known splice isoforms of Arhgap24. This band likely represents a specific degradation product of Arhgap24, perhaps due to increased turnover of the protein in undifferentiated podocytes. Confocal imaging of podocytes showed that Arhgap24 was enriched in structures at the base of the cell, and the staining intensity at these sites increased with differentiation (Figure 2D). Since Arhgap24 has been described to localize to focal adhesions (6), we verified that these structures were in fact focal adhesions by colabeling with vinculin (Figure 2E). These experiments showed that Arhgap24 is highly expressed in the focal adhesions of cultured podocytes and is upregulated as these cells are differentiated.
Arhgap24 is expressed in kidney podocytes in vivo.
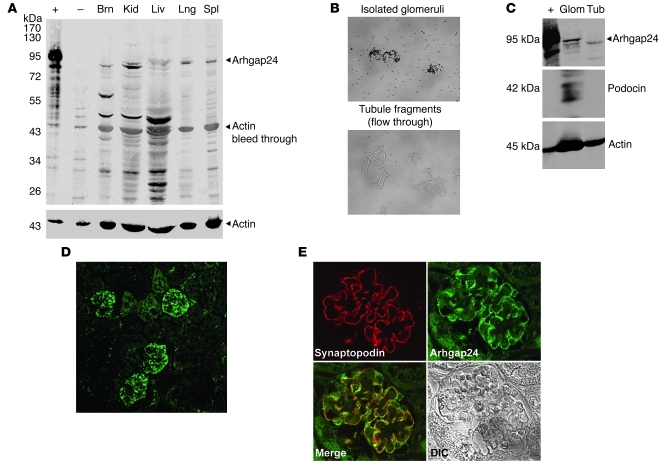
Next, we tested Arhgap24 expression and localization in murine tissues and its specific localization within the kidney. Immunoblotting of lysates from several tissues showed that the highest level of full-length Arhgap24 protein (95-kDa band) was in the kidney. Again, the approximately 50-kDa putative breakdown product was seen in lysates from the brain, kidney, and liver. We then evaluated whether Arhgap24 was expressed in the glomeruli of the kidney. We trapped magnetic particles within the glomeruli by beating heart perfusion and then used magnetic separation to isolate whole glomeruli to more than 95% purity (Figure 3B and ref. 9). Arhgap24 was enriched in the glomerular fraction (verified by podocin immunoblotting) compared with the flow through fraction that contained mostly tubule fragments (Figure 3C). Next, we stained sections of mouse kidney and detected the greatest signal within glomeruli (Figure 3D). This signal was specific since it was completely abolished by preabsorption of the antiserum with Arhgap24 antigen (Supplemental Figure 1C). Within the glomeruli, the Arhgap24 signal colocalized with the podocyte marker synaptopodin, confirming that Arhgap24 is expressed in podocytes in vivo (Figure 3E). Arhgap24 was expressed in podocytes, even at early stages of glomerular development in the embryonic mouse kidney (Supplemental Figure 2).
Figure 3. Arhgap24 is expressed in kidney podocytes in vivo.
(A) Immunoblotting of tissue lysates shows that Arhgap24 is expressed in the kidney (expected size ~95 kD). Smaller bands likely represent specific degradation products. The positive control (+) is a lysate from HEK293 cells transfected with FLAG-tagged Arhgap24. The negative control (–) is HEK293 cell lysate. Brn, brain; Kid, kidney; Liv, liver; Lng, lung; Spl, spleen. (B) Magnetic separation of glomeruli after beating heart perfusion of mice with magnetic beads results in more than 95% pure isolated glomeruli (top). The flow through consists of tubule fragments (bottom). Original magnification, ×100. (C) Arhgap24 is enriched in the glomerular (Glom) fraction that also contains the podocyte protein, podocin. Tub, tubule. (D) A low-magnification view (original magnification, ×200) of a mouse kidney stained for Arhgap24 shows that the highest signal is detected in the glomeruli. (E) Within mouse glomeruli, the Arhgap24 signal colocalizes with that of the podocyte marker synaptopodin. Original magnification, ×600.
Arhgap24 knockdown restores membrane ruffling in differentiated podocytes.
Having established that Arhgap24 is expressed in podocytes in vitro and in vivo, we next asked whether it was responsible for the dramatic decrease in membrane ruffling that we observed in differentiated podocytes (Figure 1). Membrane ruffling is dependent on the activity of the small G protein Rac1 (10). Rac1 cycles between an active, GTP-bound state and an inactive, GDP-bound state. When activated by GEFs, Rac1 interacts with multiple downstream effectors to stimulate actin nucleation and branching required for lamellipodia formation and membrane ruffling. Subsequently, Rac1 can be inactivated by GAPs that stimulate the intrinsic GTPase activity of Rac1, resulting in the hydrolysis of GTP to GDP. Previous work has shown that Arhgap24 is a GAP protein that inactivates Rac1 (6). Since Arhgap24 is upregulated in podocytes as they differentiate, it was a logical candidate to inactivate Rac1 and thereby slow membrane ruffling in differentiated podocytes.
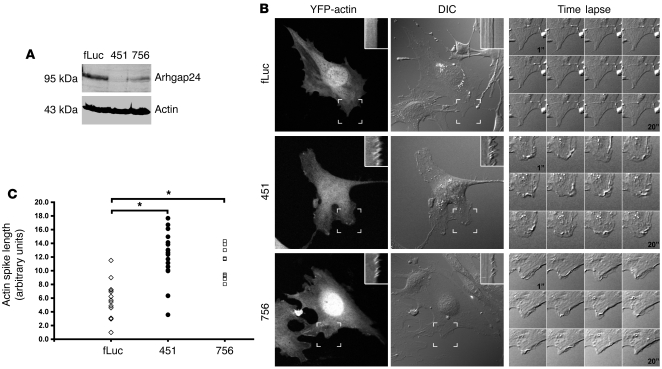
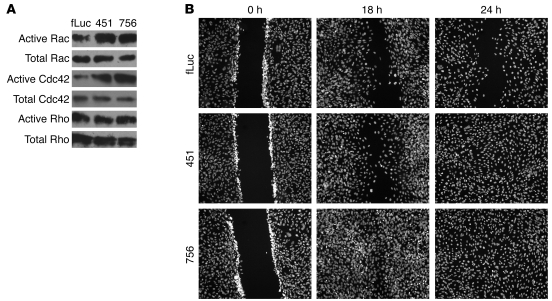
We investigated whether Arhgap24 upregulation was responsible for the reduced Rac1 activity and membrane motility of differentiated podocytes. We generated podocyte cell lines (lines 451 and 756) with 2 different Arhgap24 lentiviral knockdown constructs that coexpressed the YFP-actin reporter. Compared with that of the control vector (Fluc), the podocyte cell lines transduced with the 2 knockdown constructs resulted in approximately 85% (line 451) or 60% (line 756) downregulation of the Arhgap24 protein in differentiated podocytes (Figure 4A). After differentiation, the control and 2 Arhgap24 knockdown cell lines were assayed for their membrane motility as before. Compared with that of the control knockdown, Arhgap24 knockdown resulted in increased membrane motility in differentiated podocytes (Figure 4B and Supplemental Videos 3–5). Kymograph analysis showed that Arhgap24 knockdown significantly increased membrane motility (Figure 4C).
Figure 4. Arhgap24 knockdown in differentiated podocytes increases membrane ruffling.
(A) Two lentivirally transduced cell lines targeting different portions of the Arhgap24 transcript show marked reduction of Arhgap24 protein compared with that of an irrelevant knockdown (Fluc, 100%; line 451, 15%; line 756, 40%). Results are representative of at least 3 independent experiments. (B) Representative images of the 3 knockdown cell lines after differentiation at 37°C for 7 to 10 days. Kymographs (insets) generated from the hatched box area and time-lapse sequences show that the 2 knockdown cell lines have increased membrane ruffling compared with that of the control (also see Supplemental Videos 3–5). Original magnification, ×400; approximately ×1,000 (insets and time-lapse panels). (C) Quantitation of actin spikes in kymographs shows that both the 451 (n = 19) and 756 (n = 11) Arhgap24 cell lines have significantly greater membrane ruffling activity compared with that of the control knockdown Fluc cell line (n = 18) (*P < 0.00001). The ruffling activity is not significantly different between the 2 knockdown cell lines (P = 0.57). Group differences were analyzed by ANOVA with post-test correction.
Since Arhgap24 inhibits Rac1 and Cdc42 activity, we asked whether Arhgap24 knockdown resulted in increased levels of active Rac1 and Cdc42 in differentiated podocytes. We measured active Rac1, Cdc42, and RhoA by standard pull-down assays and found that the Arhgap24 knockdown cell lines had higher levels of active Rac1 and Cdc42 compared with those of the control knockdown cell line (Figure 5A). Interestingly, Arhgap24 knockdown did not affect RhoA activity. Another assay of Rac1 activity is epithelial monolayer wound closure. When a confluent monolayer of differentiated control knockdown podocytes was scratched/wounded, the cells migrated into the gap but did not completely close the wound in 24 hours. In contrast, both Arhgap24 knockdown cell lines showed accelerated wound closure kinetics, consistent with higher Rac1 activity (Figure 5B). Thus, Arhgap24 knockdown increases levels of active Rac1 and Cdc42. Two measures of increased Rac1 activity, membrane ruffling and epithelial wound closure, are enhanced when Arhgap24 levels are reduced in podocytes, consistent with its function as a Rac1-GAP.
Figure 5. Arhgap24 knockdown in differentiated podocytes increases active Rac1 and Cdc42 levels and accelerates epithelial monolayer wound closure.
(A) Pull down of active (GTP-bound) Rac1, Cdc42, and RhoA shows that the Arhgap24 knockdown cell lines (lines 451 and 756) have increased levels of active Rac1 and Cdc42 compared with those of the control (Fluc). However, active RhoA levels are similar across all 3 cell lines. Results are representative of 3 independent experiments. (B) Arhgap24 knockdown cells migrate and close a scratch made in a confluent monolayer faster than the control knockdown cell line. Cell nuclei were stained with Hoechst dye. Original magnification ×100.
Sequencing ARHGAP24 in patients with FSGS identifies several nonsynonymous changes in a highly conserved gene.
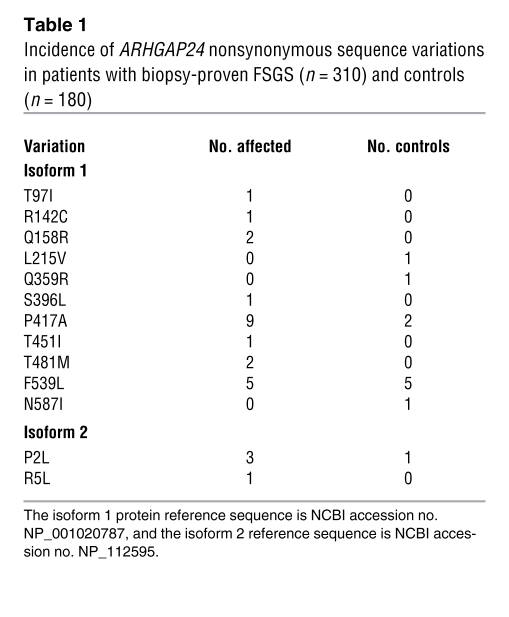
Given the podocyte-specific pattern of expression of Arhgap24 and its effects on membrane dynamics and epithelial wound healing, we next asked whether variations in ARHGAP24 were associated with kidney disease. The ARHGAP24 gene consists of at least 8 exons (depending on the splice isoform) that span approximately 500 kilobases on the long arm of human chromosome 4. Complete exon sequencing of the ARHGAP24 gene in 310 patients (620 chromosomes) with biopsy-proven FSGS and 180 controls (360 chromosomes) identified 13 nonsynonymous sequence variations in the 2 main splice isoforms of Arhgap24 (Table 1). Two of these (P417A and F539L) were found in both patients and controls and also in the database 1000 genomes ( www.1000genomes.org) (Supplemental Table 1). However, 7 of the nonsynonymous changes were only found in patients and not in any of the controls. These results suggest that variation in the ARHGAP24 gene may be linked with susceptibility to FSGS.
Table 1 .
Incidence of ARHGAP24 nonsynonymous sequence variations in patients with biopsy-proven FSGS (n = 310) and controls (n = 180)
Identification of an ARHGAP24 variation in familial FSGS.
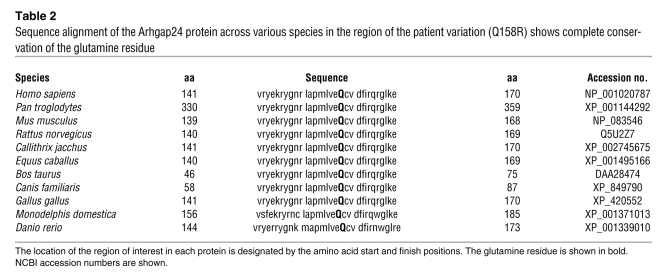
Most of the nonsynonymous changes that we detected in patients with FSGS were in the uncharacterized portions of the Arhgap24 protein. However, 1 of the sequence variations (Q158R) is located in the GAP domain of Arhgap24 close to the catalytic arginine (residue 175). This variation is predicted to result in a coding change that would substitute a positively charged arginine for a neutral polar glutamine at position 158 (isoform 1; NCBI accession no. NP_001020787) or position 65 (isoform 2; NCBI accession no. NP_112595). Remarkably, this residue is conserved in the ARHGAP24 gene across several model organisms that have been sequenced (Table 2). Of the other 2 nonsynonymous variations for which pedigree DNA was available (T97I and P417A), neither variation reliably correlated with disease status (Supplemental Figure 3).
Table 2 .
Sequence alignment of the Arhgap24 protein across various species in the region of the patient variation (Q158R) shows complete conservation of the glutamine residue
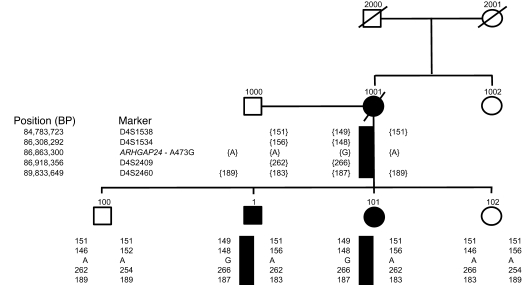
We first identified the Q158R variation in a Hispanic proband (Supplemental Figure 4). Next, we explored the association of this variation with kidney disease in his family (Figure 6). The proband (patient number 1) had elevated serum creatinine levels (16 mg/dl), and a biopsy that was performed at age 20 that showed FSGS. His sister (patient number 101) also had biopsy-proven FSGS that had progressed to end-stage kidney disease by age 12. The mother (patient number 1001) presented at a late stage and died at age 29 of renal failure. Sequencing showed that the patient’s affected sister and his mother both had the Q158R variation in the ARHGAP24 gene. In contrast, the proband’s 2 other siblings did not possess this variation. The brother (patient number 100) remains healthy, but the sister (patient number 102) had pregnancy-related proteinuria and hypertension. A biopsy was not performed on this individual, and she was lost to follow-up. However, an extended haplotype consisting of 4 flanking microsatellite markers (MSMs) showed that the haplotype of this sister was similar to that of the unaffected brother, making a disease-contributing rearrangement in this region of the genome unlikely (Figure 6).
Figure 6. Individuals with end-stage kidney disease (clinically and/or biopsy proven) are denoted by black symbols.
Deceased family members are represented by diagonal lines. Flanking MSM analysis shows that the unaffected siblings have a distinct haplotype that is different from that of the affected individuals. The columns of numbers and letters under each symbol refer to the alleles that individual carries at the given genetic markers. Where these numbers are within brackets, the haplotype is inferred. The solid black rectangle beneath individuals 1, 101, and 1001 represents the inherited disease haplotype.
In addition, the exon incorporating this variation was sequenced in an additional 554 control chromosomes (for a total of 914 control chromosomes) to decrease the probability that it represented a low-frequency noncausal variation. Altogether, we had sequenced over 900 control chromosomes across this span and did not detect the Q158R change. We also sequenced members of the affected kindred for mutations in genes known to cause hereditary FSGS and nephrotic syndrome, such as NPHS1, NPHS2, ACTN4, transient receptor potential cation channel, subfamily C, member 6 (TRPC6), phospholipase C, e1 (PLCE1), and WT1 (exons 8 and 9) and inverted formin, FH2 and WH2 domain containing (INF2) (data not shown). There were no suggestive mutations in any of these genes, ruling out known monogenic causes of FSGS in this family. Therefore, in this kindred with familial FSGS, the Q158R variation of the ARHGAP24 gene tracked with severe, early-onset kidney disease.
Arhgap24 Q158R has reduced Rac1-GAP activity.
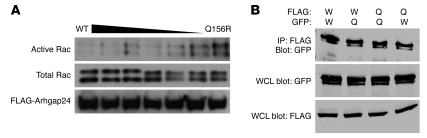
Another possibility was that a cosegregating mutation closely linked to the ARHGAP24 gene was responsible for early-onset kidney disease. We therefore tested whether the Q158R variation produced a defective form of the Arhgap24 protein. The Q158R variation is located close to the catalytically active site arginine residue of the GAP domain, and so we evaluated whether this change affected the Rac1-GAP activity of Arhgap24. For these experiments, we used the mouse Q156R protein, which corresponds to Q158R in human Arhgap24. As expected, wild-type Arhgap24 reduced the level of active Rac1 in cell lysates, showing again that it has GAP activity against Rac1 (Figure 7A). In contrast, transfection of the Q156R-mutated Arhgap24 resulted in a marked increase in the level of active Rac1. Titrating increasing levels of Q156R Arhgap24 against the wild-type protein confirmed that the Q156R mutation impairs the GAP activity of Arhgap24. Next, we tested whether the Q156R variant Arhgap24 could homodimerize and/or heterodimerize with the wild-type protein. FLAG- and GFP-tagged wild-type and Q156R versions of murine Arhgap24 were cotransfected into HEK293 cells. In bidirectional pull-down assays, we detected that both wild-type and Q156R Arhgap24 homodimerized and heterodimerized (Figure 7B). These experiments showed that the Q158R mutation reduces the enzymatic activity Arhgap24. The fact that Arhgap24 dimerizes may explain the observed dominant effect of this mutation in our family with inherited kidney disease.
Figure 7. Arhgap24 Q158R has defective Rac1-GAP activity and dimerizes with the wild-type protein.
(A) Wild-type FLAG-tagged Arhgap24 (first lane) transfected into HEK293 cells reduces active Rac1 levels compared with Q156R Arhgap24 (last lane). Titration of increasing proportions of Q156R Arhgap24 produces increased levels of active Rac1 (middle lanes). Total Rac1 and FLAG-Arhgap24 protein levels are similar across all lanes. Results are representative of 3 different experiments. (B) FLAG- or GFP-tagged wild-type (W) or Q158R Arhgap24 (Q) constructs were cotransfected into HEK293 cells. Cell lysates were immunoprecipitated with anti-FLAG antibody and then immunoblotted for GFP-tagged Arhgap24 to assess for protein dimerization. Whole cell lysates (WCLs) were immunoblotted for GFP and FLAG to ensure protein expression.
Discussion
Arhgap24, also known as FilGAP and p73RhoGAP, is the protein product of a highly conserved gene encoded on human chromosome 4. The longer isoform of Arhgap24, which we have used exclusively in this report, is highly expressed in the kidney (6). GAPs such as Arhgap24 and GEFs for small G proteins comprise a large family of proteins that have diverse patterns of spatial and temporal expression (11). Such a restricted pattern of expression may allow GAPs and GEFs to behave as cell-type specific effectors for ubiquitously expressed actin regulatory proteins. Here, we show that Arhgap24 is one such candidate for cytoskeletal regulation of the kidney podocyte.
The small G proteins, Rac1, Cdc42, and RhoA, control cell shape and motility through their effects on the actin cytoskeleton. In many systems, RhoA and Rac1 are mutually antagonistic (12, 13). However, the molecular basis of this counterregulation had been unclear until the discovery of Arhgap24. Stossel and colleagues showed that, in response to RhoA activation, Arhgap24 is phosphorylated by the RhoA effector kinase, ROCK, and this modification stimulates its GAP activity for Rac1 and Cdc42 (6). By catalyzing the conversion of Rac1 and Cdc42 to their inactive GDP-bound state, Arhgap24 inhibits cell spreading and cell motility. Recently, a similar mechanism was demonstrated for the closely related family member, Arhgap22, in melanoma cells (14). The diversity of GAPs likely ensures that specific ones may play a role in Rac1-RhoA counterregulation in different cell types and tissues.
Recent work from several groups has shown that the balance of RhoA and Rac1 signaling is carefully regulated in podocytes. Normally, the podocyte appears to be a stationary cell with predominantly active RhoA signaling. The podocyte-enriched adaptor protein synaptopodin controls a signaling module that promotes RhoA activity via 2 different mechanisms. Synaptopodin blocks Smurf1-mediated ubiquitination and subsequent proteasomal degradation of RhoA (15). Synaptopodin also disrupts IRSp53 binding to the Cdc42-Mena complex, interrupting Cdc42 signaling that opposes RhoA-mediated stress fiber formation (16). Synaptopodin itself is phosphorylated and protected from degradation by binding to 14-3-3 (17). Conversely, when it is dephosphorylated by calcineurin, synaptopodin is degraded by cathepsin L (17). The net result is that reduced synaptopodin levels lead to decreased RhoA levels and activity (15).
The RhoA-Rac1 signaling balance in podocytes is also responsive to hormonal stimulation. Binding of the vasoactive hormone angiotensin II to its receptor, angiotensin receptor type I (AT1R), leads to a calcium flux through the channels Trpc5 and Trpc6. Recent work by Tian et al. has shown that Trpc5 associates with Rac1 in membrane microdomains, while Trpc6 has a separate association with RhoA (18). In response to AT1R stimulation, Trpc5 activation leads to Rac1 activation and RhoA inactivation. Triggering Trpc6 has the opposite response, with increased RhoA activity and reduced Rac1 activity. Of note, mutations in the TRPC6 gene have already been associated with human FSGS (19). Therefore, this recent study by Tian et al. provides new insights into how signaling events downstream of the angiotensin receptor may lead to dynamic cytoskeletal reorganization in podocytes via selective modulation of RhoA and Rac1 activity through calcium signaling.
Decreased RhoA activity and increased Rac1 activity is associated with proteinuric kidney disease, consistent with the idea that an imbalance in the RhoA-Rac1 signaling balance is harmful to the podocyte. HIV infects podocytes and modulates the cytoskeleton of the cell such that patients with HIV are susceptible to collapsing FSGS (20). In podocytes, the HIV adaptor protein nef interacts with p190RhoGAP that then inactivates RhoA (21). In addition, nef binds to diaphanous interacting protein and the Rac1-GEF vav2, resulting in Rac1 activation (22). Nef also downregulates synaptopodin expression (23) and may further destabilize RhoA signaling via the synaptopodin pathway outlined above. The net effect of HIV infection is a downregulation of RhoA signaling activity and a shift toward Rac1 signaling with consequent podocyte injury.
There is further evidence from mouse models that disrupting the RhoA-Rac1 signaling balance in podocytes can cause kidney disease. Studies on mice deficient for the Rho-guanine nucleotide dissociation inhibitor, RhoGDIα (Arhgdia), demonstrate that Rac1 activation in podocytes is harmful. RhoGDIα binds to and stabilizes Rac1 and RhoA, and, in its absence, levels of these small G proteins and their activity levels are dysregulated (24). In particular, Rac1 is relatively overactivated, leading to podocyte foot process effacement and proteinuria (25, 26). Interestingly, treatment with a Rac1 inhibitor reduced proteinuria and the severity of the kidney damage in this model.
Taken together, these studies highlight that the careful control of relative RhoA and Rac1 activity is critical for the proper functioning of podocytes. However, we believe that until this report a direct regulator of Rac1 activity in response to RhoA signaling had not been described in podocytes. Here, we show that Arhgap24 is highly expressed in podocytes and inhibits Rac1-dependent membrane ruffling and epithelial wound healing. Our work predicts that Arhgap24 would enforce the normal RhoA-dependent nonmotile podocyte phenotype by inactivating Rac1. Consistent with a role for Arhgap24 in vivo, we identified a mutation in the GAP domain of Arhgap24 in a kindred with familial FSGS. This mutation reduces the Rac1-GAP activity of Arhgap24. We also show that Arhgap24 dimerizes, and this may explain the dominant pattern of inheritance of the Q158R mutation (27, 28). Our studies show that Arhgap24 is a potential candidate gene to explain a subset of inherited FSGS. Perhaps, more informatively, Arhgap24 adds to the emerging model that aberrant Rac1 activation is a key step in podocyte dysfunction, leading to proteinuric kidney disease. Since Arhgap24 is selectively expressed in podocytes, modulating its function can alter relative RhoA/Rac1 activity, with potentially minimal systemic side effects, and provides an intriguing pathway for the therapy of proteinuric kidney disease.
Methods
Podocyte cell culture.
Generation and propagation of conditionally immortalized murine podocyte cell lines has been described previously (7, 8). Briefly, podocytes were propagated on collagen I–coated dishes at 33°C (permissive temperature) in RPMI supplemented with 10% FBS and 10 U/ml of recombinant mouse IFN-γ (a gift from Robert Schreiber, Washington University School of Medicine). To induce differentiation, the medium was changed to RPMI with 5% FBS without IFN-γ, and the cells were shifted to 37°C (nonpermissive temperature) for 7 to 14 days. Under these conditions, cells underwent growth arrest, increased in size, and developed elongated cell processes.
For live cell imaging experiments, podocytes were lentivirally transduced with YFP-actin and purified by automated cell sorting. Control and knockdown cell lines were generated using a bicistronic lentiviral vector incorporating the targeting shRNA and YFP-actin expressed downstream of an internal ribosomal entry site. The control knockdown sequence targeted the firefly luciferase gene (Fluc). The 2 Arghap24 knockdown constructs targeted the sequences 5′-TTAAGGAGCTAATGAAACA-3′ (line 451) and 5′-TAACGATGGTCATAAGAAA-3′ (line 756), respectively. Stably transduced cell lines were generated by automated cell sorting for YFP expression.
RNA isolation.
Isolation of primary mouse podocytes using Dynabead perfusion and flow cytometric cell sorting has been described previously (9, 29). RNA was extracted from 6,000 primary podocytes and from cultured podocytes grown at the permissive or nonpermissive temperatures using an RNeasy Kit (Qiagen) following the manufacturer’s protocol. RNA quality was verified by gel electrophoresis and to ensure that the 260/280 nm absorbance ratio was greater than 1.8. A biotinylated cRNA target was generated and hybridized to an Affymetrix 430 v2.0 murine genome array (Affymetrix) following the manufacturer’s protocol.
Microarray analysis.
The data sets for primary isolated podocytes (GSM253173) and the undifferentiated (GSM588087) and differentiated mouse podocyte cell line (GSM253174) have been deposited in the GEO database (series GSE23856; http://www.ncbi.nlm.nih.gov/gds/). In addition to our own data, microarray data for E13.5 podocytes (GSM396532), E15.5 isolated podocytes (GSM309472), E12.5 renal vesicle (GSM144585), E15.5 S-shaped body (GSM144602), and E15.5 stage III–IV renal corpuscle (GSM144591) were obtained from the publicly available GenitoUrinary Molecular Anatomy Project ( www.gudmap.org).
Microarray data were analyzed using dChip software (30). Expression data were normalized to the median value, and model-based expression analysis (MBEI) was performed using the perfect match/mismatch (PM/MM) model. Briefly, MBEI represents a weighted average of the PM/MM differences (30). Results were filtered on gene lists for known GAPs and GEFs generated by querying the Affymetrix NetAffx database (results are deposited as a supplemental data table in the GEO database, series GSE23856; http://www.ncbi.nlm.nih.gov/gds/). GAPs or GEFs that changed at least 5 fold were chosen for manual curation (Supplemental Table 2). Of these genes, Arhgap24 was chosen for further analysis because of the magnitude of upregulation upon podocyte differentiation and quality of the probe set hybridization pattern.
Arhgap24 antiserum production.
Amino acids 390–604 of isoform 1 (NCBI accession no. NP_083546) of mouse Arhgap24 were cloned downstream of glutathione-S-transferase (GST) in the pGEX4T-1 expression vector. This portion of Arhgap24 is well conserved across species and lies downstream of the GAP domain in both isoforms of Arhgap24. GST-tagged Arhgap24 was expressed in BL21(DE3)pLysS E. coli. After induction with IPTG and sonication of bacteria, soluble GST-Arhgap24 was batch purified using glutathione-agarose beads. GST-Arhgap24 was eluted with reduced glutathione and dialyzed against PBS to remove excess glutathione. This antigen was emulsified in complete Freund’s adjuvant (Sigma-Aldrich) and was used to immunize rabbits and Armenian hamsters. Hamster handling and immunization were performed by the Washington University School of Medicine Hybridoma Center. Specificity for Arhgap24 was confirmed by immunoblotting FLAG-tagged Arhgap24 transfected into HEK293 cells. Specific signal in immunoblotting and staining experiments was confirmed by quenching of signal with antiserum preincubated with antigen.
Confocal imaging.
Podocytes grown on sterilized collagen I–coated coverslips at 33°C or 37°C were fixed with 1% PFA in PBS for 10 minutes. Cells were then blocked and permeabilized for 1 hour with PBS with 2.5% FBS and 0.1% saponin. Primary rabbit anti-Arhgap24 antiserum (1:300) in blocking buffer was applied to the cells for 1 hour at room temperature. After four 5-minute washes with blocking buffer, coverslips were mounted using ProLong Antifade mounting medium (Invitrogen) according to the manufacturer’s protocol. Images were captured with confocal settings using an Olympus FluoView FV1000 microscope. A similar protocol was used to stain Arhgap24 in formalin-fixed paraffin-embedded mouse kidney after antigen retrieval. Podocytes were stained using a mouse anti-synaptopodin monoclonal antibody (a gift from Peter Mundel, Massachusetts General Hospital, Boston, Massachusetts, USA) (31).
For wound healing experiments, differentiated knockdown podocytes were plated to confluence on collagen I–coated coverslips. A scratch was created using a sterile 200 μl pipette tip. Loosely adherent cells were washed away by 3 vigorous washes with PBS. The cells were transferred to culture medium, and, at various time points, coverslips were harvested and fixed in 4% PFA in PBS. Cells were identified by nuclear staining with Hoechst dye.
Tissue isolation and immunoblotting.
Whole mouse tissues (~200 mg) were snap frozen on dry ice and homogenized in cold lysis buffer containing 1% triton and protease inhibitors using a dounce homogenizer. Postnuclear supernatants were loaded to equalize actin levels by immunoblotting. Glomerular isolation/enrichment experiments using magnetic particles were performed as described previously (9).
Live cell imaging and kymograph analysis.
Podocytes that were lentivirally transduced with YFP-actin were cultured in glass bottom dishes at 33°C or differentiated at 37°C (nonpermissive condition) for 7 to 14 days and then imaged. Similarly, YFP-actin transduced Arhgap24 knockdown (lines 451 and 756) and control knockdown (Fluc) cell lines were differentiated on glass bottom dishes for 7 to 14 days before imaging experiments. Sequential images were captured every 15 seconds for a 20-minute duration using an Olympus FluoView FV1000 microscope. Movies were assembled using Olympus Fluoview software. The Multiple Kymograph plug-in of ImageJ ( http://rsbweb.nih.gov/ij/) was used to generate kymographs at 3 different locations of maximal membrane ruffling for each cell imaged (32). For each kymograph, up to 13 actin spikes were measured, and the average length was computed as described previously (33). A value of 1 was assigned to kymographs without a measurable actin spike. Ruffling movies were generated by H. Suleiman, and the kymographic analysis was performed in a blinded fashion by S. Akilesh. Images were assembled in Adobe Photoshop.
Sequencing.
DNA was obtained from affected (n = 310) and control (n = 180) individuals. DNA from 96 of the control individuals was purchased from the Coriell Institute. PCR primer pairs were designed to completely sequence exons and exon-intron junctions of ARHGAP24. Bidirectional sequencing using ABI Big Dye 3.1 sequencing chemistry on an ABI PRISM 3730 sequencing platform (Applied Biosystems) was performed. Sequences were aligned using Sequencher software (Gene Codes) and manually verified. The Q158R variation was determined by A.S. Shaw’s laboratory and verified by M.P. Winn’s laboratory. In addition, the exon incorporating this variation was sequenced in an additional 554 control chromosomes to rule out the possibility that it represented a low-frequency noncausal variation.
Haplotype analysis.
MSM primer sequences for markers D4S1538, D4S1534, D4S2409, and D4S2460 were obtained from UniSTS ( http://www.ncbi.nlm.nih.gov/unists) or designed with Primer3 software ( http://frodo.wi.mit.edu/primer3/). Carboxyfluorescein succinimidyl ester–labeled (FAM-labeled) oligonucleotides were synthesized by Invitrogen Life Technologies and were run on the 3730 DNA Analyzer (Applied Biosystems) and analyzed using GeneMapper Software v4.0 (Applied Biosystems). The analysis was carried out by visual inspection, assigning the most likely linkage phase by minimizing the number of recombinants in the pedigree.
Arhgap24 dimerization assay.
FLAG- or GFP-tagged wild-type or Q156R Arhgap24 were cotransfected into HEK293 cells. Cell lysates were immunoprecipitated with M2 mouse anti-FLAG antibody (Sigma-Aldrich) with protein A conjugated to sepharose beads. The immunoprecipitates were resolved by SDS-PAGE and then immunoblotted with JL-8 mouse anti-GFP antibody (Invitrogen).
Rac1, Cdc42, and RhoA pull-down assays.
The GST-tagged Pak1-PBD used for the pull-down assays was expressed in BL21(DE3) E. coli. The recombinant protein was bound to glutathione-agarose beads stored at –80°C until use. For the active Rac1 pull-down assay, HEK293T cells were seeded in 6-cm dishes and transfected at 80%–90% confluence using Lipofectamine 2000 reagent according to the manufacturer’s protocol (GIBCO BRL). Cells were transfected with FLAG-tagged wild-type or mutant Q156R Arhgap24 or a titration of the 2 constructs, keeping the total DNA transfected constant. Twenty-four hours after transfection, the cells were lysed, and a sample of the lysate was retained for measurement of total Rac1 and FLAG-Arhgap24 levels. Equal volumes of the remaining lysates were incubated with GST-PBD bound to glutathione-agarose beads. Rac1-GTP bound to the beads (active Rac1) was eluted with Laemmli sample buffer and separated on a 12% polyacrylamide gel. After transfer to a nitrocellulose membrane, active and total Rac1 were detected with the 23A8 mouse anti-Rac1 monoclonal antibody (Upstate Biotechnology). FLAG-tagged Arhgap24 was detected with the M2 mouse anti-FLAG antibody. For Cdc42 activity assays, active Cdc42 was immunoprecipitated with GST-PBD bound to glutathione-agarose beads and detected with B-8 mouse anti-Cdc42 (Santa Cruz Biotechnology Inc.). For RhoA activity assays, active RhoA was immunoprecipitated with GST-Rhotekin bound to glutathione-agarose beads and detected with 26C4 mouse anti-RhoA (Santa Cruz Biotechnology Inc.). The immunoblot signal was detected using a LI-COR Odyssey Imaging System using their infrared dye-labeled secondary reagents. Data are representative of at least 3 independent experiments.
Statistics.
Data are represented as mean ± SD. In all cases, group differences were assessed by ANOVA with post-test correction (Bonferroni-Holm). A P value of less than 0.05 was considered significant.
Study approval.
Patient and control DNA samples were obtained after written informed consent and with local Institutional Review Board approval (Washington University School of Medicine, Duke University Medical Center, Université René Descartes, Brigham and Women’s Hospital, and the NIDDK). All animal experiments were conducted with approval of the Washington University Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We are grateful for the support of Washington University George M. O’Brien Center for Kidney Disease Research (P30 DK079333) for some of the patient specimens. We would like to thank Jeffrey Miner for helpful discussions and Jiancheng Hu for technical assistance and reagents for the GTPase activity pull-down assays. This work was supported in part by the NIDDK Intramural (to J.B. Kopp) and Extramural Research Programs (RO1DK058366, to A.S. Shaw), Howard Hughes Medical Institute, and the Medical Scientist Training Program Grant to Washington University School of Medicine.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(10):4127–4137. doi:10.1172/JCI46458.
References
- 1.Tryggvason K, Patrakka J, Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med. 2006;354(13):1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- 2.Pavenstadt H, Kriz W, Kretzler M. Cell biology of the glomerular podocyte. Physiol Rev. 2003;83(1):253–307. doi: 10.1152/physrev.00020.2002. [DOI] [PubMed] [Google Scholar]
- 3.D’Agati VD. Podocyte injury in focal segmental glomerulosclerosis: Lessons from animal models (a play in five acts). Kidney Int. 2008;73(4):399–406. doi: 10.1038/sj.ki.5002655. [DOI] [PubMed] [Google Scholar]
- 4.Faul C, Asanuma K, Yanagida-Asanuma E, Kim K, Mundel P. Actin up: regulation of podocyte structure and function by components of the actin cytoskeleton. Trends Cell Biol. 2007;17(9):428–437. doi: 10.1016/j.tcb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Mundel P, Reiser J. Proteinuria: an enzymatic disease of the podocyte? Kidney Int. 2010;77(7):571–580. doi: 10.1038/ki.2009.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006;8(8):803–814. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 7.Mundel P, et al. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp Cell Res. 1997;236(1):248–258. doi: 10.1006/excr.1997.3739. [DOI] [PubMed] [Google Scholar]
- 8.Akilesh S, et al. Podocytes use FcRn to clear IgG from the glomerular basement membrane. Proc Natl Acad Sci U S A. 2008;105(3):967–972. doi: 10.1073/pnas.0711515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takemoto M, et al. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161(3):799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rottner K, Hall A, Small JV. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol. 1999;9(12):640–648. doi: 10.1016/S0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- 11.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99(2):67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 12.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116(2):167–179. doi: 10.1016/S0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 13.Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999;147(5):1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135(3):510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 15.Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8(5):485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 16.Yanagida-Asanuma E, et al. Synaptopodin protects against proteinuria by disrupting Cdc42:IRSp53:Mena signaling complexes in kidney podocytes. Am J Pathol. 2007;171(2):415–427. doi: 10.2353/ajpath.2007.070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faul C, et al. The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat Med. 2008;14(9):931–938. doi: 10.1038/nm.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian D, et al. Antagonistic regulation of actin dynamics and cell motility by TRPC5 and TRPC6 channels. Sci Signal. 2010;3(145):ra77. doi: 10.1126/scisignal.2001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winn MP, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308(5729):1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 20.Rao TK, Friedman EA, Nicastri AD. The types of renal disease in the acquired immunodeficiency syndrome. N Engl J Med. 1987;316(17):1062–1068. doi: 10.1056/NEJM198704233161705. [DOI] [PubMed] [Google Scholar]
- 21.Lu TC, et al. HIV-1 Nef disrupts the podocyte actin cytoskeleton by interacting with diaphanous interacting protein. J Biol Chem. 2008;283(13):8173–8182. doi: 10.1074/jbc.M708920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fackler OT, Luo W, Geyer M, Alberts AS, Peterlin BM. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol Cell. 1999;3(6):729–739. doi: 10.1016/S1097-2765(01)80005-8. [DOI] [PubMed] [Google Scholar]
- 23.Sunamoto M, Husain M, He JC, Schwartz EJ, Klotman PE. Critical role for Nef in HIV-1-induced podocyte dedifferentiation. Kidney Int. 2003;64(5):1695–1701. doi: 10.1046/j.1523-1755.2003.00283.x. [DOI] [PubMed] [Google Scholar]
- 24.Boulter E, et al. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol. 2010;12(5):477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Togawa A, et al. Progressive impairment of kidneys and reproductive organs in mice lacking Rho GDIalpha. Oncogene. 1999;18(39):5373–5380. doi: 10.1038/sj.onc.1202921. [DOI] [PubMed] [Google Scholar]
- 26.Shibata S, et al. Modification of mineralocorticoid receptor function by Rac1 GTPase: implication in proteinuric kidney disease. Nat Med. 2008;14(12):1370–1376. doi: 10.1038/nm.1879. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura F, et al. Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS One. 2009;4(3):e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179(5):1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takemoto M, et al. Large-scale identification of genes implicated in kidney glomerulus development and function. EMBO J. 2006;25(5):1160–1174. doi: 10.1038/sj.emboj.7601014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mundel P, Gilbert P, Kriz W. Podocytes in glomerulus of rat kidney express a characteristic 44 KD protein. J Histochem Cytochem. 1991;39(8):1047–1056. doi: 10.1177/39.8.1856454. [DOI] [PubMed] [Google Scholar]
- 32.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 33.Moeller MJ, et al. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 2004;23(19):3769–3779. doi: 10.1038/sj.emboj.7600380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.