Abstract
Proteasomes are multisubunit proteases that play a critical role in maintaining cellular function through the selective degradation of ubiquitinated proteins. When 3 additional β subunits, expression of which is induced by IFN-γ, are substituted for their constitutively expressed counterparts, the structure is converted to an immunoproteasome. However, the underlying roles of immunoproteasomes in human diseases are poorly understood. Using exome analysis, we found a homozygous missense mutation (G197V) in immunoproteasome subunit, β type 8 (PSMB8), which encodes one of the β subunits induced by IFN-γ in patients from 2 consanguineous families. Patients bearing this mutation suffered from autoinflammatory responses that included recurrent fever and nodular erythema together with lipodystrophy. This mutation increased assembly intermediates of immunoproteasomes, resulting in decreased proteasome function and ubiquitin-coupled protein accumulation in the patient’s tissues. In the patient’s skin and B cells, IL-6 was highly expressed, and there was reduced expression of PSMB8. Downregulation of PSMB8 inhibited the differentiation of murine and human adipocytes in vitro, and injection of siRNA against Psmb8 in mouse skin reduced adipocyte tissue volume. These findings identify PSMB8 as an essential component and regulator not only of inflammation, but also of adipocyte differentiation, and indicate that immunoproteasomes have pleiotropic functions in maintaining the homeostasis of a variety of cell types.
Introduction
Autoinflammatory syndrome is characterized by autoimmunity and inflammatory responses in the absence of infection (1, 2). Recent studies have revealed several genes that are etiologic in many autoinflammatory diseases (3). Initially, inherited missense mutations in TNF receptor superfamily, member 1A (TNFRSF1A), were found in several families afflicted with prolonged fevers and severe localized inflammation, now known as TNF receptor–associated periodic syndrome (4). Subsequently, mutations in the gene NLR family, pyrin domain containing 3 (NLRP3) were reported to cause familial periodic fever syndromes, including Muckle-Wells syndrome (also known as familial cold-induced autoinflammatory syndrome) and neonatal-onset multisystem inflammatory disease.
The proteasome is a multisubunit protease that degrades the majority of nonlysosomal proteins in eukaryotic cells (5). The proteasome degrades ubiquitinated proteins for rapid turnover and generates the majority of peptides presented by major histocompatibility complex class I molecules. The 26S proteasome consists of a 20S proteolytic core with 2 19S complexes. The 20S complex is composed of 28 subunits in 4 stacked heptameric rings, with 7 different α type subunits (α1–α7) in the outer rings and 7 different β type subunits (β1–β7) in the inner rings. Of the β subunits, 3 possess catalytic activity: Δ (β1), Z (β2), and X (β5). There are 3 additional catalytic β subunits, β1i (also known as immunoproteasome subunit, β type 9 [PSMB9]), β2i (PSMB10), and β5i (PSMB8; or large multifunctional peptidase 7 [LMP7]), whose expression is induced by IFN-γ. The immunoproteasome is assembled in a stepwise manner (6). For instance, β1i first enters the assembly pathway of the immunoproteasome and forms an assembly intermediate containing an α ring, β1i, β2i, β3, and β4 (7). β5i (referred to herein as PSMB8) is incorporated preferentially over β5 into intermediates that contain β1i and β2i and is required for the processing of β1i and β2i (8, 9). PSMB8 deficiency retards immunoproteasome assembly and leads to the accumulation of proteasome precursors containing unprocessed β1i and β2i (10). PSMB8-deficient mice do not exhibit any gross changes, although they have reduced MHC class I expression (11). Although a recent paper indicated that a PSMB8 inhibitor was able to suppress autoimmune responses in mice (12), PSMB8-deficient mice have a high susceptibility for experimental autoimmune encephalomyelitis, which suggests that the loss of PSMB8 in mice is involved in the progression of inflammatory responses (13). Recently, Agarwal et al. demonstrated a missense mutation in PSMB8 in patients with a variety of symptoms, including lipodystrophy and joint contractures (14), although they did not address the mechanism by which the mutated PSMB8 contributed to each symptom. Therefore, it remains unclear how the mutation in immunoproteasomes contributes to human diseases and how immunoproteasomes contribute to homeostasis in a variety of human tissues.
We examined candidate genes of 3 patients, from 2 consanguineous families, with an autoinflammatory syndrome characterized by recurrent fever, nodular erythema, and hypergammaglobulinemia together with a loss of adipose tissue in the upper part of the body (15, 16). Clinically similar cases have been categorized as Nakajo syndrome (OMIM 256040; ref. 17), and other cases with similar clinical manifestations were previously reported in Japan (18). These manifestations, characteristic of a newly recognized type of Japanese autoinflammatory syndrome with lipodystrophy (JASL), suggest that the causative genes for JASL contribute to both immunity and adipocyte metabolism. Here, we identified a missense mutation in PSMB8 in patients with JASL, which we believe to be novel. The identified PSMB8 mutation increased assembly intermediates of immunoproteasomes, resulting in reduced expression of PSMB8. Low PSMB8 expression in humans caused increased p38 phosphorylation, which resulted in increased IL-6 production. Surprisingly, siRNA-mediated downregulation of PSMB8 disturbed adipocyte differentiation both in vitro and in vivo. These data indicate that a mutation in immunoproteasome complexes causes autoinflammation through increased IL-6 production and lipodystrophy through disturbed adipocyte differentiation. Moreover, our results illustrate the crucial functions of immunoproteasomes in cellular homeostasis and provide molecular insights into immunoproteasome-mediated human diseases.
Results
Clinical phenotypes of JASL patients.
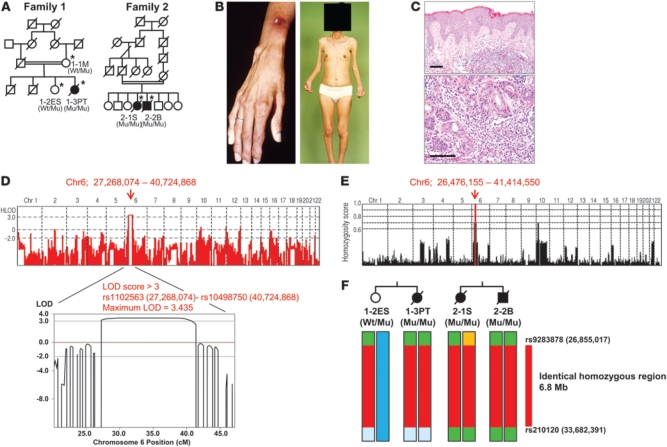
We identified 3 patients with JASL from 2 Japanese consanguineous families. The parents of the families were consanguineous, and the 2 families were not known to be related (Figure 1A). The clinical characteristics of the 3 patients are summarized in Supplemental Table 1 (supplemental material available online with this article; doi: 10.1172/JCI58414DS1). Clinical and laboratory findings of our JASL patients and previously reported Nakajo syndrome or Nakajo syndrome–like diseases are summarized in Supplemental Table 2. Patients began to present recurrent high fever (>40°C) with nodular erythema (Figure 1B) at 1 month to 3 years of age. The recurrent high fever without infectious episodes and nodular erythema on the entire body are common clinical features among JASL patients. Muscle weakness, deformities of hands (Figure 1B), and frostbitten hands were also seen in all patients. The patients began to develop partial lipodystrophy from 6 to 12 years of age (Supplemental Tables 1 and 2). The lipodystrophy was particularly prominent in the face, fingers, and upper limbs (Figure 1B), while nodular erythema developed on the skin on the entire body. Biopsy of skin lesions from patient 2-2B (from family 2) showed a massive infiltration of neutrophils and lymphocytes in the dermis (Figure 1C). The clinical and laboratory findings for JASL and other types of primary or secondary lipodystrophy are compared in Supplemental Table 3. Serum C-reactive protein, IgG, and IgA levels were very high. However, autoantibodies (including anti-nuclear antibody, dsDNA, SS-A, SS-B, and Jo-1 antibodies) were not detected, and blood glucose, total cholesterol, triglyceride, and complement levels were normal (Supplemental Table 1). Patients were treated with moderate doses of corticosteroids (10–15 mg/d), which controlled nodular erythema and high fever and reduced acute-phase reactants, but never inhibited the progression of lipodystrophy. Female patient 1-3PT (from family 1) has 1 daughter who is healthy at the age of 35 years. All patients lived over 47 years of age and died of respiratory or cardiac problems.
Figure 1. Pedigree structure and pathological features.
(A) Pedigree structures of 2 families. Black symbols denote patients with JASL; double horizontal lines denote consanguinity in a married couple; asterisks denote family members whose samples were obtained. Genotypes of PSMB8 are shown (Wt, wild-type allele; Mu, mutant allele). (B) Nodular erythema and deformity of fingers as well as partial lipodystrophy (face, upper body, distal parts of extremities) in 1-3PT. (C) Skin sections of 2-2B stained with hematoxylin and eosin. Original magnification, ×10 (top); ×20 (bottom). Scale bars: 100 μm. (D) Linkage analysis was performed using Merlin software; a linked region (rs1102563 [27,268,074] to rs10498750 [40,724,868]) with LOD score >3 on chromosome 6p (maximum LOD score, 3.435) is indicated by a red arrow. (E) Homozygosity mapping was carried out using HomozygosityMapper; homozygous regions are indicated in red. (F) Haplotype analysis around the homozygous region, carried out by SNP data, identified an identical homozygous region between rs9283878 and rs210120 (6.8 Mb; red) as a shared identical haplotype by descent in both families. PSMB8 genotypes are shown.
Genetic analysis of JASL.
Family 2 has been previously reported (15, 16). To identify candidate loci for this disease, homozygosity mapping was performed in both families. We genotyped 3 affected patients in families 1 and 2 and 1 unaffected sibling in family 1. Multipoint linkage analysis using Merlin software revealed a maximum LOD score of 3.435 at the chromosome 6p region (chr6, 27,268,074–40,724,868; Figure 1D). We then searched for consecutive homozygous SNPs by HomozygosityMapper (19) in order to identify candidate regions of homozygosity in affected but not unaffected offspring, which showed only 6p (chr6, 26,476,155–41,414,550) as a candidate region (Figure 1E) that nearly overlapped the region obtained from the linkage analysis. To further narrow these candidate regions, we identified shared haplotypes of common homozygous SNPs within and across families. Based on these approaches, we identified a single 6.8-Mb region of homozygosity on 6p22.2–6p21.31 (between rs9283878 and rs210120) shared by all patients (Figure 1F). This interval was found to contain 465 gene entries, according to the NCBI database ( http://www.ncbi.nlm.nih.gov/).
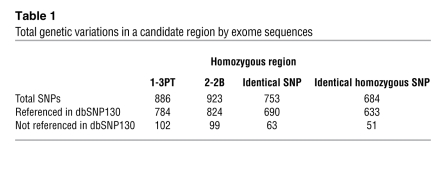
In order to identify the causative genes in the candidate region on chromosome 6, we focused on exonic and flanking intronic variants within this region. Alignment of the sequences in this region with exome analysis revealed that targeted exons (90%) had greater than 10 high-quality reads. The targeted exons with fewer than 10 high-quality reads were further analyzed by Sanger sequencing. The 684 homozygous variants in both patients were identified in this region; of these, 633 appeared in dbSNP130, and the remaining 51 were novel variants (Table 1). We found 2 variants in STK19 or PSMB8 to cause nonsynonymous substitutions; others were located in introns (not splicing-related regions) or were synonymous substitutions. In order to examine whether the 2 amino acid–altering mutations were previously identified variants in the Japanese population, we genotyped each in 312 healthy Japanese controls. The frequency of the missense mutation in STK19 was seen in 4 of 100 control alleles, which suggests that it is a benign polymorphism. In contrast, G197V of PSMB8 was not present in the 624 alleles of the 312 healthy controls (Table 2).
Table 1 .
Total genetic variations in a candidate region by exome sequences
Table 2 .
Allele frequencies of 2 candidate genes
Functional analysis of mutant PSMB8.
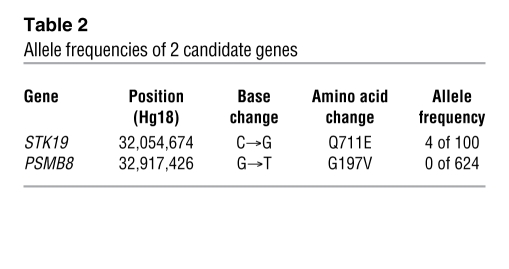
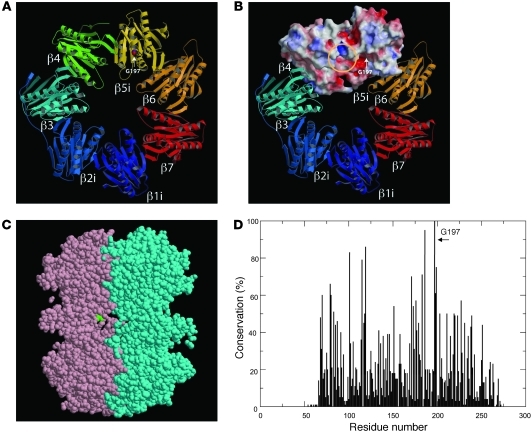
Whereas 1-3PT carried the homozygous mutation, the mother of family 1 (1-1M) and the affected patient’s healthy sibling (1-2ES) carried the heterozygous mutation (Figure 2A and data not shown). PSMB8 has 2 isoforms, and the missense mutation changed an amino acid from G to V at position 197 or 201 in the different isoforms (Figure 2B). The corresponding region was conserved among species (Figure 2C). G197 occupies a surface-exposed position in a β hairpin turn (Supplemental Figure 1, A–D). From the homology model, we can deduce that a substitution for a larger residue (V) would create some crowding, but, more significantly, would restrict the flexibility of the backbone in the region corresponding to the hairpin turn. We modeled mouse subunits β1i, β2i, and β5i (i.e., PSMB8) with the corresponding constitutive subunits and found that position 197 sat outside the subunit-subunit interface (Figure 3A) and S1 pocket (Figure 3B). We also found that G197 sat outside the β ring–β ring interface (Figure 3C). The amino acid sequence around G197 of PSMB8 was aligned with 83 sequence homologs obtained by running BLAST ( http://blast.ncbi.nlm.nih.gov/) against the protein data bank with known structure, which indicated that G197 was the most conserved residue in PSMB8, at 100% conservation among 84 proteins (Figure 3D). The structure modeling and alignment data suggest that the G197 region of PSMB8 is important for stability or folding of the protein.
Figure 2. A missense mutation in PSMB8.
(A) Sequences obtained from a healthy control with wild-type PSMB8; from 1-1M, characteristic of a person with a heterozygous PSMB8 mutation, with a single nucleotide exchange (G→T) in exon 5 of PSMB8; and 1-3PT, characteristic of a homozygous PSMB8 mutation. PSMB8 genotypes are shown. (B) 2 isoforms of PSMB8 and the location of the mutation. (C) The amino acid sequence of human PSMB8 was compared with those of Pan troglodytes, Canis lupus familiaris, Bos taurus, Mus musculus, Rattus norvegicus, and Danio rerio. Conserved residues at the mutated positions are indicated in red.
Figure 3. Structure of PSMB8.
(A) The mouse subunits β1i, β2i, and β5i (i.e., PSMB8) were modeled by the corresponding constitutive subunits using Spanner software (see Methods). G197 is depicted in space-filling representation using CPK coloring. (B) β1i, β2i, and β5i were modeled by the corresponding constitutive subunits using Spanner software. Molecular surfaces of β4 and β5i were colored according to electrostatic potential: red, white, and blue represent negative, neutral, and positive electrostatic values, respectively. The location of the S1 substrate pocket (yellow circle) and the position of G197 (arrow) are indicated. (C) The cross-section of 2 β rings of PSMB8 (pink and cyan), shown using jV software ( http://www.pdbj.org/jv/index.html). G197 (green space-filling representation) was located outside the β ring–β ring interface. (D) Sequences of PSMB8 and 83 related proteins were obtained by running BLAST against the protein data bank. These sequences were aligned using MAFFT software, after removing several short fragments that did not cover the entire aligned region. The sequence conservation was computed and plotted, demonstrating that G197 was the most conserved position (100%) in the 84 related sequences.
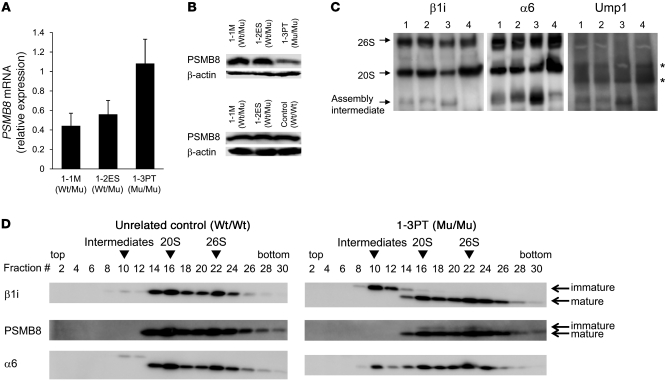
Peripheral blood B cells from 1-3PT, 1-2ES, and 1-1M and a healthy unrelated control were transformed with EBV. The mRNA level of PSMB8 in 1-3PT B cells was relatively higher than those of 1-2ES or 1-1M (Figure 4A), whereas expression of mature PSMB8 protein was much lower in 1-3PT B cells than in those from 1-2ES, 1-1M, and the unrelated control (Figure 4B). The higher level of PSMB8 mRNA expression in 1-3PT B cells relative to control cells might be due to the presence of compensatory mechanisms for low PSMB8 protein expression. Treatment of 1-3PT B cells with a proteasome inhibitor did not increase PSMB8 expression (Supplemental Figure 2A). The constitutively expressed proteasome components β1, β2, and β5 were comparable between 1-3PT B cells and those of 1-1M and 1-2ES (Supplemental Figure 2B). To examine whether the missense mutation reduced the stability of immature PSMB8, 293T cells transfected with wild-type or mutant immature PSMB8 were labeled with [35S]-methionine and cysteine, and the stability of PSMB8 was examined. The speed of degradation was comparable between the 2 PSMB8 groups (Supplemental Figure 2C).
Figure 4. Defective immunoproteasome complexes in JASL cells.
(A) PSMB8 mRNA expression in transformed B cells from 1-1M, 1-2ES, and 1-3PT were evaluated by real-time PCR. Data are representative of 4 experiments. (B) Expression of PSMB8 and β-actin in EBV-transformed B cells from 1-1M, 1-2ES, 1-3PT, and an unrelated control, evaluated by Western blotting. (C) Cell lysates from transformed B cells of 1-1M (lane 1), 1-2ES (lane 2), 1-3PT (lane 3), and unrelated healthy control (lane 4) were subjected to native PAGE and immunoblotted with anti-Ump1, anti-α6, and anti-β1i polyclonal antibodies. Arrows denote bands corresponding to the 20S or 26S proteasome, as well as intermediate complexes. Asterisks denote nonspecific bands. (D) Glycerol gradient centrifugation was performed by using extracts of transformed B cells from 1-3PT and an unrelated healthy control. Fractions were immunoblotted with anti-α6, anti-β1i, and anti-PSMB8 antibody. Arrowheads denote assembly intermediates as well as 20S and 26S immunoproteasomes. PSMB8 genotypes are shown. All data are representative of at least 4 experiments.
We then determined whether the mutation affected assembly of immunoproteasomes by immunoblotting lysates from transformed B cells using anti-β1i, anti-α6, and anti-UMP1 antibodies (20). In each blot, we found an increased proportion of an assembly intermediate (an intermediate stage of immunoproteasome before reaching full maturation; ref. 6) in cell lysates from 1-3PT compared with those from 1-1M, 1-2ES, and an unrelated healthy control (Figure 4C). The lysates from 1-1M and 1-2ES, with 1 allele of mutant PSMB8, also had more assembly intermediates than did the unrelated control (Figure 4C).
Cell lysates from control or 1-3PT B cells were separated by glycerol gradient centrifugation to determine which mature or immature immunoproteasome subunits were incorporated in assembly intermediates or in 20S or 26S proteasomes. The assembly intermediates containing immature β1i and α6 (fraction 10) were increased in 1-3PT compared with control B cells (Figure 4D). Immature PSMB8 was observed in the 20S (fraction 16) and the 26S (fraction 22) immunoproteasomes only in 1-3PT B cells (Figure 4D). Furthermore, β1i, α6, and PSMB8 in the 20S fraction were decreased in 1-3PT compared with control B cells, which suggests that the total amount of mature immunoproteasomes decreased in 1-3PT cells. Taken together, these data indicate that the mutation in PSMB8 disturbs assembly of immunoproteasomes, resulting in low levels of their expression.
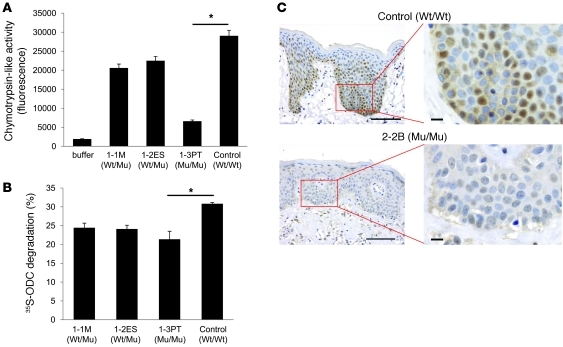
We then compared the chymotrypsin-like activities of proteasomes from 1-3PT, 1-1M, 1-2ES, and the unrelated control. We found that 1-3PT cells, with mutant PSMB8 protein, possessed reduced chymotrypsin-like activity compared with control cells (Figure 5A). Furthermore, total proteasome activity was reduced in B cells from 1-3PT compared with that of the healthy control, as determined by ornithine decarboxylase (ODC) degradation (Figure 5B). ODC is degraded by proteasomes independent of ubiquitin. Because a previous paper showed that PSMB8 is expressed in the skin (21), we next compared the expression level of PSMB8 in skin from a control and from 2-2B. Whereas PSMB8 was expressed in control skin, localized in both the nucleus and the cytoplasm, its expression was much lower in 2-2B (Figure 5C). Taken together, these data suggest that the missense mutation in PSMB8 in JASL patients reduces both protein expression and proteasomal functions.
Figure 5. A mutation in PSMB8 reduces proteasomes activity.
(A) Chymotrypsin-like activities of 26S proteasomes in transformed B cells from 1-3PT, 1-1M, 1-2ES, and unrelated control. *P < 0.01. (B) Cell extracts from transformed B cells were assayed for adenosine triphosphate–dependent protein degradation activity using 35S-labeled ODC. *P < 0.01. (C) Skin sections from a healthy control and 2-2B were stained with anti-PSMB8 mAb and hematoxylin. Scale bars: 100 μm (left); 10 μm (right). PSMB8 genotypes are shown. All data are representative of at least 4 experiments.
Accumulation of ubiquitinated proteins in cells carrying the mutant PSMB8.
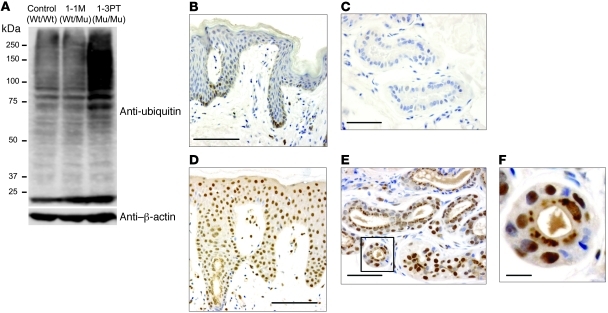
PSMB8 is one of the proteasomal components contributing to the degradation of ubiquitinated proteins. Based on the low expression of PSMB8 in B cells from 1-3PT, we first tested the expression of ubiquitin in transformed B cells from the unrelated control, 1-1M, and 1-3PT. Ubiquitin expression level was higher in B cells from 1-3PT than in those from the 2 controls (Figure 6A). We then examined by immunohistochemistry whether ubiquitin accumulates in 1-3PT cells as a result of impaired PSMB8 function. Staining of skin sections with an anti-ubiquitin antibody showed little expression of ubiquitin in healthy control skin (Figure 6, B and C). In contrast, accumulated ubiquitin was observed in the nucleus or the cytoplasm of epidermal cells from 2-2B (Figure 6D). Furthermore, a dense accumulation of ubiquitin with a punctate pattern and irregular shapes was detected in the cytoplasm of secretory cells of sweat glands and hair follicular cells (Figure 6, E and F). Taken together, our data regarding accumulation of ubiquitinated protein in transformed B cells and skin of JASL patients suggest that increased ubiquitin protein accumulation results from reduced expression of PSMB8 altering proteasomal activity, rather than from inflammation.
Figure 6. Ubiquitinated proteins accumulate in JASL cells.
(A) Expression of ubiquitin in transformed B cells from healthy control, 1-1M, and 1-3PT, evaluated by Western blotting with anti-ubiquitin and β-actin mAbs. Data are representative of 5 experiments. (B–F) Skin sections of a healthy control (B and C) and 2-2B (D–F) stained with anti-ubiquitin mAb together with hematoxylin. Original magnification, ×20 (B and D); ×40 (C and E). The boxed region in E is shown enlarged in F. Histological data are representative of 4 independent staining experiments using a biopsy sample from each patient. Scale bars: 100 μm (B and D); 50 μm (C and E); 10 μm (F).
Increased IL-6 expression in cells carrying mutant PSMB8 occurs via p38.
Hyperactivation of NLRP3 is involved in the pathogenesis of hereditary autoinflammatory syndromes (22). The activation of NLRP3 cleaves pro–caspase-1, which generates p10 caspase-1 (22). We tested whether NLRP3 inflammasomes contributed to autoinflammatory symptoms in JASL patients by measuring caspase-1 activation. B cells from the unrelated control, 1-1M, and 1-3PT did not show any activation of caspase-1 without stimulation with Alum (Supplemental Figure 3). The 3 subjects’ cells showed comparable expression of p10 of caspase-1 by stimulation with Alum (Supplemental Figure 3), which suggests that NLRP3 is not involved in autoinflammatory symptoms in JASL.
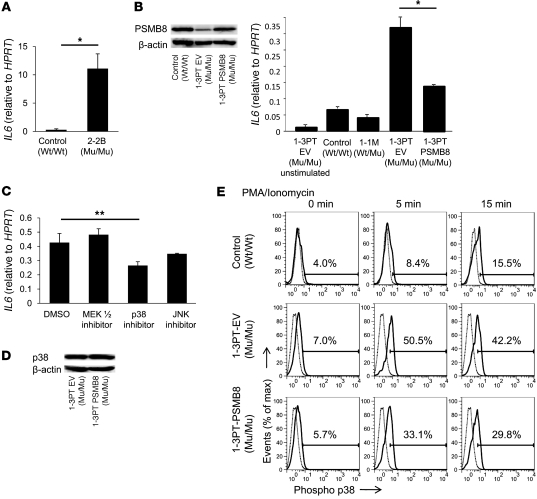
Kasagi et al. reported that serum IL-6, a crucial mediator of inflammation, was elevated in a JASL patient (17). Since the 3 JASL patients in the 2 families were deceased, we assessed expression of IL6 mRNA in skin from 2-2B, and found it to be much higher than that in control skin (Figure 7A). Next, we evaluated whether PSMB8 was involved in IL-6 regulation. Using transformed B cells from 1-3PT, we examined IL-6 expression with or without transduction by a retrovirus carrying wild-type PSMB8. The 1-3PT B cells infected with PSMB8-encoding retrovirus (referred to herein as 1-3PT–PSMB8 cells) expressed levels of PSMB8 comparable to those of control 1-3PT B cells (1-3PT–EV; Figure 7B). Expression of IL6 by PMA- and ionomycin-stimulated 1-3PT–EV cells was reduced in 1-3PT–PSMB8 cells, but still higher than that in control B cells from the unrelated healthy control or 1-1M (Figure 7B). This incomplete reduction of IL6, even by overexpression of wild-type PSMB8, might be due to the secondary effect on transformed B cells induced by long-lasting impaired immunoproteasomes functions. Since a MAP kinase pathway controls IL-6 expression (23, 24), we tested various MAP kinase inhibitors to determine whether they could suppress IL6 expression in the B cells of a JASL patient. Transformed 1-3PT B cells were cultured in the presence of inhibitors of MEK1/2, p38, or JNK. The addition of an inhibitor for p38 suppressed IL6 expression; in contrast, MEK1/2 and JNK inhibitors could not significantly suppress IL-6 expression, although the JNK inhibitor demonstrated partial suppression (Figure 7C). We next tested whether PSMB8 affects p38 phosphorylation in 1-3PT B cells stimulated with PMA and ionomycin. 1-3PT–EV and 1-3PT–PSMB8 cells expressed comparable levels of total p38 (Figure 7D). Phosphorylation of p38 in 1-3PT–EV cells was greater after stimulation with PMA and ionomycin than in 1-3PT–PSMB8 cells, although the latter was still higher than that in control B cells (Figure 7E). Again, this incomplete correction of high phospho-p38 expression, even by overexpressing wild-type PSMB8 in patient B cells, might be due to the secondary effects on transformed B cells induced by long-lasting impaired immunoproteasomes functions, although it might be difficult to compare the expression in different types of transformed B cells. Taken together, these data indicated that low PSMB8 expression in 1-3PT resulted in the upregulation of IL-6, at least partially through a p38-mediated pathway.
Figure 7. A missense mutation in PSMB8 hyperactivates B cells.
(A) IL6 expression in skin from a healthy donor and 2-2B, evaluated by real-time PCR. *P < 0.01. (B) Transformed B cells from 1-3PT were transduced with PSMB8 or a control vector. Expression of PSMB8 in healthy control, 1-3PT–EV, and 1-3PT–PSMB8 transformed B cells were evaluated by Western blotting. Expression of IL6 in PMA- and ionomycin-stimulated control, 1-1M, 1-3PT–EV, and 1-3PT–PSMB8 B cells were evaluated by real-time PCR. *P < 0.01. (C) Expression of IL6 in cells after PMA and ionomycin stimulation in the presence of MEK1/2, p38, or JNK inhibitors, evaluated by real-time PCR. **P < 0.05. (D) Expression of total p38 in 1-3PT–EV and 1-3PT–PSMB8 cells, evaluated by Western blot. (E) Phospho-p38 in healthy control, 1-3PT–EV, and 1-3PT–PSMB8 transformed B cells after PMA and ionomycin stimulation. Gray and black histograms denote cells stained with control rabbit IgG and anti–phospho-p38, respectively. Percentages within histograms denote expression of phospho-p38 in PMA- and ionomycin-stimulated cells. All data are representative of 7 experiments.
PSMB8 regulated adipocyte differentiation.
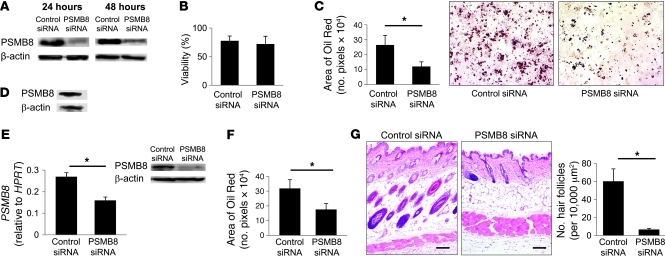
Patients with JASL gradually develop partial lipodystrophy. Therefore, we next examined the possibility that PSMB8 regulates adipocyte differentiation. The murine preadipocyte cell line 3T3-L1 highly expressed PSMB8 (Supplemental Figure 4A). In order to evaluate whether PSMB8 is involved in preadipocyte maturation, we tested the change of PSMB8 expression during adipocyte differentiation. We found that PSMB8 expression did not change during differentiation toward adipocytes (Supplemental Figure 4A). PSMB8 expression was then downregulated using a siRNA targeting PSMB8. The PSMB8 siRNA transfection efficiently downregulated PSMB8 (Figure 8A) and did not affect the viability of 3T3-L1 cells (Figure 8B). Transfected 3T3-L1 cells were allowed to differentiate into adipocytes, and cytoplasmic lipid droplets were evaluated by Oil Red staining 6–7 days after transfection. The downregulation of PSMB8 in 3T3-L1 cells inhibited adipocyte differentiation, as evidenced by the reduced number and amount of Oil Red–positive areas (Figure 8C). Downregulation of the β2i subunit did not affect adipocyte differentiation (Supplemental Figure 4B), which indicates that PSMB8-mediated proteasomal activity is required for adipocyte differentiation. We also examined the expression of PSMB8 in human preadipocytes and found high expression of PSMB8 (Figure 8D). Transfection of siRNA against PSMB8 in human preadipocytes efficiently downregulated PSMB8 (Figure 8E), which disturbed their differentiation into mature adipocytes, as evaluated by Oil Red staining (Figure 8F). Although to our knowledge, it has not been reported that animals deficient in PSMB8 exhibit lipodystrophy, we next inhibited PSMB8 expression in the skin of adult BALB/c mice by subcutaneously injecting PSMB8 siRNA. Treatment of mice with PSMB8 siRNA efficiently downregulated Psmb8, as evaluated by PCR and histological analysis (Supplemental Figure 4, C and D). Histological analysis of skin specimens showed atrophic subcutaneous adipose tissue and a decreased number of hair follicles in mice that received PSMB8 siRNA compared with control mice (Figure 8G). A smaller number of adipocytes was observed in mice that received PSMB8 siRNA, as determined by staining cells with anti-FABP4 antibody (Supplemental Figure 4E). Taken together, these data indicate that PSMB8 is critically involved in the differentiation of preadipocytes into adipocytes, which could account for the progressive lipodystrophy observed in JASL patients.
Figure 8. A missense mutation in PSMB8 blocks adipocyte differentiation.
(A and B) 3T3-L1 cells were transfected with siRNA against PSMB8 or control siRNA. (A) Expression of PSMB8 was evaluated 24 and 48 hours after transfection by Western blotting. (B) Cell viability was tested by trypan blue staining 48 hours after transfection. (C) Induction of adipocytes was evaluated by staining cells with Oil Red 6 days after induction, and Oil Red–positive cells were also quantified. *P < 0.01. (D) Expression of PSMB8 in human preadipocytes was examined by Western blotting. (E) Expression of PSMB8 or β-actin in human preadipocytes transfected with siRNA against PSMB8 or control siRNA (24 hours after transfection) was evaluated by Western blotting and quantified. *P < 0.01. (F) Human preadipocytes were transfected with siRNA against PSMB8 and induced to differentiate into adipocytes; 12 days after transfection, Oil Red–positive cells were evaluated by densitometry. *P < 0.01. (G) siRNA for PSMB8 or control siRNA was subcutaneously injected into the skin of BALB/c mice. The skin was obtained 11 days after the initial siRNA injection, and the section was stained with hematoxylin and eosin. Original magnification, ×10. Scale bars: 100 μm. Moreover, the number of hair follicles in 10,000 μm2 (10 regions) was counted. *P < 0.01. All data are representative of at least 4 experiments.
Discussion
We found patients from 2 consanguineous families characterized by nodular erythema, recurrent fever, and lipodystrophy and investigated the gene causing JASL. We found a PSMB8 homozygous missense mutation — which we believe to be novel — that increased an intermediate form of immunoproteasomes, resulting in reduced proteasomal function. The discovery of the mutation in human PSMB8 allowed us to evaluate the physiological and pathological roles of immunoproteasomes. A distinct missense mutation in PSMB8 was previously reported in patients suffering from lipodystrophy, joint contractures, and muscle atrophy (14). However, there has been no understanding of the molecular mechanisms underlying the low function of immunoproteasomes and the variety of associated symptoms. Therefore, in this study, we focused on evaluating the mechanisms underlying the 2 major symptoms of JASL, autoinflammatory responses and lipodystrophy. We found that B cells and skin from JASL patients expressed high levels of IL-6, and that JASL B cells upregulated IL-6 through the p38 pathway. High levels of IL-6 production were partially reversed by overexpression of wild-type PSMB8. We also found that downregulation of PSMB8 in mouse or human preadipocytes disturbed their differentiation toward mature cells. Furthermore, injection of siRNA targeted against PSMB8 in mouse skin reduced the number of adipocytes in their skin. These data provide mechanistic insights into autoinflammation and lipodystrophy of JASL and, furthermore, demonstrate physiological roles of immunoproteasomes in regulating both inflammation and adipocyte differentiation.
The NLRP3-mediated autoinflammatory syndrome depends on increased production of active IL-1 through caspase-1 activation (22, 25). JASL patient B cells did not show increased production of active caspase-1 compared with control B cells, which suggests that PSMB8 deficiency does not activate the NLRP3 pathway. In contrast, patients with JASL exhibited high expression of IL-6, which exerted pleiotropic effects in promoting inflammatory responses in the skin (present study) and in serum (17). Indeed, a previous paper indicating that increased expression of IL-6 causes skin inflammation (26) supports the link between increased IL-6 expression and autoinflammation in JASL patients. As for the mechanism of high IL-6 production in cells of JASL patients, our data indicated that overproduction was associated with increased phosphorylation of p38 in B cells, although it remains unclear which p38-related molecules actually promote IL-6 production. Furthermore, treatment of JASL patient B cells with a p38 inhibitor did not completely abrogate IL-6 production, which suggests that a p38-independent pathway is also involved in elevated IL-6 production in JASL. Additionally, a recent paper reported that targeted deletion of Psmb8 in mice exacerbated experimental autoimmune encephalomyelitis with an increase in oxidized proteins (13), which suggests that PSMB8 is also involved in the progression of inflammatory responses through increased accumulation of oxidized protein. It should be noted that treatment with a PSMB8 inhibitor suppressed rheumatoid arthritis in mice (12). Therefore, careful analysis of PSMB8 inhibitors could elucidate the roles of PSMB8 in distinct types of inflammatory responses. We are currently developing a knockin mouse that has the same mutation as 3 JASL patients in our study. Those mice will broaden our understanding of the roles of immunoproteasomes in the pathogenesis of autoinflammation and other inflammatory diseases.
The missense mutation in PSMB8 decreased expression of mature PSMB8, although it did not affect the stability of immature forms of the protein. We found increased levels of assembly intermediates containing immature β1i and reduced expression of 20S immunoproteasomes in JASL patient B cells. As β1i is incorporated into immunoproteasomes before incorporation of PSMB8 (7), these data suggest that the mutation in PSMB8 disturbs incorporation of PSMB8 into immunoproteasomes or facilitates separation of PSMB8 from mature immunoproteasomes. In addition, the increased levels of immature forms of PSMB8 in 20S and 26S immunoproteasomes in JASL patient B cells indicated that the mutation in PSMB8 affects its processing. Together, these data suggest that the mutation in PSMB8 leads to abnormal assembly of immunoproteasomes and reduced levels of mature immunoproteasomes. Indeed, JASL patient B cells exhibited low immunoproteasomal function compared with control cells, as evidenced by their reduced chymotrypsin-like activity and ODC degradation, although the former was more profound than the latter. Given that PSMB8 possesses chymotrypsin-like activity (6), assessing that activity might be a more sensitive measure of PSMB8 function than the ODC degradation assay, which provides a measure of total protease activity.
In our study of immunoproteasome functions in JASL patients, we detected homogenous accumulation of ubiquitin or a dense accumulation with irregular shapes in the nucleus and cytoplasm of keratinocytes, sweat gland cells, or hair follicular cells together within transformed JASL patient B cells. Although we do not have evidence that the accumulation of ubiquitin-coupled protein in JASL patients is associated with autoinflammation, the characteristic accumulation patterns might disturb cellular functions or trigger inflammatory responses. We previously reported that DNase1 deficiency can cause systemic lupus erythematosus as a result of accumulated DNA-related products (27–29). Such a defective clearance of intracellular products might be a primary cause for other immune-associated diseases. If this is the case, it would be reasonable to develop strategies for reducing or inactivating accumulated molecules, rather than suppressing the immune system in order to treat such diseases. In addition, immunoproteasomes contribute to antigen processing for MHC class I. Therefore, it will be important to analyze the contributions of mutant PSMB8 to this pathway in future studies.
We demonstrated herein that PSMB8 is involved in the differentiation of preadipocytes to adipocytes, which might contribute to lipodystrophy in JASL patients. The finding that corticosteroid therapy could completely suppress autoinflammatory responses, but not lipodystrophy progression, also suggests that the lipodystrophy in JASL patients is caused by the primary effect of impaired PSMB8 function on adipocyte differentiation, rather than a secondary effect of autoinflammation. We examined the amount of ubiquitin-coupled protein in 3T3-L1 cells in which PSMB8 was downregulated and did not observe gross accumulation of ubiquitin-coupled protein (data not shown). Therefore, we think it unlikely that nonspecific accumulation of large amounts of ubiquitin-coupled protein in preadipocytes (in which PSMB8 expression is reduced) disturbs the differentiation of preadipocytes to adipocytes. Rather, altered turnover of specific proteins that regulate adipocytes differentiation might be affected by downregulation of PSMB8. The identification of such molecules might provide new targets for therapeutic strategies in the treatment of metabolic diseases.
PSMB8-deficient mice do not spontaneously develop autoinflammatory symptoms and lipodystrophy. One possibility to explain the discrepancy between the distinct phenotypes of JASL patients and PSMB8-deficient mice is that the mutant PSMB8-mediated assembly defect in immunoproteasomes might lead to unexpected immunoproteasome activity. Reduced functions of immunoproteasomes with altered activity in JASL might contribute to some aspects of JASL phenotypes. We expect to clarify those discrepancies by establishing knockin mice that carry the JASL mutation.
Agarwal and colleagues independently published results about PSMB8 mutation in JASL-like syndrome (14), although they did not address how the mutation in PSMB8 was involved in the symptoms of JASL-like syndrome. In the present report, we characterized an autosomal-recessive mutation in JASL, which we believe to be novel, and our data demonstrated the molecular basis of autoinflammation and lipodystrophy resulting from reduced immunoproteasome functions. Given the steadily growing spectrum of physiological pathways regulated in part by proteasomes, future molecular analyses of PSMB8 are likely to further support the establishment of therapeutic strategies for treating autoinflammation and metabolic diseases.
Methods
Further information can be found in Supplemental Methods.
DNA samples.
Genomic DNA was extracted from peripheral blood and autopsy samples obtained from 2 families after obtaining informed consent.
SNP homozygosity mapping and linkage analysis.
Both affected and unaffected siblings from 2 consanguineous families were studied with the Illumina Human 610 Quad. Sample processing and labeling were performed in accordance with the manufacturer’s instructions. An average call rate greater than 99% was obtained. Homozygous regions were identified using HomozygosityMapper (19). Our laboratory has developed a graphic interface that allows for easy comparison of common homozygous regions among 3 patients from different families. For linkage analysis, multipoint LOD scores were obtained using Merlin software (30).
Flow cytometry.
Cells were resuspended in staining buffer at a density of 2 × 106 cells/ml. Cells were stained with rabbit IgG (10500; Caltag Laboratories) or polyclonal anti–phospho-p38 (4511; Cell Signaling Technology) followed by PE-conjugated anti-rabbit IgG (EI-1000; Vector Laboratories). Using the anti–phospho-p38 antibody, we did not observe phospho-p38–positive cells in the p38-deficient KOP cell line (Riken Cell Bank) or in KOP cells transfected with cDNA for human p38 (provided by S. Matsuda, Kansai Medical University, Moriguchi, Japan) or constitutively active MKK6 (provided by Y. Gotoh, University of Tokyo, Tokyo, Japan; ref. 31). However, we observed phospho-p38–positive cells in KOP cells transfected with both human p38 and constitutively active MKK6 (Supplemental Figure 5). The fluorescence intensity of 105 cells was examined using a FACSCalibur or FACS Canto II cytometer (BD Biosciences).
Assay of proteasome activity.
Transformed B cells from 1-1M, 1-2ES, 1-3PT, or a healthy unrelated control were lysed and subjected to 8%–32% glycerol gradient centrifugation. Subsequently, peptidase activity was measured using a fluorescent peptide substrate, succinyl-Leu-Leu-Vl-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-MCA), for chymotrypsin-like activity as previously reported (32).
Structure modeling.
A structural model of PSMB8 was built using Spanner ( http://sysimm.ifrec.osaka-u.ac.jp/cgi-bin/spanner), which uses a fragment assembly algorithm to produce a gapless alignment for the template. Multiple sequence alignments were computed using MAFFT (33). Electrostatic surfaces were prepared using the program ef-Surf (34).
Protein extracts, immunological analysis, and glycerol gradient analysis.
Cells were lysed in ice-cold lysis buffer (50 mM Tris–HCl, pH 7.5; 0.5% [v/v] NP-40; 1 mM dithiothreitol; 2 mM ATP; and 5 mM MgCl2), and the extracts were clarified by centrifugation at 20,000 g for 15 minutes at 41°C. The supernatants were subjected to native PAGE (7% Tris–acetate gel; Invitrogen) according to the instructions provided by the manufacturer. The separated proteins were transferred onto a polyvinylidene difluoride membrane and reacted with the indicated antibody. Anti-Ump1, anti-α6, and anti-β1i polyclonal antibodies were described previously (20, 35). ODC degradation activity was assayed as described previously (36). For glycerol gradient analysis, cell lysates were fractionated by linear glycerol density gradient centrifugation (22 hours, 100,000 g) as described previously (36).
Statistics.
Unless otherwise indicated, data are shown as mean ± SD. For all experiments, the significance of differences between groups was calculated using the Mann-Whitney U test for unpaired data. Differences were considered significant for P values less than 0.05.
Study approval.
All genetic studies were conducted after Ethical Committee approval of human genome research of the University of Tokushima and informed consent was obtained from patients, or from their relatives if patients were deceased. The study was conducted in accordance with the principles of the declaration of Helsinki. All animal research was approved by the Animal research committee of the University of Tokushima.
Supplementary Material
Acknowledgments
We thank R.N. Germain (NIAID) for discussion, M. Sugawara (Akita University) for providing information on a JASL patient, I. Inoue and A. Narita (Tokai University) for help with genetic analysis, and C. Kinouchi and C. Tomari for technical and editorial assistance. This work was supported by a Grant-in-Aid for Young Scientists (S) from the Japan Society for the Promotion of Science.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(10):4150–4160. doi:10.1172/JCI58414.
References
- 1.Masters SL, Lobito AA, Chae J, Kastner DL. Recent advances in the molecular pathogenesis of hereditary recurrent fevers. Curr Opin Allergy Clin Immunol. 2006;6(6):428–433. doi: 10.1097/ACI.0b013e3280109b57. [DOI] [PubMed] [Google Scholar]
- 2.Dinarello CA. Interleukin-1beta and the autoinflammatory diseases. N Engl J Med. 2009;360(23):2467–2470. doi: 10.1056/NEJMe0811014. [DOI] [PubMed] [Google Scholar]
- 3.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 4.McDermott MF, et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97(1):133–144. doi: 10.1016/S0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- 5.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10(2):104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 7.Nandi D, Woodward E, Ginsburg DB, Monaco JJ. Intermediates in the formation of mouse 20S proteasomes: implications for the assembly of precursor beta subunits. EMBO J. 1997;16(17):5363–5375. doi: 10.1093/emboj/16.17.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin TA, et al. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med. 1998;187(1):97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kingsbury DJ, Griffin TA, Colbert RA. Novel propeptide function in 20 S proteasome assembly influences beta subunit composition. J Biol Chem. 2000;275(31):24156–24162. doi: 10.1074/jbc.M001742200. [DOI] [PubMed] [Google Scholar]
- 10.De M, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. Beta 2 subunit propeptides influence cooperative proteasome assembly. J Biol Chem. 2003;278(8):6153–6159. doi: 10.1074/jbc.M209292200. [DOI] [PubMed] [Google Scholar]
- 11.Fehling HJ, et al. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265(5176):1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 12.Muchamuel T, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15(7):781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 13.Seifert U, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142(4):613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal AK, et al. PSMB8 encoding the beta5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am J Hum Genet. 2010;87(6):866–872. doi: 10.1016/j.ajhg.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oyanagi K, et al. An autopsy case of a syndrome with muscular atrophy, decreased subcutaneous fat, skin eruption and hyper gamma-globulinemia: peculiar vascular changes and muscle fiber degeneration. Acta Neuropathol. 1987;73(4):313–319. doi: 10.1007/BF00688252. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka M, et al. Hereditary lipo-muscular atrophy with joint contracture, skin eruptions and hyper-gamma-globulinemia: a new syndrome. Intern Med. 1993;32(1):42–45. doi: 10.2169/internalmedicine.32.42. [DOI] [PubMed] [Google Scholar]
- 17.Kasagi S, et al. A case of periodic-fever-syndrome-like disorder with lipodystrophy, myositis, and autoimmune abnormalities. Mod Rheumatol. 2008;18(2):203–207. doi: 10.1007/s10165-008-0033-4. [DOI] [PubMed] [Google Scholar]
- 18.Kitano Y, Matsunaga E, Morimoto T, Okada N, Sano S. A syndrome with nodular erythema, elongated and thickened fingers, and emaciation. Arch Dermatol. 1985;121(8):1053–1056. doi: 10.1001/archderm.121.8.1053. [DOI] [PubMed] [Google Scholar]
- 19.Seelow D, Schuelke M, Hildebrandt F, Nurnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37(Web Server issue):W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirano Y, et al. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27(16):2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dissemond J, et al. Immunoproteasome subunits LMP2 and LMP7 downregulation in primary malignant melanoma lesions: association with lack of spontaneous regression. Melanoma Res. 2003;13(4):371–377. doi: 10.1097/00008390-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 22.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nature Rev Immunol. 2010;10(3):210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 23.Thobe BM, Frink M, Choudhry MA, Schwacha MG, Bland KI, Chaudry IH. Src family kinases regulate p38 MAPK-mediated IL-6 production in Kupffer cells following hypoxia. Am J Physiol Cell Physiol. 2006;291(3):C476–C482. doi: 10.1152/ajpcell.00076.2006. [DOI] [PubMed] [Google Scholar]
- 24.Craig R, et al. p38 MAPK and NF-kappa B collaborate to induce interleukin-6 gene expression and release. Evidence for a cytoprotective autocrine signaling pathway in a cardiac myocyte model system. J Biol Chem. 2000;275(31):23814–23824. doi: 10.1074/jbc.M909695199. [DOI] [PubMed] [Google Scholar]
- 25.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*). Annu Rev Immunol. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawamura D, et al. Induction of keratinocyte proliferation and lymphocytic infiltration by in vivo introduction of the IL-6 gene into keratinocytes and possibility of keratinocyte gene therapy for inflammatory skin diseases using IL-6 mutant genes. J Immunol. 1998;161(10):5633–5639. [PubMed] [Google Scholar]
- 27.Yasutomo K, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28(4):313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 28.Maekawa Y, Yasutomo K. Defective clearance of nucleosomes and systemic lupus erythematosus. Trends Immunol. 2001;22(12):662–663. doi: 10.1016/S1471-4906(01)02084-1. [DOI] [PubMed] [Google Scholar]
- 29.Yasutomo K. Pathological lymphocyte activation by defective clearance of self-ligands in systemic lupus erythematosus. Rheumatology (Oxford). 2003;42(2):214–222. doi: 10.1093/rheumatology/keg081. [DOI] [PubMed] [Google Scholar]
- 30.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 31.Fujishiro M, et al. MKK6/3 and p38 MAPK pathway activation is not necessary for insulin-induced glucose uptake but regulates glucose transporter expression. J Biol Chem. 2001;276(23):19800–19806. doi: 10.1074/jbc.M101087200. [DOI] [PubMed] [Google Scholar]
- 32.Murata S, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316(5829):1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 33.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 34.Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20(8):1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- 35.Murata S, et al. Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J. 2001;20(21):5898–5907. doi: 10.1093/emboj/20.21.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano Y, et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437(7063):1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.