Abstract
Breast cancer is a heterogeneous disease. There is a high degree of diversity between and within tumors as well as among cancer-bearing individuals, and all of these factors together determine the risk of disease progression and therapeutic resistance. Advances in technologies such as whole-genome sequencing and functional viability screens now allow us to analyze tumors at unprecedented depths. However, translating this increasing knowledge into clinical practice remains a challenge in part due to tumor evolution driven by the diversity of cancer cell populations and their microenvironment. The articles in this Review series discuss recent advances in our understanding of breast tumor heterogeneity, therapies tailored based on this knowledge, and future ways of assessing and treating heterogeneous tumors.
Introduction
Breast cancer is a heterogeneous group of neoplasms originating from the epithelial cells lining the milk ducts. Breast tumor heterogeneity has been noted in histology and clinical outcome for a long time, and these differences have served as the basis for disease classification. More recently, the traditional, mainly pathology-driven classification has been refined and at times replaced by molecular classifications, which have the potential to combine disease mechanisms with clinical outcome measures. However, startling heterogeneity in cancer cell phenotypes accompanied by dynamic plasticity of the tumor microenvironment make tumor categorization a demanding task, especially as it relates to therapeutic responses and disease progression.
This issue of the JCI features Reviews on various aspects of heterogeneity in breast cancer as well as new approaches to evaluating and treating breast tumors based on improvements in technologies and molecular knowledge. Morag Park and José Baselga and their colleagues review the current state of breast cancer classification (1) and treatment options designed based on this knowledge (2), respectively. Max Wicha and coworkers discuss the potential role of cancer stem cells and their intricate interactions with cells constituting the microenvironment as potential mechanisms underlying intratumor heterogeneity (3). Lastly, Anne-Lise Borresen-Dale and colleagues summarize recent advances in DNA sequencing technologies and their utility for assessing genetic diversity within tumors (4).
Intertumor heterogeneity and individualized cancer treatment
Breast cancer is one of the few tumor types in which molecular classification has successfully been used for the design of individualized therapies, leading to significant improvements in disease-specific survival (5). Based on comprehensive gene expression profiling, breast tumors are classified into at least three major subtypes: luminal, human epidermal growth factor receptor 2+ (HER2+), and basal like (6, 7). Each of these subtypes has different risk factors for incidence, response to treatment, risk of disease progression, and preferential organ sites of metastases. Luminal tumors are positive for estrogen and progesterone receptors, and the majority respond well to hormonal interventions, whereas HER2+ tumors have amplification and overexpression of the ERBB2 oncogene and can be effectively controlled with a diverse array of anti-HER2 therapies. Basal-like tumors in general lack hormone receptors and HER2; thus, the majority of these tumors are also called triple-negative breast cancer (TNBC). Currently there is no molecular-based targeted therapy for TNBC, and unfortunately only approximately 20% of these tumors respond well to standard chemotherapy. Thus, developing improved treatments for TNBC is one of the highest priorities of current breast cancer research. Numerous agents are in various phases of clinical development, including several different poly(ADP-ribose) polymerase inhibitors, JAK kinase, and EGFR inhibitors as well as “revived” classical chemotherapeutic agents such as platinum salts. However, thus far none of these shows promise for treating all TNBCs, a finding that perhaps is not surprising given that several recent studies have described that even this relatively small class of breast tumors can be further divided into five or six subclasses, each with its own molecular features and unique sensitivity to therapeutic agents (8–10).
Several hypotheses have been proposed to explain the origin of intertumor heterogeneity in breast cancer, including subtype-specific tumor cell–of–origin and transforming events (11). Accordingly, luminal and HER2+ tumors may originate from luminal lineage-committed progenitors, whereas basal-like cases arise from less differentiated stem cell–like cells. However, gene expression patterns and experimental evidence in model systems imply that luminal progenitors may also serve as precursors to basal-like tumors following genetic or epigenetic event(s) that switch cellular phenotypes (12–14). For example, loss of BRCA1 or PTEN in luminal epithelial cells leads to loss of luminal differentiation, and the oncogenic transformation of these cells results in the formation of basal-like tumors (15). However, because we know that not all ER+/HER2+, or basal-like tumors, are the same, it is likely that there are multiple ways to develop each of these tumor types.
Defining the cell-of-origin and evolutionary pathway of a breast cancer in humans is a nearly impossible task, as we are rarely able to diagnose tumors at their earliest stages and follow their molecular evolution. Currently three main approaches have been used to trace the evolutionary history of human cancer. One method is to analyze tumors at the single-cell level for phenotypic traits and somatic genetic alterations, based on the assumption that some cells may be relics of the tumor’s past, and that their frequency within a tumor may reveal the probable steps of the tumor’s evolution (16). Another approach analyzes a large collection of tumors at different progression stages for molecular changes, and based on the frequency at which these are detected at a specific progression step, their probable order of events can be assembled (17, 18). Thus far these approaches have been used to map colon (19) and pancreatic cancer (20) evolution, but similar studies have not been conducted in breast tumors. Of course both of these methods are based on certain assumptions, such as frequency of somatic genetic alterations as they relate to their order, though this may be in question given a recent study describing the acquisition of several somatic genetic changes by tumor cells due to a single catastrophic rearrangement of chromosomes (21). Defining the cell-of-origin of cancer was thought to be relevant only for risk prediction and chemoprevention studies. However, if the cell-of-origin has a major influence on the evolutionary path of a tumor as it relates to the identity and frequency of acquired transforming events, then its characterization would be important in better understanding breast tumor subtypes as well.
Intratumor heterogeneity — mechanisms and consequences
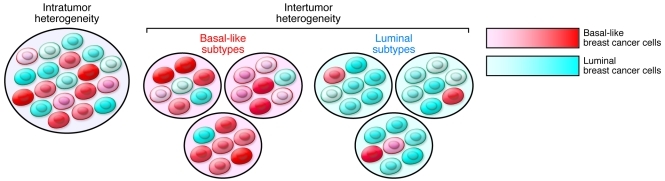
Besides the numerous differences detected between tumors, cancer cells within one tumor of a patient at any given time frequently display startling heterogeneity for various traits related to tumorigenesis, such as angiogenic, invasive, and metastatic potential (22). Some of these are due to clonal and cellular diversity for genetic and epigenetic alterations, whereas others may reflect nonhereditary mechanisms such as adaptive responses or fluctuation in protein levels and activity of signaling pathways. Intratumor heterogeneity may also underlie intertumor heterogeneity and help to explain breast tumor subtypes merely as tumors that are composed of different “mixtures” of cancer cells (Figure 1).
Figure 1. Hypothetical model explaining the origins of intertumor and intratumor heterogeneity in breast cancer.
Intratumor heterogeneity is due to the presence of cancer cells with variable phenotypes such as different degrees of basal-like and luminal features. Intertumor heterogeneity may also be explained by the presence of these different cell types within tumors at varying frequencies. Cancer cells with a basal-like phenotype predominate in basal-like tumors, whereas luminal tumors are largely composed of luminal breast cancer cells. However, due to variability in basal-like and luminal cell traits, not all basal-like and luminal tumors are the same, further contributing to heterogeneity even within tumor subtypes.
Cellular differentiation states are defined by epigenetic mechanisms including cell type–specific DNA methylation and chromatin modification patterns. Thus, it is possible that some of the epigenetic variability among tumor cells is a reflection of their differentiation states. In some cases tumors may initiate from a stem or progenitor cell, which has a preserved ability to give rise to more differentiated progeny, resulting in a heterogeneous tumor. The best example for this has been certain forms of hematopoietic malignancies, in which a leukemia stem cell has been described (23). This cancer stem cell model has been expanded to numerous tumor types including breast cancer (24), but the interpretation and validity of this model has been widely disputed. One of the defining characteristics of a cancer stem cell is the ability to initiate a tumor in xenograft transplant assays, which more or less reproduces the phenotype of the original tumor. However, this definition turned out to be very much assay dependent (25, 26) and thus may not accurately reflect the true characteristics of the cells in their physiologic environment (i.e., in patients).
In most breast tumors, cancer cells with stem cell–like and more differentiated features can be detected, but the existence of a simple differentiation hierarchy has been questioned by the high degree of genetic diversity within and between these two cell populations (27). Similarly, two recent studies described genetic heterogeneity and clonal evolution of leukemia stem (tumor-initiating) cells using single-cell transplant assays (28, 29). Consequently, clonal composition and cellular phenotypes continuously change during tumor progression, making tumors moving targets and posing a major challenge for effective cancer treatment. Better understanding the mechanisms that maintain tumor cell heterogeneity such as the cytokine signaling networks between cancer cells and the tumor microenvironment will likely aid in the design of improved therapies. An example for this is the recent identification of the IL-6/JAK/STAT3 pathway, which can support tumor cell growth and survival in an autocrine or paracrine manner, as a new therapeutic target in a subset of breast tumors (30, 31). As compounds targeting this pathway have already been in clinical trials for other indications, testing them in breast cancer can fairly easily and rapidly be accomplished.
Novel approaches for assessing heterogeneous tumors
One of the most comprehensive ways to address intratumor diversity and to identify potentially resistant clones even before they expand and lead to treatment failure is whole-genome sequencing of all individual cancer cells within tumors. Such an experiment has not and may never be performed. However, sampling of the tumor or deep sequencing of bulk tumors represent fairly close alternatives. Wigler and colleagues described one of the first such attempts in one breast tumor, in which they completely sequenced 100 tumor cells (16). Similarly, several recent studies have performed whole-genome or exome sequencing of single tumors or a panel of breast tumors (32–34). With continuous advances in DNA sequencing technologies, it is likely that deep sequencing of single cells will be achieved maybe even in a way that preserves information on the topology within tumors (e.g., location of cancer cells with specific mutations). However, the interpretation and clinical translation of these results remain a major challenge. For example, filtering out tumor-driving mutations from “passengers” is not an easy task, especially considering that the roles of driver and passenger mutations may change places as the tumor evolves. The combination of whole-genome sequencing studies with well-designed high-throughput functional screens in physiologically relevant models will likely facilitate the translation of these technical advances into clinical practice.
Conclusions
At first glimpse the tremendous heterogeneity and continuously evolving nature of tumors seems daunting and makes curing or even effectively controlling cancer a nearly impossible task. However, I firmly believe that an in-depth understanding of tumor evolution will lead to the eradication and maybe even prevention of this disease.
Acknowledgments
The author thanks Franziska Michor, Andriy Marusyk, and Ian Krop for their critical reading of this manuscript and useful discussions. K. Polyak is supported by the National Cancer Institute, US Army Congressionally Directed Research, the Avon Research Foundation, V Foundation, Breast Cancer Research Foundation, Novartis Pharmaceuticals Inc., and the Susan G. Komen Foundation.
Footnotes
Conflict of interest: K. Polyak receives research support from Novartis Oncology and is a consultant to Novartis Oncology. She also serves on the Scientific Advisory Board of Metamark Genetics Inc. and Theracrine Inc. and holds AVEO Pharmaceuticals Inc. stocks.
Citation for this article: J Clin Invest. 2011;121(10):3786–3788. doi:10.1172/JCI60534.
References
- 1.Bertos NR, Park M. Breast cancer — one term, many entities? J Clin Invest. 2011;121(10):3789–3796. doi: 10.1172/JCI57100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higgins MJ, Baselga J. Targeted therapies for breast cancer. J Clin Invest. 2011;121(10):3797–3803. doi: 10.1172/JCI57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121(10):3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russnes HG, Navin N, Hicks J, Borresen-Dale A-L. Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest. 2011;121(10):3810–3818. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez EA. Breast cancer management: opportunities and barriers to an individualized approach. Oncologist. 2011;16 suppl 1:20–22. doi: 10.1634/theoncologist.2011-S1-20. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann BD, et al. Identification of human triple–negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russnes HG, et al. Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci Transl Med. 2010;2(38):38ra47. doi: 10.1126/scitranslmed.3000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polyak K. Breast cancer: origins and evolution. . J Clin Invest. 2007;117(11):3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proia TA, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8(2):149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 15.Liu S, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105(5):1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 18.Attolini CS, et al. A mathematical framework to determine the temporal sequence of somatic genetic events in cancer. Proc Natl Acad Sci U S A. 2010;107(41):17604–17609. doi: 10.1073/pnas.1009117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105(11):4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 24.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana E, et al. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18(5):510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120(2):636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 29.Notta F, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469(7330):362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 30.Marotta LL, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24– stem cell–like breast cancer cells in human tumors. J Clin Invest. 2011;121(7):2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71(2):614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 33.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 34.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]