Abstract
A unique challenge in air pollution cohort studies and similar applications in environmental epidemiology is that exposure is not measured directly at subjects’ locations. Instead, pollution data from monitoring stations at some distance from the study subjects are used to predict exposures, and these predicted exposures are used to estimate the health effect parameter of interest. It is usually assumed that minimizing the error in predicting the true exposure will improve health effect estimation. We show in a simulation study that this is not always the case. We interpret our results in light of recently developed statistical theory for measurement error, and we discuss implications for the design and analysis of epidemiologic research.
There has been a major effort in air pollution epidemiology research to develop statistical models to predict exposures at subjects’ locations in situations where measurements at the desired locations are not available.1–7 These efforts assume that exposure predictions with less measurement error relative to the unknown true values will improve health effect estimation.8–10 We demonstrate in a simulation study that this assumption is not always true, and we interpret our results using recently developed statistical theory for measurement error resulting from spatially misaligned data.11
Mathematical framework and simulation study
Most modern statistical models for predicting long-term average air pollution concentrations are based on land-use regression. In land-use regression modeling, a linear regression model with geographic (land-use) covariates such as population density, proximity to traffic, and proximity to commercial areas is fit to monitoring data and then used to predict concentrations at subjects’ locations. Elaborations on this framework account for spatial and spatio-temporal correlation and various approaches to model selection, but land-use regression remains a central component. We focus on a pure land-use regression model in this paper.
Stochastic data-generating model
Consider an association study with the N × 1 vector of observed health outcomes Y, N × 1 vector of exposures X, and N × m matrix of covariates Z. Assume a linear regression model
| (1) |
with coefficient of interest βX and ε an N × 1 random vector with independent elements distributed as Gaussian random variables with mean 0 and variance (i.e., N(0, ).
We are interested in the situation where Y and Z are observed, but instead of X we observe the N* × 1 vector X* of exposures at various locations. N* is the number of exposure monitors. Assume that X, the subjects’ exposures, and X*, the exposure concentrations measured at the monitors, are jointly distributed as
| (2) |
In this expression, S and S* are random N × k and N* × k dimensional matrices of the k geographic covariates used in the land-use regression model observed without error, α is an unknown k × 1 vector of coefficients, and η and η* are independent vectors with elements distributed as N(0, ). The stochasticity in S and S* derives from random selection of subject and monitor locations. If the exposure model is known, it is standard practice to estimate α based on X* and then use W = Sα̂ in place of X in equation (1) to estimate βX. That is, predictions from the land-use regression model are used as estimated exposures in place of the unknown true values, a form of regression calibrat ion.12
We quantify the accuracy in approximating X by W by
where larger values correspond to less measurement error. This defines an out-of-sample measure of prediction accuracy, as it is based on prediction error at subjects’ locations, and it is not subject to bias from overfitting the exposure model to the monitoring data.13 is a random quantity that varies for each realization of the data-generating model, and we denote its expectation .
There are a number of criteria for evaluating the validity and reliability of health effect estimates. We consider bias, standard deviation, root mean squared error, and coverage probability (the proportion of 95% confidence intervals that include the true βX).14
Misspecified exposure model
We generally do not know the exact form of the exposure model and may use a misspecified model for prediction. One form of model misspecification is to omit a geographic covariate from the land-use regression model. This corresponds to observing only the N × (k − 1) and N* × (k − 1) matrices S′ and S*′ obtained by deleting the k th columns of S and S*. We then estimate the corresponding (k − 1) × 1 vector of coefficients α′ and replace X in equation (1) by W′ = S′ α̂ ′ to obtain . We denote measures of exposure prediction accuracy and as in the case of the correctly specified exposure model.
We generally expect to be larger than , which from the perspective of exposure modeling implies that the correctly specified exposure model gives better predictions than the misspecified one. It is reasonable to expect that this will also lead to improved health effect estimation. However, in the next subsection we will demonstrate a class of examples in which is consistently larger than , but β̂X has more error than as measured in terms of bias, variance, root mean squared error, and coverage probability. We emphasize that is not inflated by overfitting because it is based on the correctly specified exposure model and quantifies out-of-sample prediction accuracy at subjects’ locations.
Simulation study
We set k = 4 (three geographic covariates and an intercept) and consider scenarios with N between 100 and 10,000 subjects and N* equal to 100 monitors. We assume the three geographic covariates are independent of each other at all locations and are independent between subjects. In particular, for each subject i we assume the j th geographic covariate Sij is independently distributed as N(0,1).
Similarly we assume the are distributed as N(0,1) for j = 1,2, but the third geographic covariate for the monitoring sites is distributed as N(0,σ2) for σ2 = 0.1,1.0, or 4.0. Finally, we set α0 = 0, αj = 4 for j = 1,2,3, β0 = 1, βX = 2, σε = 25, and ση = 4, and we assume there are no additional covariates Z. Example simulation code in R15 can be found in the eAppendix (http://links.lwww.com).
The choice of σ2 controls the level of variability in the third geographic covariate at the monitoring locations. By comparing the misspecified model (i.e., the model that does not contain the third geographic covariate) to the correctly specified full model, we are able to assess the added value of including the third geographic covariate in predictions, depending on its variability. The situation with σ2 = 0.1 is of particular interest, as it represents a geographic covariate that has limited variability in the monitoring data compared with the other geographic covariates but is equally variable in the subject data where it will be used to predict exposures. This is realistic, for example, if the covariate measures near-road traffic exposure. Regulatory monitors are often sited away from roadways in order to measure background pollution levels, and so they may not span the full range of covariate values relevant for predicting exposures at subjects’ home locations – a significant fraction of which are near major roads.
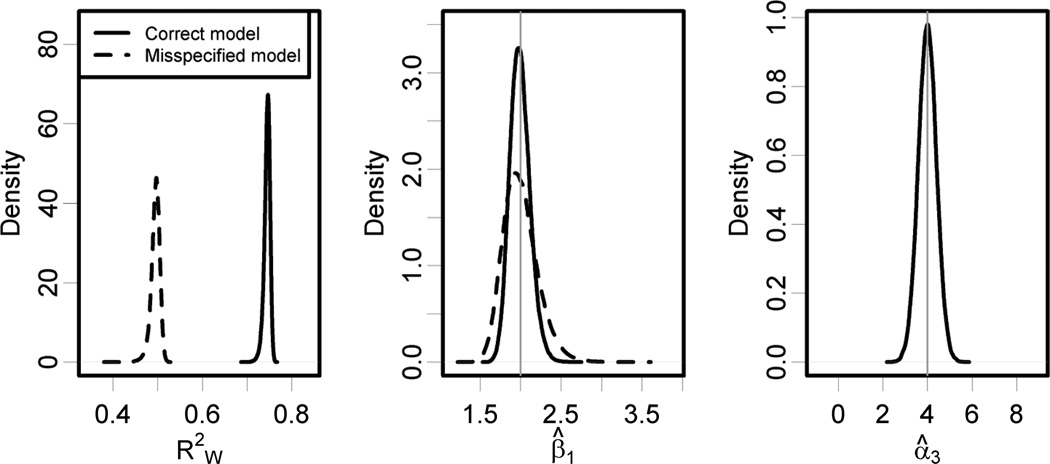
In the Table and Figure 1, we show the results from 80,000 Monte Carlo simulations with N = 10,000 subjects, N* = 100 monitoring sites, and σ2 = 1.0. The coefficient for the third geographic covariate α3 is estimated well in the full model and is statistically significant in all simulations. The corresponding exposure prediction accuracy is consistently near 0.75, compared with near 0.50 with the misspecified model. Health effect estimation efficiency is improved by using the correctly specified exposure model, which gives a standard deviation for β̂X of 0.12 compared with 0.21 for with the misspecified model. The coverage probabilities for both models are poor, as the standard error estimates fail to account for exposure measurement error. The correctly specified exposure model results in a modest improvement in coverage probability, although it also introduces slightly more bias than the misspecified model.
Table.
Results from 80,000 Monte Carlo simulations with N = 10,000 and N* = 100. The and are the out-of-sample prediction and variance of predicted exposures, respectively, averaged over 80,000 Monte Carlo simulations. The standard deviation and fraction of Monte Carlo runs statistically significant are given for estimates of α̂3 in the correctly specified exposure model only, because this parameter is not included in the misspecified model. The bias, standard deviation, root mean squared error (RMSE), and 95% confidence interval (CI) coverage are given for estimates of the health effect parameter β̂X. We also report the average estimated standard error (SE) for β̂X. The Monte Carlo standard error in estimating the bias of β̂X in all models is less than 0.001. (R code to produce these results is provided in an electronic appendix, http://links.lww.com.)
| σ2 = 1 | σ2 = 0.1 | |||
|---|---|---|---|---|
| Correct model |
Misspecified model |
Correct model |
Misspecified model |
|
| Exposure predictions | ||||
| 0.74 | 0.49 | 0.73 | 0.50 | |
| 48.5 | 32.7 | 50.2 | 32.3 | |
| Exposure model parameter estimate α̂3 | ||||
| Standard deviation | 0.41 | - | 1.37 | - |
| Statistically significant (p < 0.05) | 100% | - | 83% | - |
| Health effect parameter estimate β̂x | ||||
| Bias | −0.007 | −0.001 | −0.035 | 0.001 |
| Standard deviation | 0.12 | 0.21 | 0.23 | 0.16 |
| RMSE | 0.12 | 0.21 | 0.23 | 0.16 |
| E(SE) | 0.038 | 0.049 | 0.038 | 0.049 |
| 95% CI coverage | 45% | 35% | 26% | 46% |
Figure 1.
Results from 80,000 Monte Carlo simulations with N = 10,000, N* = 100, and σ2 = 1.0. For the correctly specified exposure model the average out-of-sample prediction accuracy is and the health effect estimation standard deviation is 0.12 with a bias of −0.007 (95% CI = −0.008 to −0.006). Corresponding statistics for the misspecified exposure model are and health effect estimation standard deviation 0.21 with a bias of (95% CI = −0.002 to 0.0006). The standard error of α̂3 for the correctly specified model is 0.41, and α̂3 is statistically significant in all simulations.
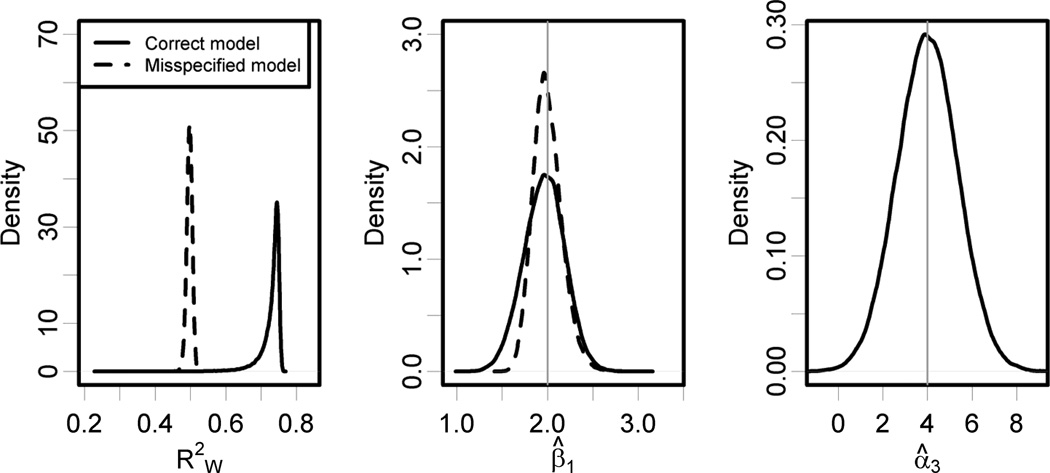
Analogous results are shown in the Table and Figure 2 for σ2 = 0.1, representing a situation where one of the geographic covariates is less variable in the distribution of monitoring locations than are the other geographic covariates. The smaller value of σ2 results in more variability in estimating α3, but this parameter is still estimated well and is statistically significant in 83% of Monte Carlo simulations. There is clear improvement in the exposure predictions from using the full model with at least 0.67 in 95% of simulations, as compared with the misspecified model with consistently near 0.50. But in this situation, the health effect estimates are more precise when we use the misspecified exposure model, with the standard deviation of equal to 0.16, compared to 0.23 for β̂X using the fully specified model. The misspecified model also results in less bias and a modest improvement in coverage probability.
Figure 2.
Results from 80,000 Monte Carlo simulations with N = 10,000, N* = 100, and σ2 = 0.1. For the correctly specified exposure model the average out-of-sample prediction accuracy is and the health effect estimation standard deviation is 0.23 with a bias of −0.035 (95% CI = −0.037 to −0.034). Corresponding statistics for the misspecified exposure model are and health effect estimation standard deviation 0.16 with a bias of 0.001 (95% CI = −0.0003 to 0.002). The density plot for shows some small outliers for the full model, but the prediction accuracy is better than for the misspecified model in all but 144 of the 80,000 simulations. The standard deviation of α̂3 for the correctly specified model is 1.37, and α̂3 is statistically significant in 83% of simulations.
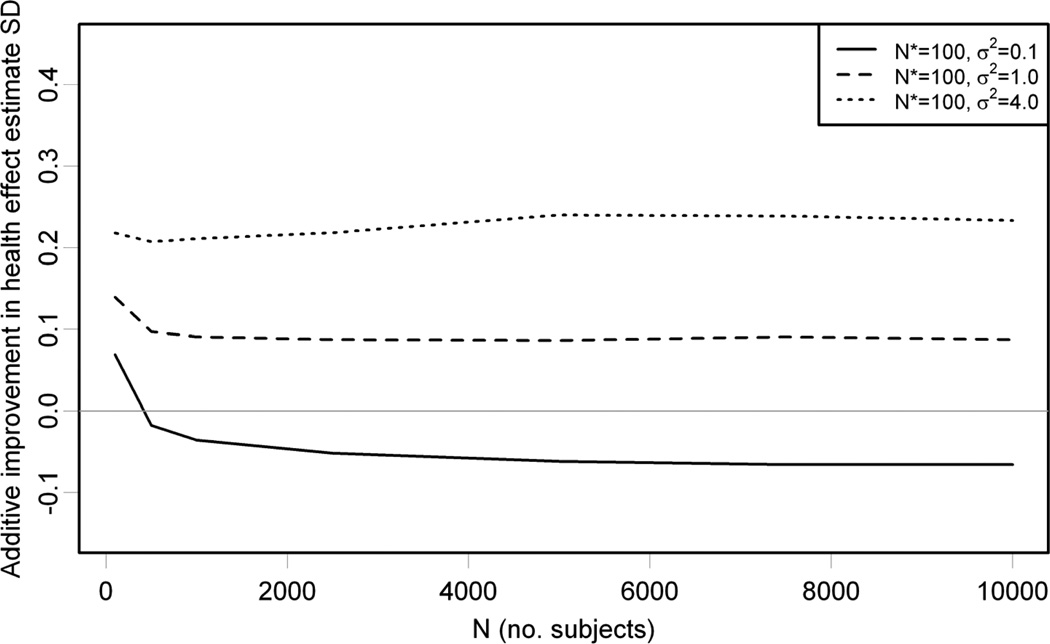
We vary the number of subjects as well as σ2and summarize the results in Figure 3 by plotting the difference between the standard deviation of , based on the misspecified exposure model, and β̂X, based on the correct exposure model, on the vertical axis against N on the horizontal axis; a positive difference indicates that the correctly specified model is more efficient. We restrict to 5,000 Monte Carlo simulations because this is sufficient to estimate the standard deviations (the biases are smaller and require more Monte Carlo simulations). The difference is positive for σ2 = 1.0 and 4.0, consistent with the prior expectation that more accurate exposure predictions result in more efficient health effect estimation. But it is negative for σ2 = 0.1 except for the case where there are only N = 100 subjects, demonstrating that in larger health studies the misspecified exposure model results in more efficient health effect estimation, even though it gives less accurate exposure predictions. For all simulations, the average out-of-sample exposure model prediction accuracies are between 0.73 and 0.75 for the correctly specified model and between 0.73 and 0.75 for the misspecified model that omits the third geographic covariate.
Figure 3.
Results from 5,000 Monte Carlo simulations with N* = 100, σ2 = 0.1,1.0,4.0, and N ranging from 100 to 10,000. The vertical axis shows the difference between standard deviation (SD) of β̂X from the misspecified and correct exposure models. A positive difference indicates that the correctly specified model is more efficient. For all values of σ2, the average exposure model prediction accuracies are between 0.73 and 0.75 and between 0.49 and 0.50 .
Theoretical interpretation in a measurement error framework
The results of our simulation study seem paradoxical in that more accurate exposure predictions do not necessarily lead to improved health effect estimation. The Table shows that for σ2 = 0.1 the correctly specified model consistently gives more variable exposure predictions and more accurate out-of-sample prediction, compared with the misspecified exposure model. However, a small part of the additional exposure variability is induced by error in estimating α3, which leads to less efficient estimation of βX. These findings can be understood in a theoretical context by referring to the statistical measurement error framework developed for this setting.11, 12
Briefly, for a fairly general class of exposure models there are two components to the measurement error. The Berkson-like component of error results from smoothing the exposure surface using a model that may not account for all sources of variation and can be thought of as the part of the true exposure that is not predictable from the model. It is similar to standard Berkson error16 in that it inflates the standard deviation of the health effect estimate and introduces little or no bias. However, it is different from Berkson error in that it is correlated in space and is not completely independent of the predicted exposures.11,12 The classical-like component comes from uncertainty in estimating the exposure model parameters. It is similar to classical measurement error in that it is a source of variability in the predicted exposures and can introduce bias in health effect estimates as well as change their standard errors. The classical-like component is also different from classical measurement error in that the additional variability from exposure model parameter estimation is shared across all prediction locations rather than being independent.11
For the simple land-use regression exposure model considered here, the Berkson-like component is pure Berkson error because there is no spatial dependence structure in η and η* and the Sij and are independent. When we use the correctly specified exposure model, the Berkson error is just η, but misspecifying the model by omitting the third geographic covariate increases the Berkson error substantially, resulting in a degradation of prediction accuracy. However, Berkson error plays the same role mathematically as the random ε in the disease model, and so its impact on the health effect estimation error diminishes for large N. On the other hand, each coefficient that needs to be estimated in the exposure model contributes to the classical-like error, and this part of the error remains important regardless of the number of subjects. In some situations, this could result in a bias-variance tradeoff because classical-like error induces bias while Berkson-like error does not.
It turns out that for σ2 = 0.1 in the monitoring data, we get relatively variable estimates of α3 when using the full exposure model, while still improving out-of-sample prediction accuracy at subjects’ locations. This results in substantial classical-like measurement error that (for sufficiently large N) is more important than the additional Berkson error that is introduced by omitting the corresponding geographic covariate. There is very little bias in any of our simulations, and so the dominant classical-like error primarily results in more variable estimates of βX.
Implications for future research
We have shown a class of examples in which more accurate exposure prediction does not lead to improved health effect estimation. It bears emphasis that this does not result from overfitting the exposure model, at least not as overfitting is traditionally understood for prediction models.13 In all cases, using the correctly specified model that includes all three geographic covariates leads to improved prediction accuracy, as measured by out-of-sample evaluated at subjects’ locations.α̂3
Our findings have important implications for the design and analysis of environmental epidemiologic studies. Development of models for exposure prediction and health effect estimation should be considered simultaneously, in contrast with the current practice of first selecting an exposure model to optimize prediction accuracy and then using the resulting predictions for health effect estimation. Recent papers that address measurement error in air pollution cohort studies represent progress in this direction.11, 12, 14, 17 Our results do not necessarily suggest employing a joint statistical estimation model for the exposure and health parameters in which the health data would influence estimation of the exposure model parameters. The issue we have highlighted relates more directly to model selection than to parameter estimation.
There is extensive literature on penalization and other methods for optimizing accuracy of prediction models,13 but these techniques are not directly applicable because better prediction accuracy may induce less precise health effect estimation. New statistical methodology is needed to select exposure models to optimize efficiency of health effects inference, perhaps involving alternative forms of penalization that account for the structure in both the monitoring and health outcome data. It is also worth exploring asymptotic methods to estimate the bias and variance of β̂X in order to select optimal geographic covariates, particularly when there is a relatively large number of monitoring locations compared with the geographic covariates.
The relative benefits of various air-pollution exposure models depend on the variability of geographic covariates in the subject population and monitor locations, and on the size of the cohort. It is evident that study design can be improved by accounting for statistical issues at the intersection of exposure prediction and health effect estimation. All else being equal, it is preferable to design an exposure monitoring campaign to maximize the variability of pertinent geographic covariates across monitor locations. An asset allocation-algorithm may be useful for optimizing the monitoring design to predict exposures in an epidemiology study with known subjects’ locations.18
We have considered only the relatively simple setting of a linear disease model with an exposure model that is land-use regression with independent geographic covariates. Even in this case we have shown that more accurate exposure prediction does not necessarily lead to improved health effect estimation. We expect that similar phenomena can occur in other settings, but further research is needed to identify general conditions and assess the implications of more complex situations.
Supplementary Material
Acknowledgements
We thank the journal editor and three anonymous referees for their valuable suggestions and Sverre Vedal for helpful comments on a draft of this manuscript.
Financial support:
Supported by the United States Environmental Protection Agency through R831697 and Assistance Agreement CR-83407101 and by the National Institute of Environmental Health Sciences through R01-ES009411 and 5P50ES015915 (Drs. Szpiro and Sheppard) and by the National Institute of Environmental Health Sciences through R01 ES017017 (Dr. Paciorek).
References
- 1.Yanosky JD, Paciorek CJ, Suh H. Predicting chronic fine and coarse particulate exposure using spatio-temporal models for the northeastern adn midwestern US. Environmental Health Perspectives. 2009;117:522–529. doi: 10.1289/ehp.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szpiro A, Sampson PD, Sheppard L, Lumley T, Adar SD, Kaufman JD. Predicting intra-urban variation in air pollution concentrations with complex spatio-temporal dependencies. Environmetrics. 2010;21(4):606–631. doi: 10.1002/env.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brauer M. How much, how long, what, and where: Air pollution exposure assessment for epidemiologic studies of respiratory disease. Proceedings of the American Thoracic Society. 2010;7:111–115. doi: 10.1513/pats.200908-093RM. [DOI] [PubMed] [Google Scholar]
- 4.Fanshawe TR, Diggle PJ, Rushton S, Sanderson R, Lurz PWW, Glinianaia SV, et al. Modelling spatio-temporal variation in exposure to particulate matter: a two-stage approach. Environmetrics. 2008;19(6):549–566. [Google Scholar]
- 5.Su JG, Jerrett M, Beckerman B, Wilhelm M, Ghosh JK, Ritz B. Predicting traffic-related air pollution in Los Angeles using a distance decay regression selection strategy. Environmental Research. 2009;109(6):657–670. doi: 10.1016/j.envres.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jerrett M, Arain A, Kanaroglou P, Beckerman B, Potoglou D, Sahsuvaroglu T, et al. A review and evaluation of intraurban air pollution exposure models. Journal of Exposure Analysis and Environmental Epidemiology. 2005;15:185–204. doi: 10.1038/sj.jea.7500388. [DOI] [PubMed] [Google Scholar]
- 7.Hoek G, Beelen R, de Hoogh K, Vienneau D, Gulliver J, Fischer P, et al. A review of land-use regression models to assess spatial variation in outdoor air pollution. Atmospheric Environment. 2008;42:7561–7578. [Google Scholar]
- 8.Jerrett M, Burnett RT, Ma R, Pope CA, Krewski D, Newbold KB, et al. Spatial analysis of air pollution mortality in Los Angeles. Epidemiology. 2005;16(6):727–736. doi: 10.1097/01.ede.0000181630.15826.7d. [DOI] [PubMed] [Google Scholar]
- 9.Kunzli N, Jerrett M, Mack WJ, Beckerman B, LaBree L, Gilliland F, et al. Ambient air pollution and atherosclerosis in Los Angeles. Environmental Health Perspectives. 2005;113(2):201–206. doi: 10.1289/ehp.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puett RC, Hart JE, Yanosky JD, Paciorek C, Schwartz J, Suh H, et al. Chronic fine and coarse particulate exposure, mortality, and coronary heart disease in the Nurses’ Health Study. Environmental Health Perspectives. 2009;117(11):1697–1701. doi: 10.1289/ehp.0900572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szpiro A, Sheppard L, Lumley T. Efficient measurement error correction for spatially misaligned data. Biostatistics. 2011 doi: 10.1093/biostatistics/kxq083. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gryparis A, Paciorek CJ, Zeka A, Schwartz J, Coull BA. Measurement Error Caused by Spatial Misalignment in Environmental Epidemiology. Biostatistics. 2009;10(2):258–274. doi: 10.1093/biostatistics/kxn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastie T, Tibshirani R, Friedman J. Elements of Statistical Learning. New York: Springer; 2001. [Google Scholar]
- 14.Kim SY, Sheppard L, Kim H. Health effects of long-term air pollution: influence of exposure prediction methods. Epidemiology. 2009;20(3):442–450. doi: 10.1097/EDE.0b013e31819e4331. [DOI] [PubMed] [Google Scholar]
- 15.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2010. ISBN 3-900051-07-0. [Google Scholar]
- 16.Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement Error in Nonlinear Models: A Modern Perspective. Second Edition. Chapman and Hall/CRC; 2006. [Google Scholar]
- 17.Madsen L, Ruppert D, Altman NS. Regression with spatially misaligned data. Environmetrics. 2008;19:453–467. [Google Scholar]
- 18.Kanaroglou PS, Jerrett M, Morrison J, Beckerman BB, Arain MA, Gilbert NL, et al. Establishing an air pollution monitoring network for intraurban population exposure assessment: A location-allocation approach. Atmospheric Environment. 2005;39:2399–2409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.