Abstract
Angiogenesis is the growth of new blood vessels from existing vasculature. Excessive vascularization is associated with a number of diseases including cancer. Anti-angiogenic therapies have the potential to stunt cancer progression. Peptides derived from type IV collagen are potent inhibitors of angiogenesis. We wanted to gain a better understanding of collagen IV structure-activity relationships using a ligand-based approach. We developed novel peptide-specific QSAR models to study the activity of the peptides in endothelial cell proliferation, migration, and adhesion inhibition assays. We found that the models produced quantitatively accurate predictions of activity and provided insight into collagen IV derived peptide structure-activity relationships.
Background
Excessive vascularization is a hallmark of many diseases including cancer, rheumatoid arthritis, diabetic nephropathy, pathologic obesity, age-related macular degeneration, and asthma. Compounds that inhibit angiogenesis represent potential therapeutics for many diseases. Judah Folkman performed pioneering research in the field of angiogenesis;1 his work lead to the identification of a number of polypeptides with anti-angiogenic activity.2 One of polypeptides called endostatin was derived from the noncollagenous (NC1) domain of collagen XVIII.3 Work led by Raghu Kalluri resulted in the development of small antiangiogenic peptides from the NC1 domain of collagen IV including canstatin,4 arrestin,5 and tumstatin.6 These collagen IV derived fragments were reviewed in the context of other angiogenesis modulating compounds.7–9 Based on these parent compounds, work in our laboratory identified more than 100 similar peptide sequences from diverse parent proteins throughout the proteome.10 The set of parent proteins included collagen IV, CXC chemokines, type I thrombospondin domain (TSP-1)-containing proteins, serpins, somatotropins, and tissue inhibitors of metalloproteinases (TIMPs). Work carried out in our group experimentally validated in vitro inibition of endothelial cell (EC) proliferation and migration by peptides derived from type IV collagens,11 thrombospondin domain-containing proteins,12, 13 and CXC chemokines.14 These studies showed that a large fraction of the peptides have antiangiogenic potential. Subsequently, our laboratory tested some of these peptides in vivo using mouse xenograft models of breast and lung cancer,15, 16 and ocular models.17 The peptides derived from type IV collagen are attractive targets because of their efficacy against multiple angiogenic properties (i.e. endothelial cell proliferation, migration, and adhesion).18
A better understanding of the structure-activity relationship of type IV collagen peptides could help us better understand the mechanism of action and produce more active peptides. For many of these peptides, the receptor had not been elucidated. When the receptor is unknown, ligand-based modeling approaches must be used. Examples of ligand-based design methods include pharamcophore modeling19–22 and quantatitive structure-activity relationship (QSAR)23–26 analysis. These methods correlate diverse aspects of molecular structure and flexibility with a quantatitive measure of activity. Some work has been done on developing peptide-specific feature sets for QSAR.27, 28 Others make use of position weight matrices to describe a family of peptides.29 Many of these methods require solving NP-hard30 problems. That means a polynomial time algorithm is not known for solving these problems. For large datasets, these methods must resort to using inexact approaches and heuristics.
To continue developing the type IV collagen-derived peptides, we aimed to (i) develop techniques for computationally efficient, peptide-specific, QSAR analysis, (ii) enable predictions of peptide activity, and (iii) gain a better understanding of the structure-activity relationship of collagen IV derived peptides. In this work, we described several novel peptide-specific QSAR methods that helped us address these aims. We formulated the models using convex optimization in a way that could be solved quickly to global optimality. We used experimentally-determined activity data from collagen IV peptides to develop individual models for endothelial cell proliferation, migration, and adhesion. We validated the QSAR models by making activity predictions and performing experiments for an external set of peptides. The activity of the external set of peptides was verified by endothelial cell proliferation, migration, adhesion, and tube formation assays.
Results
Peptide activity in vitro using EC proliferation, migration, and adhesion assays
This study is based on a libary of 23 collagen IV derived peptides. The founding peptide 0 (SP2000)10 was found as a homolog of tumstatin6 in the human proteome. These peptides consisted of a series of truncations and selected amino acid substitutions designed to improve translational potential. In Table 1 we present the activity of the 23 (21 training + 2 external verification) peptides in endothelial cell proliferation (at 100μM), migration (at 50μM), and adhesion (at 100μM). Peptide concentrations were chosen to provide diversity in activity measurements. All experiments were performed in duplicate and the result of each experiment was the average of three replicates on the same plate. Activity measurements are given as a percentage of the vehicle control.
Table 1. The compound database.
A dataset of 23 collagen IV derived compounds tested for endothelial cell proliferation (at 100 mg/ml), migration (at 50 mg/ml), and adhesion (at 100 mg/ml) inhibition. The table gives the mean % inhibition for each assay and the standard error of the mean (SEM). The screening of all compounds was done with n=2 and normalized to a vehicle control. We use the single letter X to represent L-α-amino-n-butyric acid (Abu). Peptides 27 and 35 were held out as an external validation set.
| Ref. | Compound | Structure | Proliferation | Migration | Adhesion |
|---|---|---|---|---|---|
| 0 | SP2000 | LRRFSTMPFMFCNINNVCNF | 67.55 ± 2.58 | 91.02 ± 0.63 | 96.25 ± 0.34 |
| 2 | SP2002 | LRRFSTMPFMFGNINNVGNF | 25.71 ± 0.78 | 87.90 ± 0.70 | 97.80 ± 0.15 |
| 4 | SP2004 | LRRFSTMPFMF********* | 2.97 ± 2.62 | −14.20 ± 7.85 | −2.60 ± 0.32 |
| 6 | SP2006 | LRRFSTMPFMFXNINV**** | 59.70 ± 4.10 | 26.55 ± 3.15 | 95.62 ± 1.22 |
| 7 | SP2007 | LRRFSTMPFMFX******** | 4.85 ± 1.62 | −24.38 ± 5.87 | 11.04 ± 1.81 |
| 8 | SP2008 | LRRFSTMP************ | 7.02 ± 0.69 | −20.09 ± 4.67 | −0.14 ± 0.00 |
| 9 | SP2009 | ************NINNVXNF | 24.25 ± 12.25 | 31.65 ± 0.85 | 70.78 ± 0.05 |
| 10 | SP2010 | ********FMFXNINNVXNF | 28.30 ± 3.39 | −16.60 ± 9.83 | 66.18 ± 1.83 |
| 11 | SP2011 | ****STMPFMFXNINNVXNF | 13.83 ± 7.64 | −24.46 ± 6.89 | 6.82 ± 2.87 |
| 12 | SP2012 | LRRFSTMPFMFXNINNVXNF | 69.72 ± 2.25 | 96.97 ± 0.19 | 98.46 ± 0.32 |
| 13 | SP2013 | LNRFSTMPF*********** | 6.35 ± 4.17 | −16.05 ± 0.69 | 0.32 ± 0.86 |
| 14 | SP2014 | LRRFSTNLPFNLF******* | 2.95 ± 3.01 | −22.77 ± 1.28 | −24.59 ± 15.37 |
| 15 | SP2015 | LRRFSTMPAMFXNINNVXNF | 65.40 ± 1.20 | 99.75 ± 0.25 | 95.60 ± 0.31 |
| 16 | SP2016 | LRRFSTMPFAFXNINNVXNF | 63.60 ± 0.35 | 99.60 ± 0.40 | 98.33 ± 1.14 |
| 17 | SP2017 | LRRFSTMPFMAXNINNVXNF | 65.10 ± 0.57 | 99.60 ± 0.40 | 99.27 ± 0.21 |
| 20 | SP2020 | **********FXNINNVXN* | 20.55 ± 0.46 | 40.35 ± 3.45 | 77.51 ± 2.36 |
| 21 | SP2021 | **********FXNIN***** | 8.17 ± 2.27 | −23.68 ± 0.59 | 3.45 ± 2.25 |
| 22 | SP2022 | LRRFSTMPFMFSNINNVSNF | 50.47 ± 0.66 | 92.56 ± 0.97 | 97.95 ± 1.27 |
| 23 | SP2023 | LRRFSTMPFMFANINNVANF | 48.61 ± 0.53 | 99.59 ± 0.13 | 98.19 ± 0.03 |
| 24 | SP2024 | LRRFSTMPFMFININNVINF | 73.20 ± 0.78 | 92.92 ± 1.78 | 98.58 ± 0.12 |
| 25 | SP2025 | LRRFSTMPFMFTNINNVTNF | 59.10 ± 0.85 | 96.08 ± 0.15 | 98.69 ± 0.08 |
| 27 | SP2027 | LRRFSTMPFMFVNINNVVNF | 62.20 ± 0.64 | 98.78 ± 0.14 | 95.12 ± 0.62 |
| 35 | SP2035 | LRRFSTMPFAFININNVINF | 46.58 ± 4.23 | 69.33 ± 0.14 | 94.80 ± 0.93 |
Modeling overview
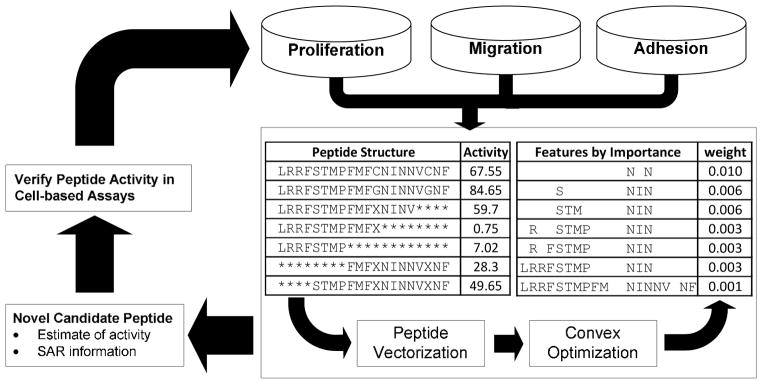
In Figure 1, we outline the peptide modeling procedure. The methods are based on data that associates peptide features with a quantitative activity score (e.g., endothelial cell (EC) proliferation inhibition activity). Peptides are converted into unique sparse vector of features. For example, Figure 2 shows the vectorization of the short peptide LRRFSTMPFMF. In the simplest methodology that we consider, each feature uniquely identifies an amino acid at a single position. We use convex optimization to select features that differentiate highly active and inactive peptides. We formulate the convex optimization objective in a way that can be solved quickly to global optimality.
Figure 1. Peptide optimization overview.
An overview of the peptide optimization framework. The procedure is based on data that associates peptides with an activity score (e.g., endothelial cell proliferation inhibition activity). The peptides are converted into unique sparse vectors. We use convex optimization to select features that differentiate high activity peptides from low performing peptides. The selected features can be used to help understand the structure activity relationships of the peptides. New peptides can be synthesized based on the SAR information. A feedback loop can be created by adding new experimentally tested peptides to the database.
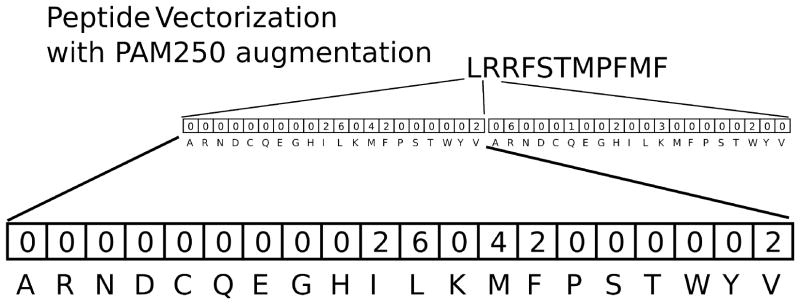
Figure 2. Peptide vectorization.
Each peptide is converted into a sparse vector which uniquely maps specific amino acids to positions in the peptide. The mapping is augmented by the PAM250 amino acid substation matrix. PAM matrices are based on the empirical mutation rate of amino acids in evolutionarily related proteins. For example, the figure shows the vectorization of the peptide LRRFSTMPFMF. The first amino acid leucine (L) can mutate to isoleucine (I), methionine (M), phenylalanine (F), and valine (V) at a rates greater than expected by chance. The weights assigned to these amino acids are given by log odds ratio in the PAM250 matrix. All other amino acids mutate from leucine at a lower rate than expected by chance. As a result, their value is set to zero. The PAM matrix gave us a principled way to associate common amino acids based on their chemical and structural properties.
Peptide-specific QSAR Method Comparison
We developed four approaches to model the data in Table 1 and learn about the structure-activity relationship of type IV collagen peptides. The approaches were based on the least absolute shrinkage and selection operator (Lasso).31 The approaches differed in the features that they consider and the weight assigned to training examples. The specific details of these approaches can be found in the Materials and Methods section.
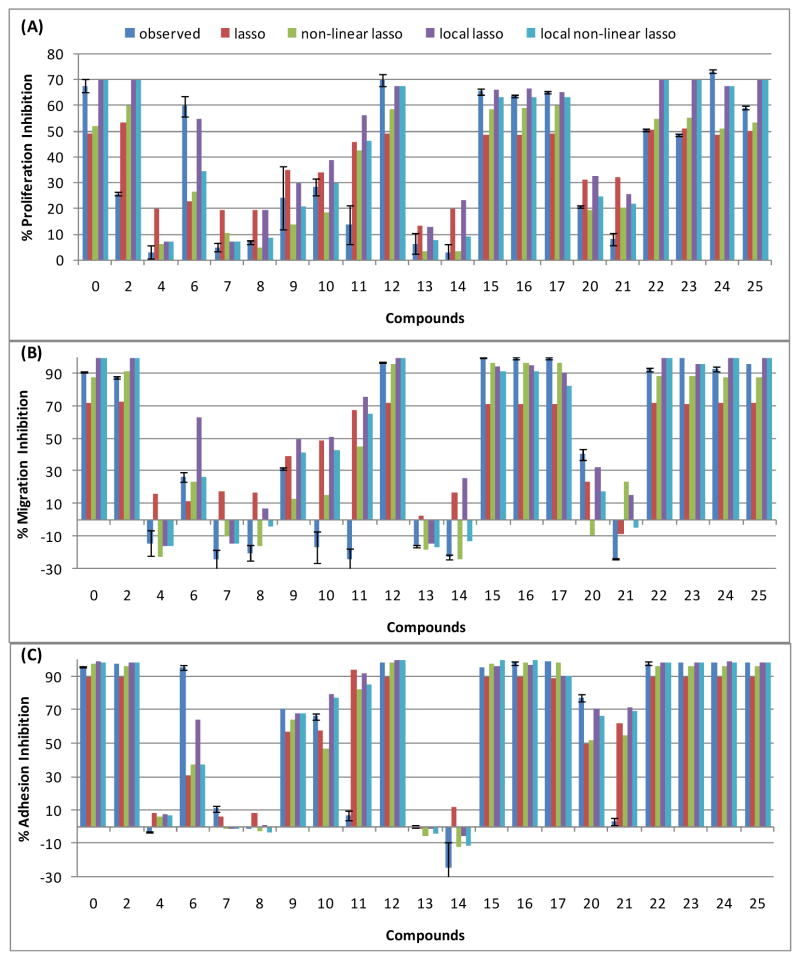
In Table 2, we compared four methods for their ability to predict peptide efficacy. We compared each of these methods to a naive featureless method that always predicted the average activity from the training set. The methods were evaluated on three datasets that measured the ability of peptides to inhibit endothelial cell proliferation (A), migration (B), and adhesion (C). To compare these approaches, we took a leave-one-out cross validation (LOOCV) approach. The concept of LOOCV is that we use all but a single peptide to train the model. We then use that model to predict the efficacy of the single peptide, which was left out. This allowed us to compute the error between predicted and observed activity measurements. To determine which methods were statistically superior to others, we conducted t-tests for all pairs of methods based on their squared test errors. Significantly low test errors indicate better performance. The table gives the p-value associated two-tailed paired t-test. At the 0.05 level, all of the models had lower error than the naive featureless method. Also, the non-linear Lasso method had significantly less error than the Lasso method. These results held over all three datasets. Based on these results, the rest of the study was performed using non-linear Lasso. In Figure 3, we show the observed and leave-one-out predictions for each method for all peptides in the dataset in endothelial cell proliferation, migration, and adhesion assays. The figure illustrates that no single method had the least error in all trials, and that the predictive performance is good even in cases where percent inhibition is negative as seen in the migration and adhesion datasets.
Table 2. Comparison of algorithms for predicting peptide efficacy.
We tested 5 methods for their ability to predict peptide efficacy. A complete description of each method can be found in Materials and Methods. The methods were evaluated on three datasets that measured the ability of peptides to inhibit endothelial cell proliferation (A), migration (B), and adhesion (C). For each method and dataset, we compute LOOCV test error. To determine which methods were superior to others, we conducted t-tests for all pairs of methods based on their squared test errors. Significantly low test errors indicate better performance. The table gives the p-value associated two-tailed paired t-test. At the 0.05 level, the naive featureless method had significantly higher error than all other methods. Also, the non-linear Lasso method had significantly less error than the Lasso method.
| (A) Proliferation Models | Lasso | non-linear Lasso | local Lasso | local non-linear Lasso | featureless |
|---|---|---|---|---|---|
| Lasso | - | 0.004 | 0.580 | 0.218 | 0.003 |
| non-linear Lasso | - | 0.496 | 0.815 | 0.001 | |
| local Lasso | - | 0.343 | 0.018 | ||
| local non-linear Lasso | - | 0.007 | |||
| featureless | - | ||||
| (B) Migration Models | Lasso | non-linear Lasso | local Lasso | local non-linear Lasso | featureless |
| Lasso | - | 0.034 | 0.340 | 0.000 | 0.000 |
| non-linear Lasso | - | 0.179 | 0.651 | 0.000 | |
| local Lasso | - | 0.034 | 0.000 | ||
| local non-linear Lasso | - | 0.000 | |||
| featureless | - | ||||
| (C) Adhersion Models | Lasso | non-linear Lasso | local Lasso | local non-linear Lasso | featureless |
| Lasso | - | 0.033 | 0.205 | 0.098 | 0.017 |
| non-linear Lasso | - | 0.862 | 0.432 | 0.004 | |
| local Lasso | - | 0.740 | 0.005 | ||
| local non-linear Lasso | - | 0.007 | |||
| featureless | - |
Figure 3. Quantitative predictions of peptide activity using non-linear Lasso.
The observed and predicted activity of the 21 training peptides screened in endothelial cell (A) proliferation, (B) migration, and (C) adhesion assays. Compounds are given in Table 1. Predictions are made using LOOCV to assess the generalization error of the method. Predictions are shown for the four methods described in this Materials and Methods. The results imply an average error of between 14-20% depending on the assay.
QSAR analysis for type IV collagen derived peptides
In the previous sections we make extensive use of leave-one-out cross validations to estimate generalization error. We concluded from these analyses that non-linear Lasso had statistically lower generalization error than Lasso. Low generalization error is an indication that the features used in the models may be useful for understanding the structure-activity relationship of type IV collagen peptides.
In this section and unlike the previous sections, we train models for endothelial cell proliferation, migration, and adhesion based on all of the data in Table 1 except for the external validation set consisting of 27 and 35. The models are structured such that important features receive high weight. The model features (first column) and weights (second column) are given in decreasing order in Table 3. The features are indicated for each row by the change in sequence from the preceding row. The weights were determined using the non-linear Lasso method (as described in Materials and Methods). We analyse these features for QSAR analysis. This approach gives us a way of indirectly identifying putative pharmacophores for the collagen-IV derived peptides.
Table 3. Model details for proliferation, migration, and adhesion inhibition.
Feature selection using the non-linear Lasso method and trained on all of the data from Table 1. The table gives the features and weights selected for the (A) proliferation, (B) migration, and (C) adhesion inhibition models, respectively. The features are indicated for each row by the change in sequence from the preceding row. For each model, pairs of amino acid features (summarized in the first column) were given a weight by a linear model (shown in the second column). Asterisks indicate a preference for a missing amino acid. A full length amino acid sequence (i.e., SP2012) is given at the base of each model for reference.
| (A) Proliferation Inhibition Model | (C) Migration Inhibition Model | ||
| NN | 0.009 | ||
| NIN | 0.009 | ||
| NIN | 0.009 | ||
| L NIN | 0.009 | ||
| LR NIN | 0.009 | ||
| LR F NIN | 0.009 | ||
| LRRF NIN | 0.009 | * N | 0.018 |
| LRRF NIN | 0.009 | * N N | 0.018 |
| LRRF NIN X | 0.003 | ** N N | 0.018 |
| LRRF NIN XN | 0.003 | ** NV N | 0.018 |
| LRRF NIN VXN | 0.003 | ** NVXN | 0.018 |
| LRRF NINNVXN | 0.003 | ** A NVXN | 0.016 |
| LRRF P NINNVXN | 0.001 | ** A NVAN | 0.016 |
| LRRF MP NINNVXN | 0.001 | F* * NVAN | 0.013 |
| LRRF MP NINNVXN | 0.001 | R F* * I NVAN | 0.013 |
| (B) Adhesion Inhibition Model | RR F* *NI NVAN | 0.013 | |
| X * | 0.034 | RRF F* *NI NVAN | 0.013 |
| L X I * | 0.017 | RRF F* *NINNVAN | 0.013 |
| LR XNI * | 0.017 | LRRF F* *NINNVAN | 0.013 |
| LRR XNI * | 0.017 | LRRF F* *NINNVAN | 0.011 |
| LRR XNIN * | 0.017 | LRRF F* *NINNVANF | 0.011 |
| LRRF XNIN * | 0.017 | LRRF F* *NINNVXNF | 0.007 |
| LRRF * XNIN N* | 0.011 | LRRF F* *NINNVXN* | 0.006 |
| LRRF ** XNIN N* | 0.011 | LRRF F* *NINNVXN* | 0.006 |
| LRRF*** XNINN N* | 0.011 | LRRF F* SNINNVXN* | 0.006 |
| LRRF*** XNINNV N* | 0.011 | LRRF F* SNINNVSN* | 0.006 |
| LRRF*** XNINNVXN* | 0.011 | LRRF T F* ININNVSN* | 0.002 |
| LRRF**** XNINNVXN* | 0.011 | LRRFST F* ININNVSN* | 0.002 |
| LRRF**** XNINNVXN* | 0.011 | LRRFST FM *NINNVSN* | 0.002 |
| LRRF**M* XNINNVXN* | 0.009 | LRRFST FMF*NINNVSN* | 0.002 |
| LRRF**MP XNINNVXN* | 0.009 | LRRFS* FMF*NINNVSN* | 0.001 |
| LRRF**MP **XNINNVXN* | 0.003 | LRRFS** FMF*NINNVSN* | 0.001 |
| LRRF**MP ***NINNVXN* | 0.003 | LRRFS***FMF*NINNVSN* | 0.001 |
| LRRF**MP ***NINNVXN* | 0.003 | LRRFS***FMF*NINNVXN* | 0.001 |
| LRRF**MPF***NINNVXN* | 0.002 | LRRF****FMF*NINNVXN* | 0.001 |
| LRRF**MPF**XNINNVXN* | 0.002 | LRRFS***AMF*NINNVXN* | 0.001 |
| LRRF**MPF**XNINNVXN* | 0.001 | LRRFST**AMF*NINNVXN* | 0.001 |
| LRRF**MP****NINNVXN* | 0.001 | LRRFST**AMF*NINNVXN* | 0.001 |
| LRRFSTAPFMFXNINNVXNF | weights | LRRFSTAPFMFXNINNVXNF | weights |
When multiple amino acids are viable options in a position, they are shown in decreasing order of importance. In the migration model (Table 3, C) in the 18th position, L-α-amino-n-butyric acid (indicated by X) is preferred with a weight 0.018 over alanine with a weight of 0.016. The proliferation model (Table 3, A) makes it clear that there are important regions on the N-terminus (LRRF) and the C-terminus (NINNVXN). In the adhesion model (Table 3, B), the highly weighted asterisks in the 20th position indicates that truncation of the phenylalanine may improve the anti-adhesion activity of the peptide. Like the proliferation model, the regions on the N-terminus (LRRF) and C-terminus (NINNVX) are selected. Unlike the proliferation model, the L-α-amino-n-butyric acid in the 12th position is one of the most important features for anti-adhesion activity. The migration model (Table 3, C) highlights the C-terminal (ANINNVXN) as a useful indicator of anti-migration activity; however for full anti-migration activity the LRRF sequence is also required. From all three models we found that both the C-terminal sequence LRRF and the N-terminal sequence XNINNVXN are required for full activity.
Structural association
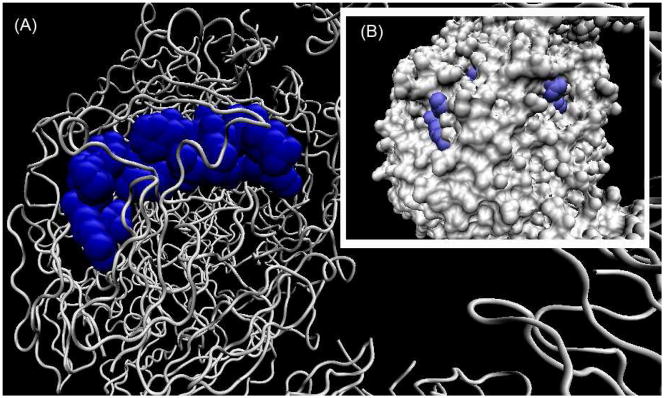
We examined the structure of peptide 0 as it exists in the native type IV collagen NC1 domain (pdb:1T60). In Figure 4, we show the conformation of the peptide in the native protein. By computing the solvent accessible surfaces of the protein, we found two exposed regions corresponding to the N-terminal (LRR) and C-terminal (INN). These regions correlate with the peptide motifs needed for anti-angiogenic activity.
Figure 4. Solvent accessible surfaces of the peptide 0 in non-collagenous (NC1) domain of collagen IV.
(A) the location of the peptide 0 in the NC1 domain of collagen IV. (B) the solvent exposed surfaces of peptide 0. The regions at the N-terminus and C-terminus are solvent accessible.
Experimental model validation
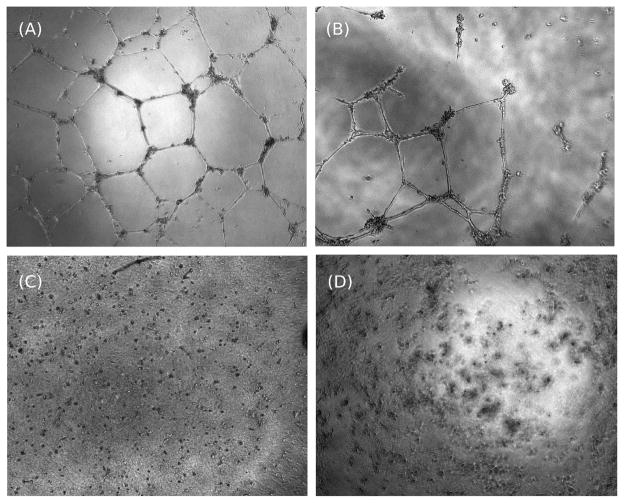
Two peptides, 27 and 35, were held out as an external validation set. Models for proliferation, migration, and adhesion were trained using all other peptides from Table 1. Based on these models, peptides 27 and 35 were predicted to have similar activity. They were predicted to have 54.15, 93.35, and 97.54 percent proliferation, migration, and adhesion inhibition, respectively. Based on the experimentally determined activities given in Table 1 and predicted activities, R2 values on the external validation set were 0.84, 0.85, and 0.99 for the proliferation, migration, and adhesion models, respectively.32 From the R2 values on the external validation set, we could conclude that the models were predictive for anti-angiogenesis phenotypes. In Figure 5, endothelial cell tube formation assays at 100μM confirmed the potency of peptides 27 (Figure 5, C) and 35 (Figure 5, D), relative to a vehicle control (Figure 5, A) and a weaker peptide 8 (SP2008) (Figure 5, B).
Figure 5. Endothelial tube formation assay.
Endothelial cell tube formation assays are useful indicators of angiogenesis potential. (A) Tube formation for the positive control (vehicle control). Entothelial cell tube formation without an added compound. HUVECs form robust tube structures (B) Endothelial cell tube formation with the addition of 100μM of 8. The figure shows only partial inhibition of tube structures. (C) 100μM of 27 completely inhibits the formation of tube structures. (D) 100μM of 35 completely inhibits the formation of tube structures.
Discussion and Conclusions
Type IV collagens are basement membrane proteins that are essential for binding cells to the extracellular matrix.33 Type IV collagen derived peptides have proven to be effective inhibitors of angiogenesis.34 Using the models trained using the data from Table 1, we found a pair of regions namely LRRF at the C-terminus and XNINNVXN at the N-terminus are needed for full activity. This pair of important regions indicates that secondary structure or multiple binding sites may be important for the endothelial cell proliferation, migration, and adhesion inhibition activity of type IV collagen derived peptides. These results are consistent with a previous study on the tumstatin peptide by Eikesdal et al..35 They found that the mutations to the NINN region resulted in a significant change in EC proliferation inhibition. These results also indicate that truncations to the 20-mer peptide with the exception of the phenylalanine in the 20th position would be detrimental to the activity of the collagen IV derived peptides.
In this article, we describe four novel peptide-specific QSAR approaches. We compared these approaches by testing their ability to predict the outcome of in vitro experiments. The comparison indicated that one approach called non-linear Lasso had statistically lower generalization error than Lasso (Table 2). We showed the individual predictions made by this approach in Figure 3. We found that the predictions made using the all four approaches were statistically significant compared to a method based on naive predictions. These results gave us confidence in the utility of the peptide-specific QSAR models. We analyzed the features of these models to learn about the structure-activity relationship of collagen IV derived peptides. By analysing the structure of the collagen IV NC1 domain, we found that the solvent accessible regions of the peptide in the parent protein correlated with the motifs needed for anti-angiogenic activity.
Materials and methods
Peptide dataset
All peptides were synthesized by New England Peptide with at least 95% purity evaluated using both HPLC and MALDI by the manufacturer. Table 1 gives the compound structures in terms of the one letter amino acid codes. Truncated amino acids are indicated by asterisks. The error in the activity measurements was based on two biological replicates each derived from the mean of three technical replicates. The data are shown as percent inhibition relative to a vehicle control. A single dose was selected for each dataset that produced a diverse set of activities for the candidate peptides. Proliferation and adhesion measurements were taken at a peptide concentration of 100μM, while migration measurements were taken with a compound dose of 50μM.
Cell culture
Human umbilical vein endothelial cells (HUVEC) were purchased from Lonza and were grown under the manufacturer’s recommendation using Endothelial Basal Media (EBM-2) supplemented with the Bullet Kit (EGM-2, Lonza). Cells of passages 2–7 were used for experiments. Cells were grown at 37°C in a humidified incubator with 5% CO2.
Proliferation assays
Colorimetric WST-1 reagent (Roche, IN) was used to perform the proliferation assays. HUVECs were plated in 96-well plates at a 2000 cell/well density. Peptides at 100 μM in fully supplemented media were added to the adherent cells and incubated for 72 hours. WST-1 reagent was added in serum free media for four hours and the color intensity was measured at 450 nm with Victor-V plate reader (Perkin Elmer, MA).
Migration assay
The effect of the migration inhibition of the peptides on the cells was determined using electrical impedance measurements with a continuous and real time migration assay (RT-CIM, ACEA Biosciences, CA). The top compartment of the CIM plate was coated with fibronectin (20μg/ml) and 45,000 HUVEC/well were added either in the presence or absence of the peptide at 50 μM. Fully supplemented media was added to the bottom compartment serving as chemoattractant. The migration of the cells is measured by the integrated sensors in the bottom side of the porous membrane which divides the two chambers. This technology allows for easy quantification of cell migration by monitoring the cell index (derived from the measured impedances).
Adhesion assay
The adhesion inhibitory potential of the peptides was also measured using RT-CIM technology. In this instance single compartment E-plates (ACEA, Biosciences,CA) were used, in which 25,000 HUVEC/well were plated in the presence or absence of the peptides at 100 μM and the adhesion measured by the changes in the cell index amplitude for 3 hours.
Tube formation
Tube formation assay was performed by following the published protocol by Arnaoutva et. al.36 Briefly, 96 well plates were coated with Geltrex, Reduced Growth Factor Basement Membrane Matrix (Invitrogen, CA) (50μl/well) and incubated at 37ºC for 30 minutes to allow gelation to occur. HUVECs were added to the top of the gel at a density of 15,000 cells/well in the presence or absence of the peptide (100 μM). The positive control included the same amount of solvation vehicle (i.e., DMSO) as the experimental condition. Cells were incubated at 37ºC with 5%CO2 overnight and pictures were captured with a CCD Sensicam camera mounted on a Nikon inverted microscope.
Peptide-specific QSAR approaches
We took as input a set of peptide sequences along with an experimentally measured efficacy for each peptide. The method returned a model which could be used to predict the efficacy of hypothetical peptides from the same class. The method worked by converting each peptide sequence into an input space of amino acids and positions. Those were the explanatory variables in the peptide-specific QSAR modeling framework. A weight for each feature was learned using non-negative Lasso regression37 with the peptide efficacies as response variables. The scaling term for the L1-norm regularizer was determined using leave-one-out cross validation. Despite evaluating many features, the use of L1-norm regularization allowed the model to avoid over-fitting. The convex nature of the optimization problem allowed the method to quickly reach the globally optimal solution without a combinatorial search of input space. The software which was implemented in Matlab using CVX38 is freely available upon request.
Lasso with an amino acid substation matrix
Without loss of generality, we describe the method in terms of the 20 common amino acids. Given m peptides of length n, let pij be amino acid j in peptide i. Let r be a list of all 20 natural amino acids. Let S be a 20 by 20 amino acid association matrix, in this study we use the PAM250 matrix,39 such that S(a,b) gives the association between amino acid a and b. We use the PAM250 matrix as a principled approach to give weight to amino acids with similar biochemical properties. Let A be an m by 20n matrix that encodes the amino acid sequences, such that
| (1) |
Let b be a vector of length m representing the activity of each peptide. In this study, the quantitative measure of activity is given by percent endothelial cell proliferation, migration, or adhesion inhibition. Our goal is to learn values in the weight vector x of length 20n. The values in the weight vector x correspond to the relative importance of the features considered in the model. Using this formulation, we solve the standard Lasso objective subject to x ≥ 0. Lasso is composed of the least-squares objective regularized by the L1 norm of the weight vector. The parameter λ influences the sparsity of the weight vector x
| (2) |
Non-linear Lasso
In the previous section, we described the linear version of Lasso using only the input space described in A. As an alternate approach, we expand on the input space given in the previous approach to a feature space consisting of pairs of features. Let A′ be an m by (20n)2 matrix. Although the number of features is large, we use sparse matrices to eliminate unused variables and reduce the problem size. We make use of aggressive regularization to avoid over-fitting. The Lagrange multiplier λ is selected automatically by leave-one-out cross validation. We use the objective from equation (2) except that we make use of A′ and the x vector is of length (20n)2.
Locally-weighted methods
We extend both linear and non-linear Lasso to construct locally-weighted variants of both methods. The idea is that we will weight training examples in A by their proximity to the vectorized peptide y to be predicted. The intuition is that we prefer to make smaller training errors for points close to the test point y. The weight w assigned to each training example in A is given in equation (3).
| (3) |
The weighted objective for the linear version of Lasso is given in equation (4).
| (4) |
Statistical significance and cross validation
To evaluate the quality of the predictions given by the peptide-specific QSAR approaches, we perform leave-one-out cross validation. For each of the m peptide examples, we split the examples into a test set containing the ith peptide and a training set containing all other peptides. We use the training set of peptides to obtain the weight vector x. Let pi be the vector of length 20n that encodes the ith peptide. The predicted activity qi for the ith peptide is given by
| (5) |
The statistical significance of the predictions is determined by comparing the set of residuals generated using our model predictions with residuals generated using naive model predictions. We test the null hypothesis that the residuals between the observed and predicted values are equal to the residuals between the observed and naive model predictions (i.e., a model that always predicts the mean training efficacy). The alternative hypothesis is that the residuals between the observed and predicted values are less than the residuals between the observed and naive model predictions. We generate a p-value for each model using a one-sided paired t-test. We used R2 as a metric of model performance on the external validation set.
| (6) |
In this metric, experimentally observed values y are compared with predicted values ŷ relative to the mean observed value from the training set ȳ.
Acknowledgments
The work was supported by NIH grants R01 CA138264, R01 HL101200, and U54RR020839.
Abbreviations
- QSAR
quantitative structure activity relationship
- NC1
non-collagenous domain
- CXC
n-terminal cysteine-x- cysteine domain
- TSP-1
thrombospondin 1 domain
- TIMP
tissue inhibitor of metalloproteinases
- EC
endothelial cell
- NP-hard
non-deterministic polynomial-time hard
- Lasso
least absolute shrinkage and selection operator
- LOOCV
leave-one-out cross validation
- HUVEC
human umbilical vein endothelial cells
- L1-norm
one norm
- PAM250
250% point accepted mutation matrix
Footnotes
Authors’ contributions
CGR designed the method, performed the analysis, and wrote the paper. EVR, JEK and NBP performed the in vitro experiments. JSB and ASP motivated the problem, provided guidance for the analysis and manuscript. All co-authors edited the paper.
Competing Interests
The authors’ declare no competing interests.
References
- 1.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 3.O’Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 4.Kamphaus GD, Colorado PC, Panka DJ, Hopfer H, Ramchandran R, Torre A, Maeshima Y, Mier JW, Sukhatme VP, Kalluri R. Canstatin, a novel matrix-derived inhibitor of angiogenesis and tumor growth. Journal of Biological Chemistry. 2000;275:1209. doi: 10.1074/jbc.275.2.1209. [DOI] [PubMed] [Google Scholar]
- 5.Nyberg P, Xie L, Sugimoto H, Colorado P, Sund M, Holthaus K, Sudhakar A, Salo T, Kalluri R. Characterization of the anti-angiogenic properties of arresten, an [alpha] 1 [beta] 1 integrin-dependent collagen-derived tumor suppressor. Experimental cell research. 2008;314:3292–3305. doi: 10.1016/j.yexcr.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeshima Y, Sudhakar A, Lively JC, Ueki K, Kharbanda S, Kahn CR, Sonenberg N, Hynes RO, Kalluri R. Tumstatin, an endothelial cell-specific inhibitor of protein synthesis. Science. 2002;295:140. doi: 10.1126/science.1065298. [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nature Reviews Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 8.Mundel TM, Kalluri R. Type IV collagen-derived angiogenesis inhibitors. Microvascular research. 2007;74:85–89. doi: 10.1016/j.mvr.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyberg P, Xie L, Kalluri R. Endogenous inhibitors of angiogenesis. Cancer research. 2005;65:3967. doi: 10.1158/0008-5472.CAN-04-2427. [DOI] [PubMed] [Google Scholar]
- 10.Karagiannis ED, Popel AS. A systematic methodology for proteome-wide identification of peptides inhibiting the proliferation and migration of endothelial cells. Proc Natl Acad Sci U S A. 2008;105:13775–13780. doi: 10.1073/pnas.0803241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karagiannis ED, Popel AS. A theoretical model of type I collagen proteolysis by matrix metalloproteinase (MMP) 2 and membrane type 1 MMP in the presence of tissue inhibitor of metalloproteinase 2. J Biol Chem. 2004;279:39105–39114. doi: 10.1074/jbc.M403627200. [DOI] [PubMed] [Google Scholar]
- 12.Karagiannis ED, Popel AS. Anti-angiogenic peptides identified in thrombospondin type I domains. Biochem Biophys Res Commun. 2007;359:63–69. doi: 10.1016/j.bbrc.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karagiannis ED, Popel AS. Peptides derived from type I thrombospondin repeat-containing proteins of the CCN family inhibit proliferation and migration of endothelial cells. Int J Biochem Cell Biol. 2007;39:2314–2323. doi: 10.1016/j.biocel.2007.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karagiannis ED, Popel AS. Novel anti-angiogenic peptides derived from ELR-containing CXC chemokines. J Cell Biochem. 2008;104:1356–1363. doi: 10.1002/jcb.21712. [DOI] [PubMed] [Google Scholar]
- 15.Koskimaki JE, Karagiannis ED, Rosca EV, Vesuna F, Winnard PT, Jr, Raman V, Bhujwalla ZM, Popel AS. Peptides derived from type IV collagen, CXC chemokines, and thrombospondin-1 domain-containing proteins inhibit neovascularization and suppress tumor growth in MDA-MB-231 breast cancer xenografts. Neoplasia. 2009;11:1285–1291. doi: 10.1593/neo.09620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koskimaki JE, Karagiannis ED, Tang BC, Hammers H, Watkins DN, Pili R, Popel AS. Pentastatin-1, a collagen IV derived 20-mer peptide, suppresses tumor growth in a small cell lung cancer xenograft model. BMC Cancer. 2010;10:29. doi: 10.1186/1471-2407-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cano Mdel V, Karagiannis ED, Soliman M, Bakir B, Zhuang W, Popel AS, Gehlbach PL. A peptide derived from type 1 thrombospondin repeat-containing protein WISP-1 inhibits corneal and choroidal neovascularization. Invest Ophthalmol Vis Sci. 2009;50:3840–3845. doi: 10.1167/iovs.08-2607. [DOI] [PubMed] [Google Scholar]
- 18.Rosca EV, Koskimaki JE, Rivera CG, Pandey NB, Tamiz AP, Popel AS. Anti-angiogenic peptides for cancer therapeutics. Curr Pharm Biotechnol. 2011;12:1101–1116. doi: 10.2174/138920111796117300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA. PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des. 2006;20:647–671. doi: 10.1007/s10822-006-9087-6. [DOI] [PubMed] [Google Scholar]
- 20.Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ. PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008;36:W223–228. doi: 10.1093/nar/gkn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Güner OF. Pharmacophore perception, development, and use in drug design. Intl Univ Line; 1999. [Google Scholar]
- 22.Mason JS, Morize I, Menard PR, Cheney DL, Hulme C, Labaudiniere RF. New 4-point pharmacophore method for molecular similarity and diversity applications: overview of the method and applications, including a novel approach to the design of combinatorial libraries containing privileged substructures. Journal of medicinal chemistry. 1999;42:3251–3264. doi: 10.1021/jm9806998. [DOI] [PubMed] [Google Scholar]
- 23.Blankley C. Quantitative structure-activity relationships of drugs. Academic Press; New York: 1983. Introduction: A review of QSAR methodology. [Google Scholar]
- 24.Dudek AZ, Arodz T, Galvez J. Computational methods in developing quantitative structure-activity relationships (QSAR): a review. Combinatorial Chemistry; High Throughput Screening. 2006;9:213–228. doi: 10.2174/138620706776055539. [DOI] [PubMed] [Google Scholar]
- 25.Hansch C, Leo A, Hoekman D. Exploring QSAR.:. Fundamentals and applications in chemistry and biology. An American Chemical Society Publication; 1995. [Google Scholar]
- 26.Kubiny H. Variable selection in QSAR studies. I. An evolutionary algorithm. Quantitative Structure Activity Relationships. 1994;13:285–294. [Google Scholar]
- 27.Lin ZH, Long HX, Bo Z, Wang YQ, Wu YZ. New descriptors of amino acids and their application to peptide QSAR study. Peptides. 2008;29:1798–1805. doi: 10.1016/j.peptides.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhou P, Chen X, Shang Z. Side-chain conformational space analysis (SCSA): a multi conformation-based QSAR approach for modeling and prediction of protein-peptide binding affinities. J Comput Aided Mol Des. 2009;23:129–141. doi: 10.1007/s10822-008-9245-0. [DOI] [PubMed] [Google Scholar]
- 29.Doytchinova IA, Blythe MJ, Flower DR. Additive method for the prediction of protein-peptide binding affinity. Application to the MHC class I molecule HLA-A* 0201. Journal of proteome research. 2002;1:263–272. doi: 10.1021/pr015513z. [DOI] [PubMed] [Google Scholar]
- 30.Finn P, Halperin D, Kavraki L, Latombe JC, Motwani R, Shelton C, Venkatasubramanian S. Geometric manipulation of flexible ligands. Applied Computational Geometry Towards Geometric Engineering. 1996:67–78. [Google Scholar]
- 31.Tibshirani R. Regression Shrinkage and Selection Via the Lasso. Journal of the Royal Statistical Society, Series B. 1994;58:267–288. [Google Scholar]
- 32.Hawkins DM, Basak SC, Mills D. Assessing model fit by cross-validation. Journal of chemical information and computer sciences. 2003;43:579–586. doi: 10.1021/ci025626i. [DOI] [PubMed] [Google Scholar]
- 33.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microscopy research and technique. 2008;71:357–370. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeshima Y, Manfredi M, Reimer C, Holthaus KA, Hopfer H, Chandamuri BR, Kharbanda S, Kalluri R. Identification of the anti-angiogenic site within vascular basement membrane-derived tumstatin. J Biol Chem. 2001;276:15240–15248. doi: 10.1074/jbc.M007764200. [DOI] [PubMed] [Google Scholar]
- 35.Eikesdal HP, Sugimoto H, Birrane G, Maeshima Y, Cooke VG, Kieran M, Kalluri R. Identification of amino acids essential for the antiangiogenic activity of tumstatin and its use in combination antitumor activity. Proceedings of the National Academy of Sciences. 2008;105:15040. doi: 10.1073/pnas.0807055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnaoutova I, George J, Kleinman HK, Benton G. The endothelial cell tube formation assay on basement membrane turns 20: state of the science and the art. Angiogenesis. 2009;12:267–274. doi: 10.1007/s10456-009-9146-4. [DOI] [PubMed] [Google Scholar]
- 37.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B (Methodological) 1996;58:267–288. [Google Scholar]
- 38.Grant M, Boyd S. Graph implementations for nonsmooth convex programs. Recent advances in learning and control. 2008:95–110. [Google Scholar]
- 39.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences: CABIOS. 1992;8:275. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]