Abstract
Tumor-specific promoters that limit transgene expression to tumors play a vital role in cancer gene therapy. Although tumor specific, the human Survivin promoter (pSurv) is a poor activator of transcription. A bi-directional Two Step Transcriptional Amplification (TSTA) system was designed to enhance expression of the therapeutic gene TNF-alpha Related Apoptosis Inducing Ligand (TRAIL or TR) and the reporter gene Firefly Luciferase (FL) from pSurv. An adenoviral vector carrying the enhanced targeting apparatus (Ad-pSurv-TR-G8-FL) was tested for efficiency and specificity of gene expression in cells and in living animals. Compared to the one-step systems (Ad-pSurv-FL or Ad-pSurv-TR), the bi-directional TSTA system showed 10-fold higher expression of both the therapeutic and the reporter gene and their expression correlated in cells (R2=0.99) and in animals (R2=0.67). Noninvasive quantitative monitoring of magnitude and time variation of TRAIL gene expression was feasible by bioluminescence imaging of the transcriptionally linked FL gene in xenograft tumors following intratumoral adenoviral injection. Moreover, the TSTA adenovirus maintained promoter specificity in non-target tissues following tail-vein administration. These studies demonstrate the potential of the bi-directional TSTA-system to achieve high levels of gene expression from a weak promoter, while preserving specificity and the ability to image expression of the therapeutic gene noninvasively.
Introduction
Transcriptionally targeted cancer gene therapy exploits the fact that cancer cells tend to activate or over-express many genes that are important for uncontrolled proliferation and cell survival1. Promoter elements of such genes are therefore engaged to achieve cancer-specific expression of a therapeutic gene and killing of tumor cells. Survivin, a member of the inhibitor of apoptosis (IAP) family that counteracts cell death and controls mitotic progression, is over-expressed in common cancers but not in normal adult tissues2. Survivin mRNA or protein over-expression was demonstrated in tumors of the lung, breast, colon, ovaries, skin etc3–5. Recent studies demonstrated the ability of the human Survivin promoter to specifically target tumor cells in lung, breast cancer and glioma model6–8.
Among the various tumor-specific cytotoxic genes being explored, Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL/Apo2L) has shown a lot of potential as a therapeutic agent for cancer9–11. TRAIL, a type II transmembrane protein belonging to the TNF-family of cytokines, can rapidly induce apoptosis in a wide variety of tumorigenic or transformed cells via interaction with the death receptors DR4/TRAIL-R1 and DR5/TRAIL-R214. Although normal tissues including lymphocytes, spleen etc. seems to contain mechanisms that protect them from apoptosis induction by TRAIL, recent reports have raised concerns of potential hepatotoxicity due to systemically administered recombinant TRAIL in humans15,16. Other disadvantages of the recombinant protein include a high dose requirement and a short half-life. This has led to a growing interest in TRAIL-mediated gene therapy to circumvent these problems. Intratumoral delivery of TRAIL via a replication-competent Ad5-TRAIL adenovirus showed significant antitumor activity17,18. However, the wide spectrum of cell types infected by adenovirus necessitates a requirement for targeting. Therefore, controlling the expression of TRAIL by the tumor-specific human Survivin promoter can render the therapeutic regimen safe.
However, a major drawback of most tumor-specific promoters is their poor transcriptional efficiency. Consequently, in gene therapy applications, they often fail to produce optimum levels of the therapeutic protein, resulting in poor treatment outcome. We have previously demonstrated the utility of the Two-Step Transcriptional Amplification (TSTA) system for augmenting the transcriptional activity of weak promoters like the prostate-specific promoter PSA or the Vascular Endothelial Growth Factor (VEGF) promoter21–24. We rationalized that the transcriptional amplification power of the TSTA system will enable us to improve the therapeutic efficacy of the TRAIL gene therapy regimen in two ways: firstly by producing multiple copies of TRAIL from the Survivin promoter and secondly by minimizing the viral dose.
An ideal gene therapy paradigm is one which apart from achieving good expression of the therapeutic transgene allows one to monitor gene delivery and duration of transgene expression. In the past we reported a novel bi-directional promoter system based on the TSTA strategy that is capable of driving two individual genes simultaneously and in a highly correlated fashion26. The bi-directional TSTA system is extremely useful in gene therapy applications for determining the spatio-temporal distribution of the delivered therapeutic gene, indirectly, by studying the expression of a transcriptionally coupled reporter gene. The goal of the current study was to employ the system to amplify transgene expression from the weak Survivin promoter, and to enable imaging of the amplified expression by the use of non-invasive imaging modalities. We demonstrate for the first time that a bi-directional TSTA based adenovirus can amplify expression of the TRAIL gene from the weak Survivin promoter in cell culture and in living animals and the increased expression can be imaged non-invasively in living animals over a period of time.
Results
The 977bp human Survivin promoter is considerably weak in activity compared to the constitutive CMV promoter
The luciferase reporter assay was employed to quantify the activity of the human Survivin promoter. The 977bp long Survivin promoter (pSurv) was employed to drive the expression of firefly luciferase reporter gene in the basic PGL3 backbone. Although active in different cancer cells, Survivin promoter activity is significantly weak when compared to that of the robust and constitutive human Cytomegalovirus (pCMV) promoter in different cancer cell lines (0.5–3% of that of the pCMV promoter) (Table 1).
Table 1. Comparing activity of the Survivin promoter with the viral CMV promoter in different cancer cell lines.
pCMV-FL and pSurv-FL are PGL3 plasmids where the FL reporter gene is driven by the CMV or Survivin promoter respectively. Firefly activity obtained in different cell lines normalized to microgram protein is given as mean values ± SD. Sensitivity Index = (RLUpSurv-FL/RLUpCMV-FL)×100
| Cancer Cell Line | pCMV-FL RLU/µg protein |
p-Surv-FL RLU/µg protein |
Sensitivity Index % pCMV |
|---|---|---|---|
| HCT116 | 92±1.2×104 | 42.4±8.6×102 | 0.46 |
| HT29 | 54.5±8.8×102 | 21±5.8×101 | 3.85 |
| WiDr | 8.85±4.2×105 | 37.6±4.8×102 | 0.42 |
| HT1080 | 8.11±2.1×104 | 68.4±8.8×101 | 0.84 |
| SKOV3 | 21±2.2×103 | 11±3.2×101 | 0.53 |
Schematic representation of pSurv based one-step and TSTA systems
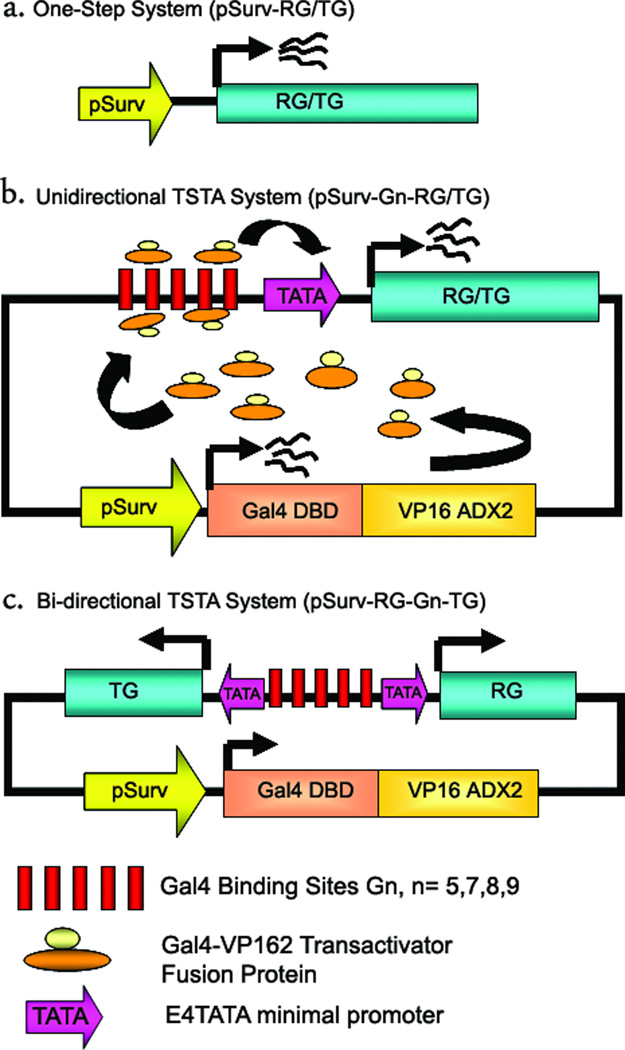
In the current study, TSTA was employed to amplify the strength of the Survivin promoter. For convenience, we will refer to the system where pSurv directly drives the firefly luciferase or TRAIL gene expression as the one-step systems (pSurv-RG/TG) (Fig 1a). Construction of the uni-directional TSTA plasmid is explained in the schematic in (Fig 1b). Broadly, all TSTA plasmids comprised of two main segments-a) an activator segment and b) effector segment cloned into a single plasmid. The activator segment composes of the transactivator fusion protein GAL4-VP2 comprised of the 147 amino-acid Gal4-DNA binding domain (DBD) of the yeast transcription factor Gal4 fused to two tandem copies of the 42 amino-acid transactivation domain (AD) of Herpes Simplex Virus VP16 protein. Transcription of GAL4-VP2 in cells and tissues is under the direct control of pSurv. The effector segment comprised of 17bp-tandem repeats of Gal4-binding sites (Gn) followed by a TATA-box carrying minimal promoter from the adenoviral E4 gene. The minimal promoter controlled the expression of either a reporter gene (RG) (firefly luciferase) or a therapeutic gene (TG) (TRAIL) in the uni-directional TSTA system (pSurv-Gn-RG/TG). The TSTA system works in two steps. In the first step, pSurv drives expression of the recombinant transactivator fusion protein Gal4-VP16. In the next step, this fusion protein binds to the Gal4-responsive promoter in the effectors segment and facilitates transcription of the target gene. The bi-directional single plasmid TSTA plasmid is exactly similar in construction to the uni-directional plasmid, except that in this case instead of one gene a unique Gal4-VP2 responsive bi-directional promoter is employed to express two independent transgenes simultaneously. Of the two transgenes, one is the firefly luciferase (FL) reporter gene and the other is the human TRAIL (TR) therapeutic gene. This bi-directional promoter shown in (Fig 1c) was engineered by placing multiple copies of Gal4-binding sites in between two copies of E4TATA minimal promoters. The two minimal promoters, oriented in two opposite directions, are responsible for expressing their respective downstream genes.
Fig 1. Schematic representations of the different Survivin targeted systems under investigation.
(a). This construct describes the simplified one-step system where the human Survivin promoter (pSurv) drives the expression of either a reporter gene (RG) or a therapeutic gene (TG) directly. (b) This demonstrates the working principle of the uni-directional TSTA system. It is a single plasmid system where pSurv is employed to drive the expression of the transactivator fusion protein Gal4-VP2 (Gal4DBD-VP16ADX2) composed of a Gal4 DNA binding domain fused in frame to two copies of the VP16 transactivation domain. This protein in turn induces expression of a target gene (either RG or TG) under the control of multiple Gal4-binding sites (Gn, where “n” could be 5, 7, 8 or 9) and an E4TATA minimal promoter. The use of the GAL4-VP2 fusion helps in amplifying the level of the target protein in the targeted cells. (c) This represents the bi-directional TSTA system. Unlike the uni-directional TSTA system, the bi-directional system can drive both the TG and the RG, simultaneously and in a highly correlated fashion. This is achieved by the binding of the Gal4-VP2 protein to multiple Gal4-binding sites situated in between two E4TATA minimal promoters, each responsible for driving a downstream gene. Thus this system is helpful in indirectly imaging the expression of a delivered TG by monitoring the level of expression of the transcriptionally coupled RG.
Two Step Transcriptional Amplification (TSTA) system can increase gene expressions from the weak Survivin promoter in cell culture
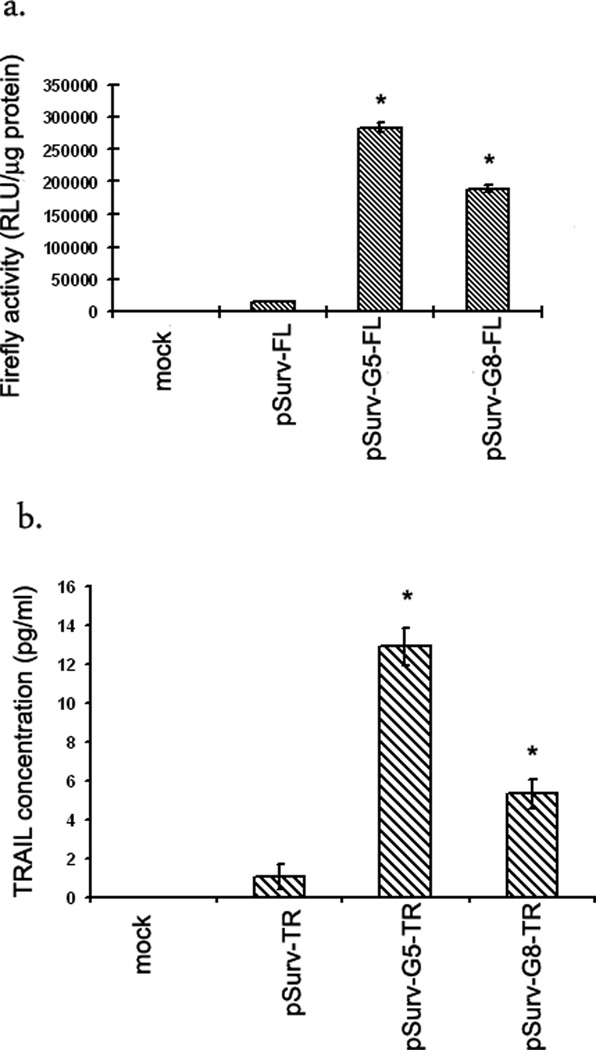
HCT116 cells were transfected with either the One-step (pSurv-FL or pSurv-TR) or the uni-directional TSTA system (pSurv-Gn-FL/TR, n=5 or 8). The pSurv-G5-FL system amplifies expression of the firefly luciferase reporter gene by 20-fold compared to the one-step pSurv-FL system (p<0.001)(Fig 2a). Similarly, the pSurv-G5-TR construct shows a 12-fold higher expression of the TRAIL gene compared to the corresponding one-step system (p<0.001)(Fig 2b). Although on increasing the number of Gal4-binding sites from 5 to 8, the uni-directional TSTA system suffered a loss in the level of amplification (30% for FL and 58% for TR), overall gene expression is still significantly higher than the corresponding one-step systems (14-fold for FL and 5-fold for TR) (p<0.001 in all cases).
Figure 2. Amplification of gene expression from the Survivin promoter by the unidirectional TSTA system in cell culture.
HCT116 cells were transiently transfected with (i) TE buffer (mock), (ii) one-step system (pSurv-FL or pSurv-TR), (iii) G5-based unidirectional TSTA system (pSurv-G5-FL or pSurv-G5-TR) and (iv) G8-based unidirectional TSTA system (pSurv-G8-FL or pSurv-G8-TR). The cells were harvested 24 h after transfection and assayed for (a) FL activity by luminometer and (b) TRAIL expression by Eliza. The error bars represent SEM for triplicate measurements. All values have been normalized for transfection efficiency. * indicates p<0.05 comparing one-step systems vs unidirectional TSTA systems.
Amplification of expression of two independent genes from the Survivin promoter by the bi-directional TSTA required additional Gal4-binding sites
Since the ultimate goal in gene therapy is to be able to express both TG and RG simultaneously, we wanted to optimize the bi-directional TSTA system to achieve robust gene expression. From our previous studies we know that the performance of the TSTA system can depend largely on: 1) number of Gal4 binding sites, and 2) orientation of the genes. Each of these variables can play an important role in modulating gene expression.
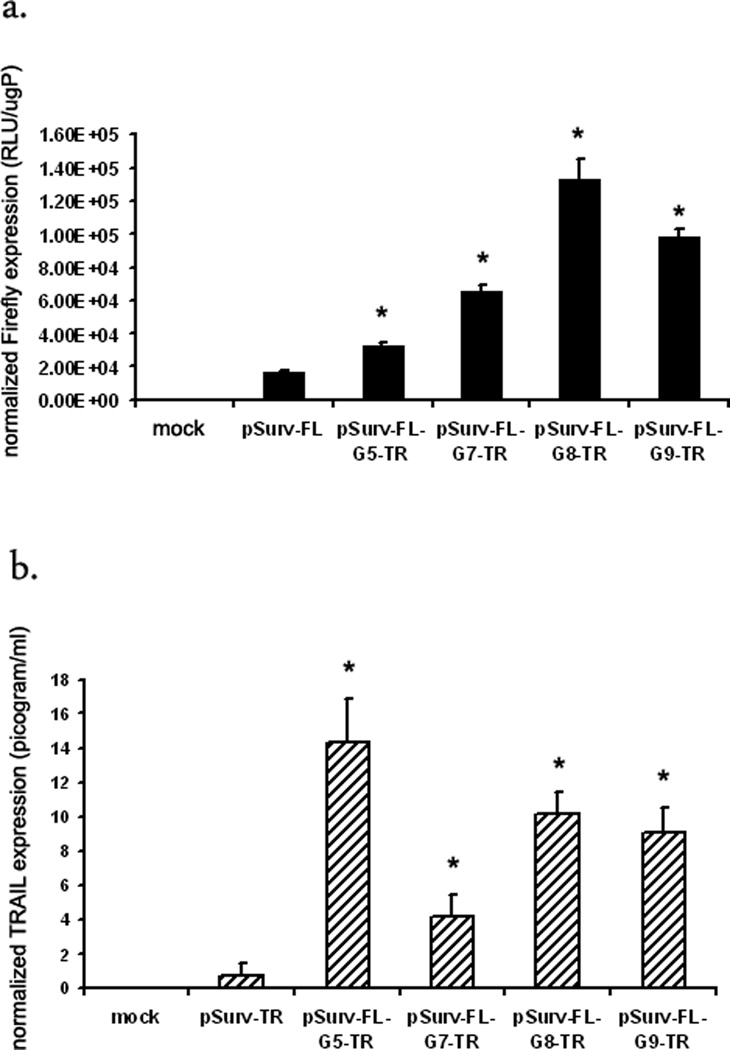
Firstly, to determine the optimum number of Gal4-binding (Gn) sites we built several constructs where Gn varied from G5 to G7, G8 or G9 (Fig 3a). HCT116 cells transfected with the pSurv-FL or pSurv-TR were used as positive controls. Bi-directional plasmid with G5 shows a 12 fold higher expression of the TR gene compared to pSurv-TR, but only a 2-fold higher expression of the FL gene compared to pSurv-FL. On varying Gn from G7–9, expression of FL gene increases from 2-fold in case of G5 system to approximately 7-fold for G7, G8 and G9 systems. However, the fold amplification of the TR gene dropped from 12-fold in case of the G5 system to 5-fold for G7, and 9-fold for G8 and G9 based systems (Fig 3b). Thus, the bi-directional TSTA system requires a higher number of Gal4-binding sites, either 8 or 9 copies of Gal4-binding sites compared to the uni-directional TSTA system in order to obtain significant amplification of gene expressions from a weak promoter.
Figure 3. A bi-directional TSTA system with 8-Gal4-binding sites can amplify expression of both FL and TR genes from the Survivin promoter.
To assess the optimum number of total Gal4-binding sites, 0(G5), 2(G7), 3 (G8) or 4 (G9) additional Gal4-binding sites were inserted in the vicinity of the E4TATA promoter (on the –ve strand) in the bi-directional TSTA system. HCT116 cells transfected with either the different bi-directional TSTA system or the one-step system were harvested, lysed and assayed for (a) FL activity or (b) TRAIL concentration. The error bars represent SEM for triplicate measurements. All values have been normalized for transfection efficiency. * indicates p<0.05 comparing all TSTA systems vs one-step systems.
Orientation of the genes can affect fold amplification in G5-based systems but not in G8-based Systems
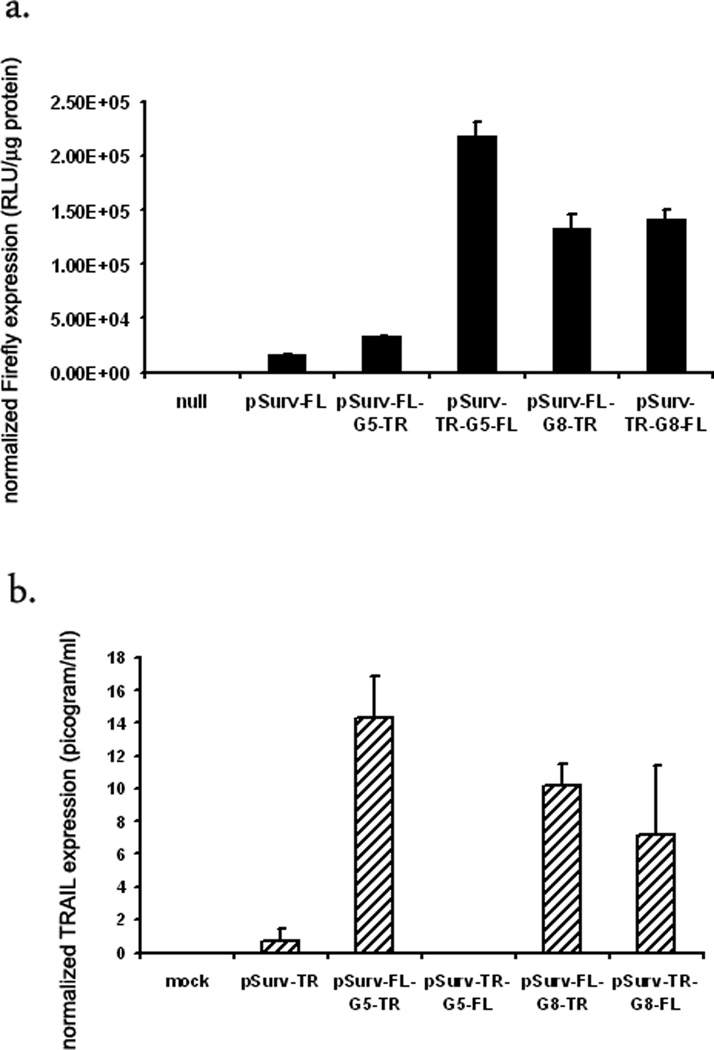
The data in Fig 3a suggests that directionality of the genes around the Gal4-binding sites might effect gene expression. To answer the effect of directionality on TSTA mediated gene amplification, we reversed the orientations of FL and TR genes in the G5 and G8-based bi-directional TSTA systems. The G5-based TSTA system demonstrates a directionality bias. In HCT116 cells FL gene expression is 11-fold higher than the one-step system in one orientation (pSurv-TR-G5-FL), but only 2-fold in the other (pSurv-FL-G5-TR) (Fig 4a). Similarly, expression of TR gene is 12-fold higher in one orientation (pSurv-FL-G5-TR) but almost undetectable in the other orientation (pSurv-TR-G5-FL) (Fig 4b). However, directionality of the genes did not compromise amplification in case of the G8-based systems. As shown in (Fig 4a), regardless of its orientation, the FL gene shows a 7-fold amplification compared to the one-step system. Similarly, there is no significant difference in TR expression levels between the two orientations and the fold amplification is approximately 8-fold.
Fig 4. Fold amplification is independent of the orientation of the genes in the G8-based bi-directional TSTA system.
Two different TSTA systems were examined for any directional bias that might affect the amplification of genes. Accordingly, FL and TR gene positions were swapped in case of both G5- and G8-based bi-directional TSTA systems and compared with the original orientation. HCT116 cells were then transfected with i) TE buffer, ii) one-step system (pSurv-FL or pSurv-TR, G5-based bi-directional TSTA systems iii) pSurv-FL-G5-TRand iv) pSurv-TR-G5-FL (switched iii), G8-based bi-directional TSTA systems v) pSurv-TR-G8-FL and vi) pSurv-FL-G8-TR (switched v). (a) FL activity or (b) TRAIL concentration. The error bars represent SEM for triplicate measurements. All values have been normalized for transfection efficiency. * indicates p<0.05 comparing all TSTA systems vs one-step systems.
Bi-directional TSTA Adenovirus showed amplified expression of FL and TR genes compared to the corresponding one-step Adenoviruses in cell culture
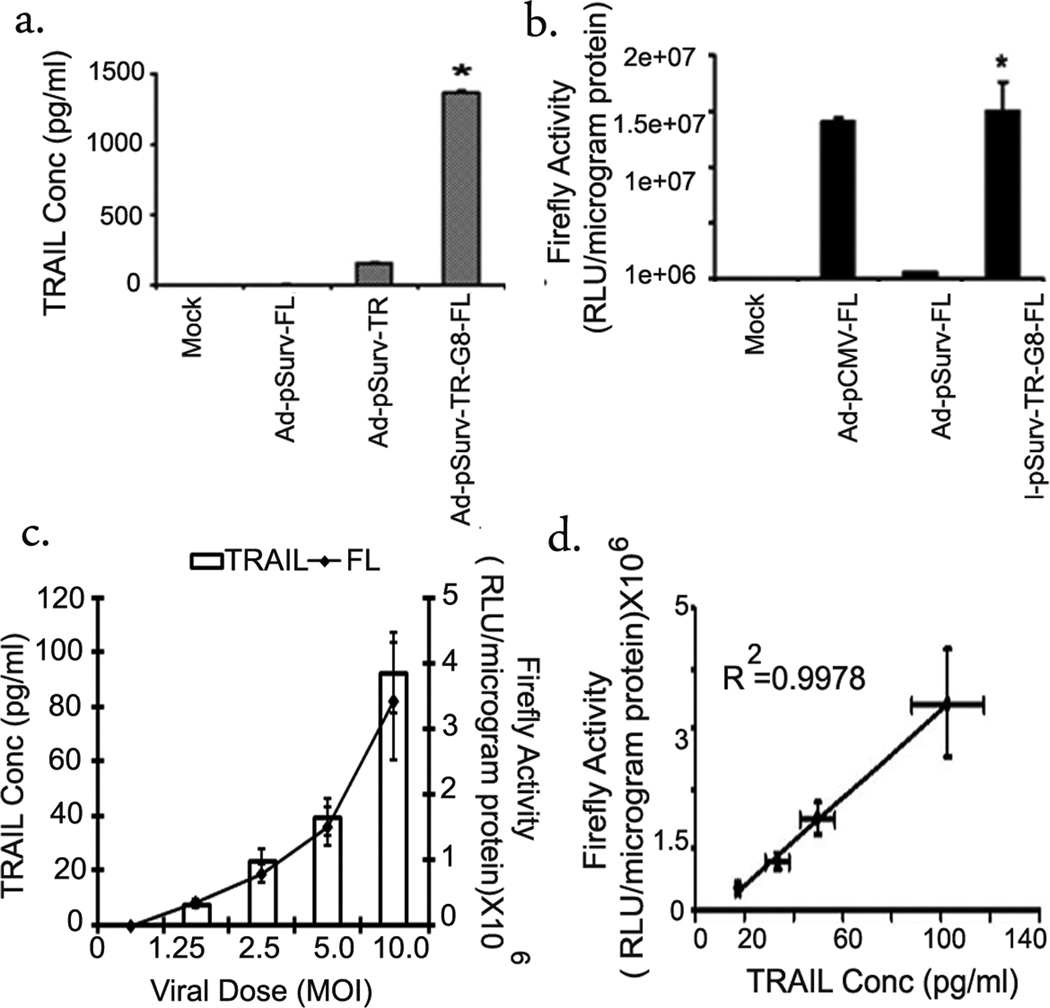
The different systems were cloned into Ad5 genome for future cell and animal studies. HCT116 cells were transduced with either the one-step viruses Ad-pSurv-FL or Ad-pSurv-TR or the bi-directional TSTA virus Ad-pSurv-TR-G8-FL at MOI 15. Forty-eight hours post-infection cell lysates were assayed for firefly luciferase expression using the luminometer and for TRAIL expression using ELISA. TRAIL gene expression is about 34-fold higher in cells treated with the TSTA adenovirus as opposed to the corresponding one-step adenovirus (Ad-pSurv-TR) (1103.33±237.1 vs 32.3±6.2 pg/ml) (p<0.001) (Fig 5a). Like TRAIL, FL gene expression was also 24-fold higher compared to the Ad-pSurv-FL treated cells 48 hours post transfection (1500±26×105 vs 63±3.4 ×105 RLU/µg protein)(p<0.001) (Fig 5b). The bi-directional system was designed to achieve correlated expression of the two genes flanking the bi-directional promoter cassette. According to our results, HCT116 cells infected with increasing doses of the Ad-pSurv-TR-G8-FL adenovirus, show linear increments in expression of both the FL and TRAIL genes (Fig 5c). Importantly, the level of expression of the two genes are highly correlated (R2= 0.99) in cell culture (Fig 5d).
Fig 5. Bi-directional TSTA armed Survivin promoter targeted adenovirus showed amplified expression of FL and TR genes in cell culture.
HCT116 cells infected with i) Media only (mock), ii) Ad-pSurv-FL, iii) Ad-pSurv-TR or iv) Ad-pSurv-TR-G8-FL at MOI 15 were harvested after 48hrs and cells lysates were assayed for TRAIL production (a) or FL activity (b). The error bars represent SEM for triplicate measurements. * indicates p<0.05 comparing TSTA virus vs one-step virus. (c) Linear increase in expression of the FL (line) and TR (bar) genes with increasing dose of bi-directional TSTA adenovirus (Ad-pSurv-TR-G8-FL) (MOI 0–10). The results were obtained 48hrs after infection in HCT116 cells. The error bars indicate SEM for triplicate measurements. (d) Correlation between FL and TR expression in HCT116 cells infected with different doses of Ad-pSurv-TR-G8-FL. The correlation is R2=0.99.
Bi-directional TSTA system showed increased reporter gene expression compared to the one-step system in colorectal cancer xenograft tumors in living animals
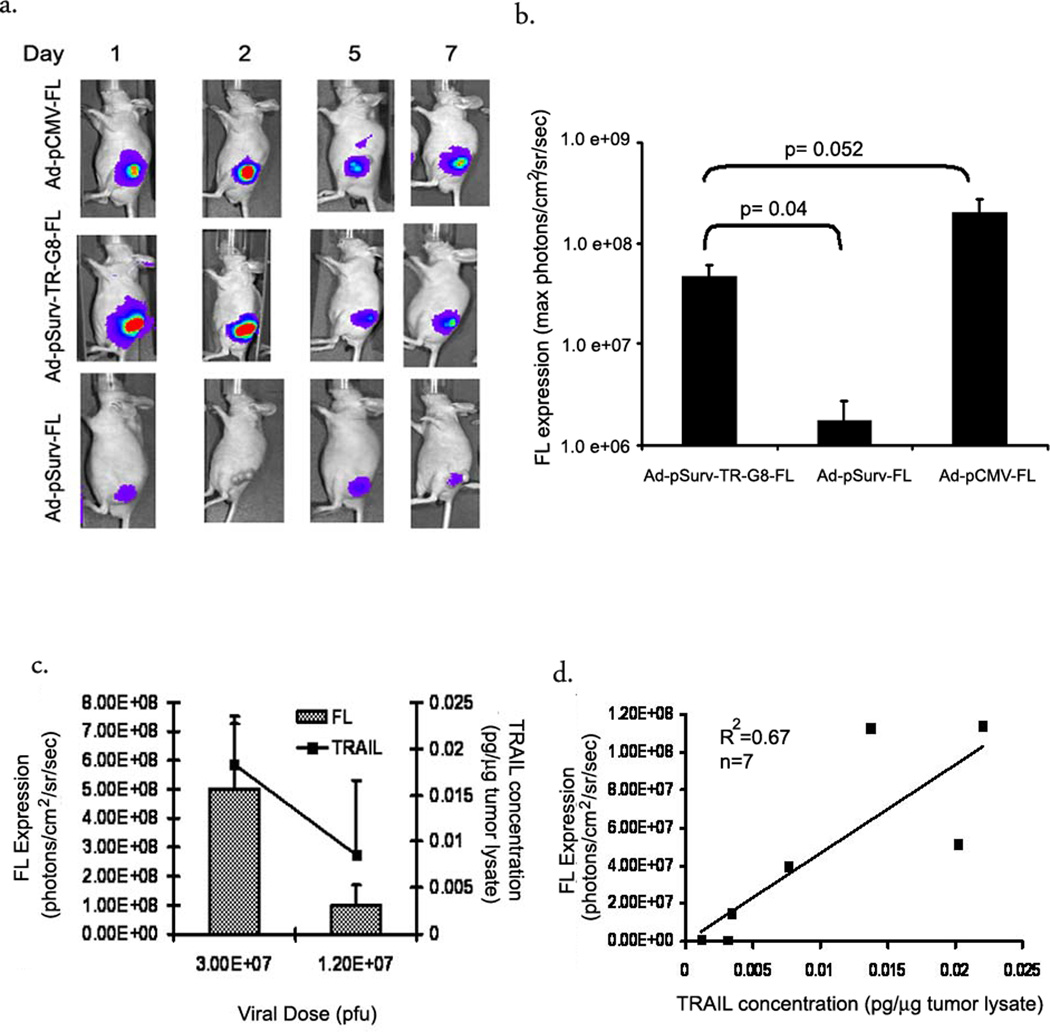
To test the ability of the bi-directional TSTA system for increasing gene expressions in tumor models, we implanted HCT116 xenografts in nude mice. Once the tumors were approximately 0.5mm in diameter, they were injected intratumorally with Ad-pSurv-TR-G8-FL bi-directional TSTA virus (2×107 pfu). One-step adenovirus, Ad-pSurv-FL and Ad-pCMV-FL virus was used as positive controls. All animals were scanned for firefly luciferase activity at different time points post-injection and ROIs were drawn around the tumor to analyze the maximum radiance (Fig 6a and b). TSTA virus injected tumors show a 10-fold high expression of FL luciferase gene compared to the one-step infected tumors at 24hrs (40.3±13.6×106 vs 4±2.4×106 photons/cm2/sec/sr) (p=0.05). Moreover, this amplified expression is only 5-fold lower than that obtained with Ad-pCMV-FL (p=0.052). TSTA adenovirus infected tumors show peak FL expression at 48 hours, with rapid reduction in levels of expression subsequently and by day 7 they are almost equal to background. Within 36hrs of intratumoral injection, firefly expression is visible in the liver and the expression increases gradually over time in Ad-pCMV-FL treated animals (data not shown). Interestingly, no such expression is detected in animals injected with the Survivin promoter targeted adenoviruses during similar time points.
Fig 6. Nonivasive bioluminescence imaging of therapeutic gene expression from the Survivin promoter following intratumoral adenoviral administration.
(a) Optical CCD imaging of a mouse carrying HCT116 xenografts infected with i) Ad-pCMV-FL (top, n=5), ii) Ad-pSurv-TR-G8-FL (middle, n=6) and iii) Ad-pSurv-FL (bottom, n=4). HCT116 xenografts grown on nude mice were injected intratumorally with 2×107 pfu of the different adenoviruses and the animals were imaged for a week. Shown here are images of a representative animal from each group. All images have been adjusted to the same scale. (b) ROIs were drawn around the tumor in these images and photon emission was quantified using the Living Image software. Plot shows a comparison of the intratumoral luciferase signal across the three groups. The error bars represent SEM. (c) Plot shows dose dependent expression of both genes following intratumoral injections of the bi-directional TSTA adenoviral vector. Tumors were injected with either 1.2 or 3.0×107 pfu of the adenovirus (n=3 per group). Firefly expression was detected in live animals by bioluminescence imaging while intratumoral TRAIL concentration was determined by ELIZA ex-vivo. (d) Correlation of FL and TR gene expression from 7 mice injected intratumorally with different doses of Ad-pSurv-TR-G8-FL. Plot of TRAIL expression (picogram/microgram of tumor lysate) measured on tumor lysates ex-vivo versus FL activity expressed as maximum (photons/cm2/sec/sr) obtained from ROIs drawn over the tumors on mouse images non-invasively (R2=0.67, n=7 mice).
Expressions of both Fl and TR genes are highly correlated using the bi-directional TSTA system in living animals
To test if the gene expression still correlated in vivo, we injected tumors with two different doses of the TSTA adenovirus (1.2 and 3×107 pfu). Forty-eight hours later animals were first imaged for firefly expression in the Ad-pSurv-TR-G8-FL injected tumors. After bioluminescence imaging, animals were sacrificed, tumors excised and ex-vivo analysis of TR expression in the tumor lysates were performed by ELISA. The gene expression in the tumors is dose dependent (Fig 6c). Importantly, there is a significant correlation between in vivo firefly activity and ex-vivo TR expression in tumors (r2=0.68, n=7, p=0.02) (Fig 6d).
The TSTA adenovirus maintains promoter specificity in non-cancerous organs like liver compared to the CMV promoter
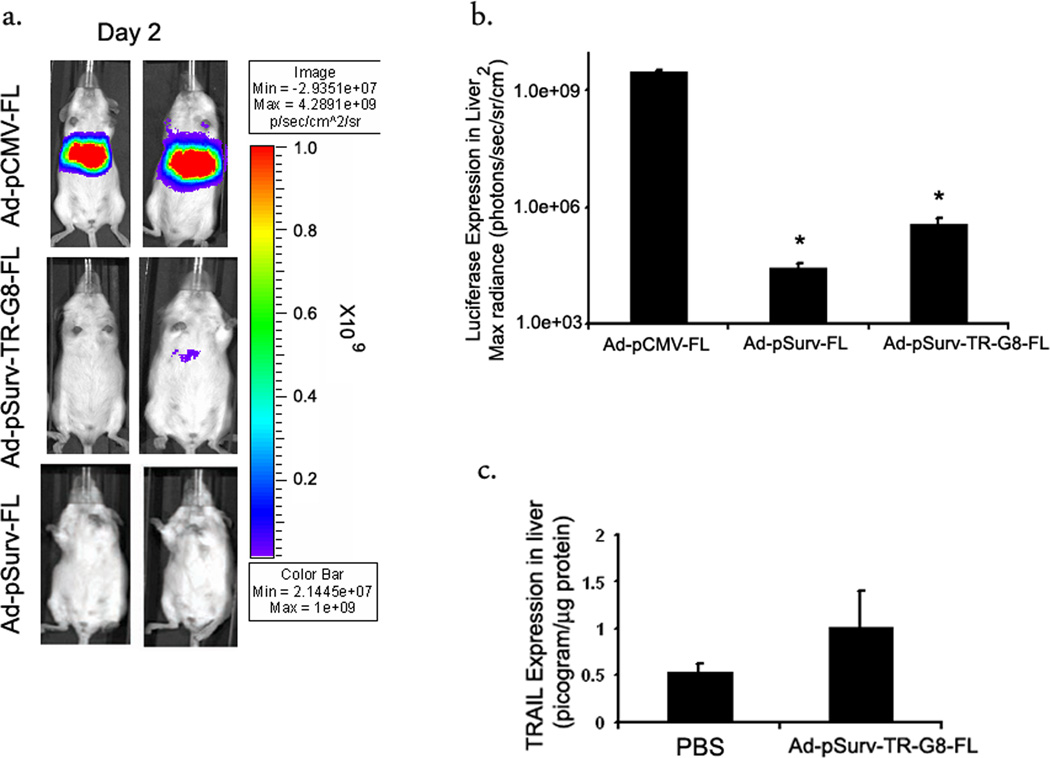
To examine the effect of the TSTA system on Survivin promoter specificity, Ad-pCMV-FL, Ad-pSurv-FL and Ad-pSurv-TR-G8-FL were injected systemically via the tail vein in healthy FVB mice (n=4 each group, 2×107 pfu of virus per animal). Mice were imaged for firefly expression 48 hours later (Fig 7a). Mice injected with Ad-pCMV-FL viruses showed an intense signal in the liver, 12.5±1.8×108 photons/sec/sr/cm2. Interestingly, both the Survivin targeted adenoviruses showed significantly low hepatic firefly luciferase signal when compared to Ad-pCMV-FL (p< 0.001). In some cases, expression could be detected in the spleen also. Although low, the expression from livers injected with the TSTA adenovirus, Ad-pSurv-TR-G8-FL (15.5±8.6 ×104 photons/sec/sr/cm2) was higher than the one-step Ad-pSurv-FL (0.83±0.36 ×104 photons/sec/sr/cm2) (Fig 7b). However, this difference was not statistically significant (p=0.14). Livers were then excised, homogenized and lysates assayed for TRAIL expression by Eliza. TRAIL expression in the TSTA adenovirus injected liver was not significantly different from a normal, PBS-injected liver (p=0.06) (Fig 7c).
Fig 7. TSTA enhanced Survivin promoter maintains specificity in vivo following systemic administration.
(a) Optical CCD imaging of FVB mice systemically injected with i) Ad-pCMV-FL (top), ii) Ad-pSurv-TR-G8-FL (middle) and iii) Ad-pSurv-FL (bottom). Female FVB mice (6 weeks old) were injected via tail vein with 2×107 pfu of the different adenoviruses in 100 µl of PBS and the animals were imaged with D-luciferin. Shown here are images of two representative animals from each group 48hours post injection. All images have been adjusted to the same scale. (b) ROIs were drawn around the liver area in these images and photon emission was quantified using the Living Image software. Plot shows a comparison of luciferase signal across the three groups 48hrs post injection. The error bars represent SEM, n=4 per group. * indicates p<0.05 comparing mice injected with Ad-pSurv-FL or Ad-pSurv-TR-G8-FL vs Ad-pCMV-FL injected animals (c) Livers were excised from the animals injected with Ad-pSurv-TR-G8-FL adenovirus post-imaging on day 2 and TRAIL assay was performed on the liver lysates (n=4). Livers from PBS-injected animals were used as negative controls (n=3). Plot shows the corresponding TRAIL concentration as measured by ELISA, mean ±SD.
Discussion
One of the main goals for all cancer therapies is the selective targeting and killing of tumor cells, thereby increasing the therapeutic ratio. Both chemotherapy and radiotherapy causes dose limiting toxicities in normal tissues, which eventually reduce their clinical effectiveness29. Unlike conventional therapeutic approaches, in cancer gene therapy suitable strategies can be employed to target the therapeutic transgene directly to tumor cells, thereby avoiding normal tissue toxicity.
Controlled regulation of transgene expression is now playing a major role in targeted cancer gene therapy strategies. Tumor-specific promoters are ideal in this regard as they should be highly active in tumor cells with little or no activity in normal cells. They are more generalizeable and safer than tissue specific promoters, as in the case of the latter, gene expression can occur in normal tissues such as brain, liver etc causing catastrophic effect. The promoter of the human Survivin gene has emerged as a favorable promoter of choice for gene therapy applications as it possesses the much coveted “Tumor on” and “Liver off” profile3,7,30–32. Survivin promoter targeted infectivity enhanced conditionally replicative adenoviruses have shown tumor regression in mouse models of human cancers like breast, mesothelioma, cholangiosarcoma etc8,33,34. In our hands, Survivin promoter provided the maximum specificity in non-tumor tissues compared to the widely employed constitutive human CMV promoter or tumor-specific human Telomerase promoter (hTERT) (data not shown). However, Survivin promoter was also a poor activator of transcription when compared to the CMV promoter, a benchmark promoter in the field. Hence the TSTA system was employed to enhance activity of the Survivin promoter.
There are many different strategies to increase promoter strength, but they each have their advantages and disadvantages20. Strategies such as eliminating extra sequences from a natural promoter or introducing activating point mutations in the promoter sequences are useful but cannot be universally employed. Chimeric promoters designed by combining the transcription regulatory elements from different promoters or enhancer elements from the CMV promoter or other stronger viral promoters are often employed in these approaches. However, it was reported that these transcriptional regulator elements can often destroy promoter specificity in non-target tissues35. Recombinant transcriptional activator approaches like the TSTA systems are by far more generalizable and has shown amplification of gene expressions from a wide variety of promoters without affecting their specificity.
The TSTA system employs the transactivator fusion protein Gal4-VP16 to amplify expressions of tissue specific promoters23,24,36,37. The potent transactivation domain of VP16 enhances transcription from the promoter by multiple ways- 1) by recruiting38 and stabilizing39 the transcriptional machinery on the promoter by interacting with different transcriptional co-activators like TBP, TFIID, TFIIA, TFIIH40–42 and with the Mediator (TRAP-SMCC-ARC) complex43 2) by relieving nucleosome-mediated repression44 and iii) by facilitating chromatin remodeling45. When the chimeric activator binds to Gal4-binding sites located upstream of the TATA box of a promoter they stimulate transcription in a synergistic fashion45,46. Earlier studies with uni-directional TSTA system showed that a combination of 5-Gal4-binding sites and two activation domains of VP16 protein is optimum for achieving maximum amplification22,47. Accordingly, in the current study the uni-directional TSTA system also amplified expression of both the genes 15–20-fold from the human Survivin promoter. The robust expression was now 4.3–11.5% of that obtained from the CMV promoter in the same cells. Interestingly, increasing the number of Gal4-binding sites from 5 to 8 resulted in a reduction in the level of expression. Despite earlier reports of a potential synergism between the different units of Gal4-VP16 bound to multiple Gal4-binding sites, studies also showed that multimerizing the VP16-activation domains can inhibit the degree of activation due to steric hindrance causing decreased interaction between the domains and their targets. In fact, linkers that relaxed the steric hindrance by spacing apart the VP16 activation domains achieved a higher level of activation39. Therefore it is possible that multiple VP2 units recruited on closely spaced 8-Gal4 binding sites interfered with each other, thereby diminishing the level of expression compared to the 5-Gal4-binding site templates. However, in case of the bi-directional TSTA system, 5-Gal4 binding sites showed a distinct directional bias. Although strong (~15–20-fold), the amplification effect was concentrated in one direction only. This is in accordance with our previous results where we showed that only one of the genes was expressed strongly by the bi-directional TSTA system26. That this was not gene specific was proved by switching around the orientation of the genes 180°. A possible reason could be the asymmetric positioning of the two E4TATA promoters around the 5X-Gal4 binding sites. While one is only 21 bp apart, the other one is 60 bp downstream of the Gal4-binding sites. Thus it is possible that the VP16 activation domains are unable to activate a promoter farther away and hence the distant promoter failed to show any gene amplification. Although earlier studies have demonstrated that VP16 can activate transcription strongly from promoters as far as 77 bp downstream39,48, there is also substantial evidence that indicates that position of the TATA box from the Gal4-binding sites do effect VP16 mediated transactivation44,49. This also explains why the introduction of 3 or 4 additional Gal4-binding sites in the vicinity of the compromised promoter that equalized the distances between the two TATA boxes (~28-bp apart) abolished the directional bias. However future studies should be performed to confirm this. The two minimal promoters seem to share the available transcriptional machinery, since an increase in expression from one direction dropped the expression from the other side, till they reached equilibrium.
In the current study, a replication incompetent adenoviral vector was developed to deliver the transgenes intratumorally. Adenoviral vectors are one of the most popular vehicles for gene transfer in clinical trials. However, the therapeutic outcome is often marred by the dose-related toxicity effects related to the virus. Intratumoral administration helps to maximize the infection ability of adenoviruses and limits exposure of the virus to the systemic circulation. As demonstrated in mouse xenograft models, the newly developed TSTA adenovirus results in efficient gene expression from the weak Survivin promoter following intratumoral administration. In living animals, the expression level was almost as high as that obtained from the very strong CMV promoter. This data indicated that the amplification system will be immensely helpful in achieving desirable therapeutic outcome with a lower viral dose in clinical trials. Unfortunately, although gene expression could be followed over time by this process, it was difficult to compare the results due to huge variability in viral retention in the tumors following intratumoral administration. Viruses are known to disseminate quickly from the site of injection into systemic circulation following injection50. Adenovirus vectors in systemic circulation are well known for their natural hepatotropism which causes majority of the delivered viral dose to get sequestered in the liver. We therefore investigated whether amplification of the promoter activity by the TSTA system will negatively affect the specificity of the promoter. Although, the Survivin promoter-targeted TSTA adenovirus, when delivered systemically, showed a basal expression in the tumor-free liver, the expression was not significantly different compared to the promoter only system. Therefore, the TSTA system was capable of amplifying the strength of the promoter without affecting its specificity. We are currently testing the vector in animals with hepatic metastases to assess the ability of the targeted virus to distinguish between normal and tumor tissues in the same organ following systemic delivery (Ray et al. unpublished data).
The therapeutic protein TRAIL is known to have a soluble form. Therefore, TRAIL concentration in tumor lysates may not be a true representation of the absolute expression level of the cytokine. Measurements can be further confounded in living animals by the presence of endogenous TRAIL. Despite all these factors, we were able to achieve a good correlation between TRAIL concentration in crude tumor lysates and firefly luciferase gene expression. This very high degree of correlation is an important finding of the current paper because it lays the foundation for using similar vectors in the future to monitor the expression of any therapeutic gene in human gene therapy trials. In the future we can couple the therapeutic gene to a PET reporter gene such as Herpes Simplex Virus Thymidine kinase51 or the Sodium Iodide Transporter52 or a multimodality reporter gene for human applications.
In conclusion, we demonstrated the potential of a novel and readily generalizable gene therapy vector system that will not only allow one to monitor the level of expression of a delivered therapeutic gene, but can also amplify the expression of the transgene in a tumor-specific pattern. Such a vector system will be instrumental in achieving high therapeutic efficacy in future gene therapy applications.
Materials and Methods
Cell lines and media
Colorectal cancer cell lines HCT116 and HT29 were obtained from ATCC while WiDr cells are a kind gift from Dr. Anna Wu (School of Medicine, UCLA). Human fibrosarcoma cell line HT1080 and human ovarian carcinoma cell line SKOV3 were obtained from ATCC. All cells were grown in their ATCC recommended growth media (McCoy’s 5A for HCT116 and HT29, MEM with Earles Salt, 1mM Sodium Pyruvate and 1XNon-essential Amino Acid for WiDr and regular DMEM for HT1080 and SKOV3 cells) supplemented with 10% Fetal Bovine Serum and 1% Pennicillin/Streptomycin antibiotic mix. 293HEK cells for adenoviral packaging was purchased from ATCC and maintained in hi-glucose DMEM supplemented with FBS and penicillin/streptomycin.
Plasmid construction
All plasmids were constructed using standard PCR and cloning technology and sequenced by Sequetech Inc, Mountain View, CA. Constructing Survivin One-Step Plasmids pSurv-FL and pSurv-TR: To construct the one-step pSurv-FL plasmid, the 977bp human Survivin promoter (nucleotides 1824–2800, GenBank Accession Number U75285) was amplified from the pSRVN-luc plasmid (a kind gift from Dr. Mien Chi-hung, MD Anderson Cancer Center, Texas) using a forward primer with a BglII site (Fwd: 5’ ttaaagatctgccatagaaccagagaagtg, Tm 59°) and reverse primer with a HindIII site (Rev: 5’ ttaaaagcttccacctctgccaacgggtcccgc, Tm 68°). The 977 bp fragment was then inserted in the basic PGL3 backbone (bPGL3) (Promega) between sites BglII and HindIII sites. To construct the pSurv-TR plasmid, the human TRAIL gene was amplified from the hTRAIL plasmid from Invivogen Corporation. The 842bp hTRAIL ORF was excised by restriction enzymes NcoI and NheI and ligated to pSurv-FL plasmid digested at NcoI/XbaI sites.
Constructing Survivin uni-directional TSTA plasmids, pSurv-G5-FL, pSurv-G5-TR, pSurv-G8-FL and pSurv-G8-TR
All TSTA plasmids were constructed based on the PBCVP2-G5L plasmid22. In this plasmid, the Gal4-VP2 fusion protein carrying the 1–147 amino acid long GAL4 DNA-binding domain fused in frame to 2 tandem repeats of the N-terminal portion of the VP16 activation domain from amino acids 413–454, together with a SV40-polyA tail was inserted between sites NheI and NotI. The 977bp Survivin promoter replaced the PBC promoter between sites MscI and NheI in the above plasmid. This plasmid had a Firefly Luciferase reporter gene between sites BglII and SalI (pSurv-G5-FL and pSurv-G8-FL). For the TRAIL plasmids (pSurv-G5-TR and pSurv-G8-TR), the human TRAIL gene with its SV40-polyA tail was amplified from the Invivogen vector using primers with inserted restriction enzyme sites BglII and SalI and the 1.13kb fragment replaced the FL gene in all the above mentioned plasmids. Forward primer with BglII: 5’aattagatctgccaccatggctatgatggaggtccagg3’, Reverse primer with SalI: 5’acgcgtcgacaattaacatttaaatggatct3’. To construct the G5-based plasmids, the Gal4-responsive promoter carrying five tandem repeats of the 17bp Gal4-binding sites fused to the E4TATA minimal promoter was inserted between sites KpnI and BglII in all the plasmids right before the downstream genes22. To make the G8-based plasmids, three additional tandem repeats of the Gal4-binding sites were added to the G5-promoter. To do this modification, annealing primers coding for 3 copies of the 17-bp long Gal4-bindings sites in tandem were annealed together and digested with KpnI restriction enzyme, and inserted at the same site in the G5-based plasmids. Anxc5’attggtacctcggaggacagtactccgctcggaggacagtactccgctcggaggacagtactccgctggtaccaatt Rev: 5’Aattggtacctcggaggacagtactccgctcggaggacagtactccgctcggaggacagtactccgctggtaccaatt (Underlined KpnI sequence)
Constructing Survivin bi-directional TSTA plasmids, pSurv-TR-G5-FL, pSurvFL-G5-TR, pSurv-TR-G8-FL and pSurvFL--G8-TR
All bi-directional TSTA plasmids were constructed based on the uni-directional TSTA plasmids. To build these plasmids, a E4TATA promoter together with a downstream gene was amplified using standard PCR techniques and inserted between sites BglII and SalI on the “+”strand. A second E4TATA promoter and its downstream gene was similarly amplified and inserted between sites KpnI and MscI on the “−” strand.
Adenovirus Construction
All adenoviruses are first generation E1, E3 deleted. The bi-directional TSTA adenovirus, Ad-pSurv-FL-G8-TR was cloned by Vector Biolabs Inc, Pennsylvania. The control viruses, Ad-pCMV-FL, Ad-pSurv-FL and Ad-pSurv-TR were constructed using the Adeasy™ kit from Stratagene according to the manufacturer’s guidelines. In short, all gene sequences were inserted in the pShuttle intermediate plasmid and then recombined into the adenoviral backbone in bacteria. Viruses were later packaged and amplified in 293HEK cells and titers determined by plaque assay.
Cell transfections and Enzyme Assays
Cells were plated in 12-well plates in respective cell media. Transient transfections were performed 24h later by Lipofectamine200Tm transfection reagent (Invitrogen) as per the manufacturer’s protocol. Each transfection mix contained 2µg of the plasmid and 5µl of Lipofectamine. A renilla luciferase construct, pCMV-hRL (Promega), was also transfected to control for transfection efficiency. After 24 h of incubation the cells were harvested and the cell lysates were used for FL, renilla luciferase and TRAIL assays.
Viral infection of Cells
1×105 HCT116 or WiDr cells were plated in 24 well plates. Adenoviruses were diluted in serum free Optimem media (Invitrogen) and added to the cells 24hr post plating. 48hrs post-infection cells were collected and assayed.
Luciferase assay
All bioluminescent assays were performed in a TD 20/20 luminometer (Turner Designs, Sunnyvale, CA) with an integration time of 10 sec. The protein content of the cell lysates was determined with Bio-Rad protein assay system (Bio-Rad) in a Beckman DU-50 spectrophotometer (Beckman Instruments) and the luminescence results reported as relative light units (RLU) per microgram protein normalized to Renilla luciferase activity. FL assays were carried on using the Luciferase Assay kit from Promega. Renilla luciferase assays were performed as described earlier27,28.
Elisa for determination of TRAIL production
Cell lysates were assayed for TRAIL by the Quantikine Human TRAIL/TNFSF10 Immunoassay kit from R&D systems, MN (DTRL00).
Animal Tumor Model and Intratumoral Gene Transfer
Animal experiments were carried out in accordance with the institutional guidelines of Stanford University. Six week old female nude mice were purchased from Charles Rivers Laboratory. A total of 5×106 HCT116 cells were suspended in 100 µl Media: BD-Matrigel (1:1) mixture and implanted in the shank of the animal. When tumors were 0.5 mm in diameter, viruses were injected intratumorally. Viruses were first diluted in 100 µl of PBS and then injected slowly into the tumors.
Systemic Administration of Adenoviruses for Liver Specificity Studies
Different adenoviruses were suspended in PBS, pH 7.4, to obtain the final concentration of virus (2×108 pfu/ml). Female FVB mice (4–6 weeks) were obtained from Charles Rivers Laboratory. Mice were injected systemically via tail vein with 100 µl of the viral stock (2×107 pfu/animal). There were three treatment groups: Ad-pCMV-FL, Ad-pSurv-FL and Ad-pSurv-TR-G8-FL and 4 animals per group.
Bioluminescence Imaging
The mice were anesthetized (ketamine–xylazine, 4:1) and imaged every alternate days, using D-Luciferin (Xenogen, Alameda, CA) as the substrate (3 mg/mouse). D-Luciferin was injected 10 min before imaging following which animals were placed in a light-tight chamber. Photons emitted from within the animal and transmitted through the tissues were collected with a cooled charge-coupled device (CCD) camera (IVIS Spectrum; Xenogen). All images were acquired for 1 to 60 secs and region of interests (ROIs) were drawn around the tumor or the liver areas in the images to determine the bioluminescence signal (photons/sec/cm2/steradian (p/sec/cm2/sr)).
Ex-vivo Analysis of TRAIL Gene Expression
Animals were sacrificed at different time points and the tumor or liver tissues were harvested. Tissues were subsequently lysed in 1X Passive lysis buffer (Promega) and homogenized thoroughly with a sonicator. Samples were then centrifuged at 14,000 rpm for 10 minutes and supernatant collected. TRAIL concentration in the lysate was determined using the Elisa kit and the results were reported as picogram per microgram of protein lysate.
Statistical testing
Linear regression analysis was performed to assess the linear relationship between two variables. The strength of correlation between them was quantified in terms of the square of Pearson product-moment correlation coefficient (R2). The significance of correlation was obtained by performing Student’s t test against the null hypothesis that the correlation coefficient (R) is zero. A probability value <0.05 was considered statistically significant. All cell culture and mouse group comparisons were performed with a student’s t test. Values of p≤0.05 were considered statistically significant.
Supplementary Material
Acknowledgement
We acknowledge our funding sources NIH Grants NCI SAIRP R24 CA92865 and R01 CA82214 (SSG). We would also like to thank Dr. Carmel Chan for his suggestions, Dr. Ian Chen and Dr. Olivier Gheysens for their help with the animal studies.
Abbreviations
- bp
base-pairs
- FL
firefly luciferase
- TR
Tumor Necrosis Factor Related Apoptosis Inducing Ligand (TRAIL)
- G
Gal4-binding Sites
- E4TATA
TATA box carrying minimal promoter of adenovirus E4 gene
- pCMV
cytomegalovirus promoter
- pSurv
human Survivin promoter
- CCD
charge-coupled device
Reference
- 1.Lo HW, Day CP, Hung MC. Cancer-specific gene therapy. Adv Genet. 2005;54:235–255. doi: 10.1016/S0065-2660(05)54010-0. [DOI] [PubMed] [Google Scholar]
- 2.Chen JS, et al. Cancer-specific activation of the survivin promoter and its potential use in gene therapy. Cancer Gene Ther. 2004;11:740–747. doi: 10.1038/sj.cgt.7700752. [DOI] [PubMed] [Google Scholar]
- 3.Lu B, et al. Evaluation of tumor-specific promoter activities in melanoma. Gene Ther. 2005;12:330–338. doi: 10.1038/sj.gt.3302385. [DOI] [PubMed] [Google Scholar]
- 4.Chen WC, Liu Q, Fu JX, Kang SY. Expression of survivin and its significance in colorectal cancer. World J Gastroenterol. 2004;10:2886–2889. doi: 10.3748/wjg.v10.i19.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 6.Li B, et al. A survivin-mediated oncolytic adenovirus induces non-apoptotic cell death in lung cancer cells and shows antitumoral potential in vivo. J Gene Med. 2006;8:1232–1242. doi: 10.1002/jgm.953. [DOI] [PubMed] [Google Scholar]
- 7.Van Houdt WJ, et al. The human survivin promoter: a novel transcriptional targeting strategy for treatment of glioma. J Neurosurg. 2006;104:583–592. doi: 10.3171/jns.2006.104.4.583. [DOI] [PubMed] [Google Scholar]
- 8.Zhu ZB, et al. Incorporating the survivin promoter in an infectivity enhanced CRAd-analysis of oncolysis and anti-tumor effects in vitro and in vivo. Int J Oncol. 2005;27:237–246. [PubMed] [Google Scholar]
- 9.Oikonomou E, et al. Newly established tumourigenic primary human colon cancer cell lines are sensitive to TRAIL-induced apoptosis in vitro and in vivo. Br J Cancer. 2007;97:73–84. doi: 10.1038/sj.bjc.6603835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashkenazi A, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley SK, et al. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J Pharmacol Exp Ther. 2001;299:31–38. [PubMed] [Google Scholar]
- 12.Pitti RM, et al. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 13.Wiley SR, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 14.LeBlanc HN, Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 15.Jo M, et al. Apoptosis induced in normal human hepatocytes by tumor necrosis factor-related apoptosis-inducing ligand. Nat Med. 2000;6:564–567. doi: 10.1038/75045. [DOI] [PubMed] [Google Scholar]
- 16.Ganten TM, et al. Preclinical differentiation between apparently safe and potentially hepatotoxic applications of TRAIL either alone or in combination with chemotherapeutic drugs. Clin Cancer Res. 2006;12:2640–2646. doi: 10.1158/1078-0432.CCR-05-2635. [DOI] [PubMed] [Google Scholar]
- 17.Griffith TS, et al. Adenoviral-mediated transfer of the TNF-related apoptosis-inducing ligand/Apo-2 ligand gene induces tumor cell apoptosis. J Immunol. 2000;165:2886–2894. doi: 10.4049/jimmunol.165.5.2886. [DOI] [PubMed] [Google Scholar]
- 18.Kagawa S, et al. Antitumor activity and bystander effects of the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) gene. Cancer Res. 2001;61:3330–3338. [PubMed] [Google Scholar]
- 19.Sadeghi H, Hitt MM. Transcriptionally targeted adenovirus vectors. Curr Gene Ther. 2005;5:411–427. doi: 10.2174/1566523054546189. [DOI] [PubMed] [Google Scholar]
- 20.Nettelbeck DM, Jerome V, Muller R. Gene therapy: designer promoters for tumour targeting. Trends Genet. 2000;16:174–181. doi: 10.1016/s0168-9525(99)01950-2. [DOI] [PubMed] [Google Scholar]
- 21.Iyer M, et al. Bioluminescence imaging of systemic tumor targeting using a prostate-specific lentiviral vector. Hum Gene Ther. 2006;17:125–132. doi: 10.1089/hum.2006.17.125. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, et al. Molecular engineering of a two-step transcription amplification (TSTA) system for transgene delivery in prostate cancer. Mol Ther. 2002;5:223–232. doi: 10.1006/mthe.2002.0551. [DOI] [PubMed] [Google Scholar]
- 23.Iyer M, et al. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci U S A. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. Noninvasive indirect imaging of vascular endothelial growth factor gene expression using bioluminescence imaging in living transgenic mice. Physiol Genomics. 2006;24:173–180. doi: 10.1152/physiolgenomics.00308.2004. [DOI] [PubMed] [Google Scholar]
- 25.Penuelas I, Haberkorn U, Yaghoubi S, Gambhir SS. Gene therapy imaging in patients for oncological applications. Eur J Nucl Med Mol Imaging. 2005;32 Suppl 2:S384–S403. doi: 10.1007/s00259-005-1928-3. [DOI] [PubMed] [Google Scholar]
- 26.Ray S, et al. Novel bidirectional vector strategy for amplification of therapeutic and reporter gene expression. Hum Gene Ther. 2004;15:681–690. doi: 10.1089/1043034041361271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhaumik S, Gambhir SS. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc Natl Acad Sci U S A. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhaumik S, Lewis XZ, Gambhir SS. Optical imaging of Renilla luciferase, synthetic Renilla luciferase, and firefly luciferase reporter gene expression in living mice. J Biomed Opt. 2004;9:578–586. doi: 10.1117/1.1647546. [DOI] [PubMed] [Google Scholar]
- 29.Robson T, Hirst DG. Transcriptional Targeting in Cancer Gene Therapy. J Biomed Biotechnol. 2003;2003:110–137. doi: 10.1155/S1110724303209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoff-Khalili MA, et al. Preclinical evaluation of transcriptional targeting strategies for carcinoma of the breast in a tissue slice model system. Breast Cancer Res. 2005;7:R1141–R1152. doi: 10.1186/bcr1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rein DT, et al. Evaluation of tissue-specific promoters in carcinomas of the cervix uteri. J Gene Med. 2004;6:1281–1289. doi: 10.1002/jgm.606. [DOI] [PubMed] [Google Scholar]
- 32.Zhu ZB, et al. Transcriptional targeting of tumors with a novel tumor-specific survivin promoter. Cancer Gene Ther. 2004;11:256–262. doi: 10.1038/sj.cgt.7700679. [DOI] [PubMed] [Google Scholar]
- 33.Zhu ZB, et al. Targeting mesothelioma using an infectivity enhanced survivin-conditionally replicative adenoviruses. J Thorac Oncol. 2006;1:701–711. doi: 10.1097/01243894-200609000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu ZB, et al. Survivin promoter-based conditionally replicative adenoviruses target cholangiocarcinoma. Int J Oncol. 2006;29:1319–1329. [PubMed] [Google Scholar]
- 35.Hurtado Pico A, et al. Viral and nonviral factors causing nonspecific replication of tumor- and tissue-specific promoter-dependent oncolytic adenoviruses. Mol Ther. 2005;11:563–577. doi: 10.1016/j.ymthe.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 36.Dzojic H, Cheng WS, Essand M. Two-step amplification of the human PPT sequence provides specific gene expression in an immunocompetent murine prostate cancer model. Cancer Gene Ther. 2007;14:233–240. doi: 10.1038/sj.cgt.7701007. [DOI] [PubMed] [Google Scholar]
- 37.Hattori Y, Maitani Y. Two-step transcriptional amplification-lipid-based nanoparticles using PSMA or midkine promoter for suicide gene therapy in prostate cancer. Cancer Sci. 2006;97:787–798. doi: 10.1111/j.1349-7006.2006.00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 39.Ohashi Y, et al. Modulating the potency of an activator in a yeast in vitro transcription system. Mol Cell Biol. 1994;14:2731–2739. doi: 10.1128/mcb.14.4.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingles CJ, et al. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 41.Stringer KF, Ingles CJ, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 42.Liljelund P, Ingles CJ, Greenblatt J. Altered promoter binding of the TATA box-binding factor induced by the transcriptional activation domain of VP16 and suppressed by TFIIA. Mol Gen Genet. 1993;241:694–699. doi: 10.1007/BF00279913. [DOI] [PubMed] [Google Scholar]
- 43.Naar AM, et al. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 44.Croston GE, Laybourn PJ, Paranjape SM, Kadonaga JT. Mechanism of transcriptional antirepression by GAL4-VP16. Genes Dev. 1992;6:2270–2281. doi: 10.1101/gad.6.12a.2270. [DOI] [PubMed] [Google Scholar]
- 45.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Lin YS, Carey MF, Ptashne M, Green MR. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 47.Sato M, et al. Optimization of adenoviral vectors to direct highly amplified prostate-specific expression for imaging and gene therapy. Mol Ther. 2003;8:726–737. doi: 10.1016/j.ymthe.2003.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagmann M, Georgiev O, Schaffner W. The VP16 paradox: herpes simplex virus VP16 contains a long-range activation domain but within the natural multiprotein complex activates only from promoter-proximal positions. J Virol. 1997;71:5952–5962. doi: 10.1128/jvi.71.8.5952-5962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dion V, Coulombe B. Interactions of a DNA-bound transcriptional activator with the TBP-TFIIA-TFIIB-promoter quaternary complex. J Biol Chem. 2003;278:11495–11501. doi: 10.1074/jbc.M211938200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Wang H, Li CY, Yuan F. Effects of rate, volume, and dose of intratumoral infusion on virus dissemination in local gene delivery. Mol Cancer Ther. 2006;5:362–366. doi: 10.1158/1535-7163.MCT-05-0266. [DOI] [PubMed] [Google Scholar]
- 51.Gambhir SS, et al. Imaging transgene expression with radionuclide imaging technologies. Neoplasia. 2000;2:118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niu G, et al. Multimodality noninvasive imaging of gene transfer using the human sodium iodide symporter. J Nucl Med. 2004;45:445–449. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.