Abstract
The enzyme FrbF from Streptomyces rubellomurinus has attracted significant attention due to its role in the biosynthesis of the antimalarial phosphonate FR-900098. The enzyme catalyzes acetyl transfer onto the hydroxamate of the FR-900098 precursors cytidine 5′-monophosphate-3-aminopropylphosphonate and cytidine 5′-monophosphate-N-hydroxy-3-aminopropylphosphonate. Despite the established function as a bona fide N-acetyltransferase, FrbF shows no sequence similarity to any member of the GCN5-like N-acetyltransferase (GNAT) superfamily. Here, we present the 2.0 Å resolution crystal structure of FrbF in complex with acetyl-CoA, which demonstrates a unique architecture that is distinct from those of canonical GNAT-like acetyltransferases. We also utilized the co-crystal structure to guide structure-function studies that identified the roles of putative active site residues in the acetyltransferase mechanism. The combined biochemical and structural analyses of FrbF provide insights into this previously uncharacterized family of N-acetyltransferases and also provide a molecular framework toward the production of novel N-acyl derivatives of FR-900098.
Keywords: Antibiotics, Crystallography, Drug Design, Enzyme Structure, Protein Structure, Acetyltransferase, Malaria
Introduction
Malaria represents one of the most significant threats to modern global health (1). Although uncomplicated malaria can be treated with antimalarials such as chloroquine and sulfadoxine/pyrimethamine, decades of monotherapeutic application have led to the emergence of resistance in the causative Plasmodium falciparum parasite (2, 3). The emergence of resistant strains can be minimized through combination therapies that utilize multiple drugs with independent modes of action and different targets in the parasite (4). Consequently, new antimalarials that can supplemental existing compounds present a focus of significant research interest (5).
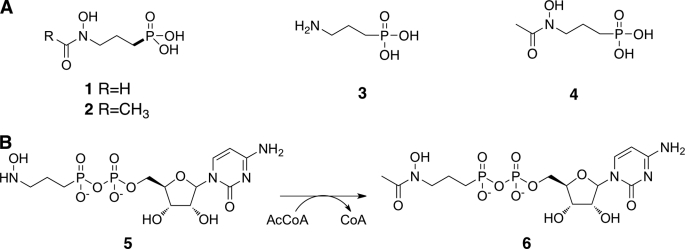
The phosphonate fosmidomycin and its N-acetylated analog FR-900098 (Fig. 1A, Compounds 1 and 2) were first identified from Streptomyces rubellomurinus as broad-range antibiotics effective against Gram-negative bacteria (6, 7). These compounds were later discovered to be potent inhibitors of 1-deoxy-d-xylulose-5-phosphate reductoisomerase, the first committed step in the non-mevalonate isoprenoid biosynthesis pathway (8). The discovery of the non-mevalonate pathway in malarial parasites resulted in the application of both fosmidomycin and FR-900098 as attractive antimalarial drugs (9).
FIGURE 1.
Clinically relevant phosphonates and overall mechanism of FrbF. A, chemical structures of the antimalarial phosphonates fosmidomycin (1) and FR-900098 (2) and the precursor H3APn (3). B, FrbF catalyzes the N-acetylation of the CMP conjugate of H3APn.
Although fosmidomycin and FR-900098 differ only by the presence of an additional methyl decoration on the hydroxamate, FR-900098 has significantly more potent activity both in vitro and in vivo (10, 11). Consequently, significant research efforts have focused on the characterization of FR-900098 and its derivatives. We have identified the FR-900098 biosynthetic cluster and heterologously expressed the cluster in both Streptomyces lividans and Escherichia coli, allowing for characterization of each step in the biosynthetic pathway (12). More recently, the results of our whole cell feeding studies and cluster deletion analysis suggest that the enzyme FrbF catalyzes the acetyl transfer onto the hydroxylamine of the FR-900098 precursor N-hydroxy-3-aminopropylphosphonate (H3APn)4 (Fig. 1A, Compound 3) to generate the final FR-900098 product (13).
Characterization of FrbF and FrbG activity using purified recombinant enzyme demonstrated that these enzymes act on CMP-conjugated substrates, with acetyl transfer occurring after hydroxylation to generate CMP-5′-FR-900098 (Fig. 1B) (14). As FrbF provides a potential branching point for generation of fosmidomycin derivatives with alternative N-acyl moieties, the enzyme has been a target of interest for biochemical analysis. In addition, although FrbF is functionally analogous to GCN5-like N-acetyltransferases (GNATs) (15), FrbF does not demonstrate any sequence similarity to other members of this highly conserved superfamily. Rather, the enzyme demonstrates sequence similarities to a class of putative aminoglycoside 3-N-acetyltransferases that have not yet been kinetically or structurally characterized.
To provide a framework for understanding the mechanism for acetyl transfer within this uncharacterized N-acetyltransferase family, we carried out kinetic characterization of FrbF with multiple coenzyme A and phosphonate substrates. We also determined the co-crystal structure of FrbF in complex with acetyl-CoA and carried out mutational analysis of putative active site residues. These biochemical and structural biological data provide a molecular basis for future engineering experiments targeted at expanding the donor and substrate scope for FrbF with the goal of producing various N-acyl derivatives of fosmidomycin.
EXPERIMENTAL PROCEDURES
Materials
E. coli BL21(DE3) and pET-28a were purchased from Novagen (Madison, WI). Kanamycin, isopropyl 1-thio-β-d-galactopyranoside, NAD, NADP, NADH, and NADPH were obtained from Sigma-Aldrich. Other salts and reagents were purchased from either Fisher or Sigma-Aldrich. Nickel-immobilized metal affinity resin was purchased from GE Healthcare. Amicon® Ultra-15 filter devices were purchased from Millipore. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA). Kits for plasmid purification and gel and column purification of DNA fragments were obtained from Qiagen (Valencia, CA). DNA oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA).
Protein Expression and Purification
The construction of an expression vector for the heterologous production of hexahistidine-tagged FrbF in E. coli has been described previously (13). E. coli expression strain BL21(DE3) was transformed with this expression vector, and single colonies of transformed E. coli were used to inoculate 5 ml of LB medium supplemented with kanamycin (50 μg/ml). Five hours following inoculation, the small-scale culture was added to 1 liter of LB medium containing kanamycin (50 μg/ml) and ZnCl2 (0.5 mm) and grown at 37 °C. When the A600 of the culture reached 0.6, protein expression was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside, and the cells were further grown for 18 h at 30 °C. For selenomethionine-labeled FrbF, a 200-ml starter culture was inoculated in minimal medium (6 g/liter Na2HPO4, 3 g/liter KH2PO4, 1 g/liter NH4Cl, and 0.5 g/liter NaCl) supplemented with 2 mm MgSO4, 0.5% glucose, and 0.05% glycerol and grown at 37 °C overnight. The starter culture (8 ml) was then used to inoculate 800 ml of the same supplemented minimal medium also containing AMP (1 μg/ml) and thiamine (1 μg/ml). Following growth at 37 °C to an A600 of 1.0, the temperature was reduced to 25 °C, and 5 ml of amino acid mixture (14 mg/ml each lysine, threonine, and phenylalanine; 7 mg/ml each leucine, isoleucine, and valine; and 8.4 mg/ml selenomethionine) was added. Twenty minutes later, protein expression was induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 18 h. Bacterial cells were pelleted by centrifugation (4000 rpm for 30 min) and resuspended in 100 mm KCl, 20 mm Tris-HCl (pH 8.3), 10% glycerol, and protease inhibitor mixture (2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 0.3 μm aprotinin, 130 μm bestatin, and 1 μm leupeptin). Resuspended cells were disrupted by multiple passes through an Avestin EmulsiFlex-C5 French press cell, and insoluble aggregates and cellular debris were removed by centrifugation (15,000 rpm for 1 h).
Recombinant proteins were purified from the clarified supernatant by virtue of the N-terminal polyhistidine tag using a nickel affinity resin column (GE Healthcare) charged with cobalt chloride coupled to a BioLogic LP FPLC system (Bio-Rad). After loading the protein onto the column from the clarified lysate at a flow rate of 10 ml/min, nonspecific binding proteins were removed using wash buffer containing 10 mm imidazole, 0.5 m NaCl, and 15% glycerol in 20 mm Tris-HCl. Elution was then carried on an imidazole gradient from 10 to 200 mm over a period of 10 min, followed by an additional 5 min at 200 mm imidazole. Diafiltration in an Amicon 10-kDa cutoff centrifugal filter unit was used for buffer exchange into 50 mm HEPES (pH 7.25) and subsequent concentration of the protein. Following concentration, the cleavable polyhistidine tag was removed using thrombin (1 unit/mg of protein; GE Healthcare). The protein was further purified to remove the cleaved tag and thrombin contaminants by anion exchange chromatography (5-ml HiTrap Q, GE Healthcare) using an isocratic gradient of 100 mm to 1000 m KCl in buffer containing 100 mm Tris-HCl (pH 8.5) over 20 column volumes and size exclusion chromatography (Superdex 75 16/60, GE Healthcare) in 100 mm KCl and 20 mm HEPES (pH 7.5) prior to crystallization.
Crystallization and X-ray Data Collection
Fractions containing pure FrbF were pulled and concentrated to 15 mg/ml prior to crystallization. Crystals of FrbF·acetyl-CoA were grown using the hanging drop vapor diffusion method. A 2-μl drop containing the preincubated sample (15 mg/ml FrbF in 100 mm KCl, 10 mm HEPES (pH 7.5), and 10 mm acetyl-CoA) was added to 2 μl of precipitant (0.2 m MgCl2, 0.1 Tris (pH 7.7), 20–25% PEG 4000, and 20% glycerol) and equilibrated over a well containing the precipitant solution at 8 °C. Crystals grew within 2–3 weeks and reached a maximum size of 0.2 × 0.1 × 0.1 mm. Crystals were vitrified by direct immersion into liquid nitrogen prior to data collection.
Crystals of FrbF occupy space group P2, with unit cell parameters a = 128.8, b = 35.7, and c = 136.1 Å and β = 117.6° and contain four molecules of the complex in the crystallographic asymmetric unit. Diffraction data were collected to a limiting resolution of 2.0 Å at an insertion device line (Life Sciences Collaborative Access Team Sector 21-ID-D, Advanced Photon Source, Argonne National Laboratory) and integrated using MOSFLM (16) and scaled using SCALA (17) from the CCP4 package suite (18). A 5-fold redundant data set was collected to a limiting resolution of 2.0 Å (overall Rmerge = 0.107, I/σ(I) = 2.4 in the highest resolution shell). Crystallographic phases were determined from anomalous diffraction data collected on crystals of selenomethionine-labeled FrbF. Heavy atom positions were determined using HySS, resulting in identification of 14 of 20 SeMet positions (figure of merit = 0.32). Phases were subsequently improved by density modification and 4-fold non-crystallographic symmetry averaging to yield a final figure or merit of 0.79 (19). Model building was initially guided using the atomic coordinates of the Bacillus subtilis yokD gene product (Protein Data Bank code 2NYG) and then subjected to automated rebuilding using ARP/wARP (20). The remainder of the model was fitted using COOT (21, 22) and further improved by rounds of refinement with REFMAC (23, 24). During the final stages of refinement, the structure was refined using BUSTER-TNT (25). Cross-validation used 5% of the data in the calculation of the free R-factor (26). Ligand atoms and solvent molecules were built into the model only after the free R-factor dropped below 0.30. The stereochemistry of the models was routinely monitored throughout the course of refinement using PROCHECK (27). Crystal parameters, data collection parameters, and refinement statistics for each of the structures are summarized in Table 2. The refined coordinates have been deposited in the Protein Data Bank (code 3SMA).
TABLE 2.
Data collection, phasing, and refinement statistics
| FrbF·acetyl-CoA | SeMet-labeled FrbF | |
|---|---|---|
| Data collection | ||
| Space group | P2 | P2 |
| Cell dimensions | a = 128.8, b = 35.7, c = 136.1 Å; β = 117.6° | a = 129.3, b = 36.5, c = 136.8 Å; β = 117.7° |
| Resolution (Å)a | 50–2.0 (2.11–2.0) | 50–3.0 (3.11–3.0) |
| Total reflections | 397,116 | 134,515 |
| Unique reflectionsa | 74,767 | 22,839 |
| Rsym (%) | 10.7 (47.5) | 11.4 (30.0) |
| I/σ(I) | 11.9 (2.4) | 15.5 (3.9) |
| Completeness (%) | 98.8 (96.0) | 97.5 (84.3) |
| Redundancy | 5.3 (5.2) | 5.9 (5.0) |
| Wilson B-value | 24.3 | 24.6 |
| FOM/FOM DMb | 0.32/0.79 | |
| Refinement | ||
| Resolution (Å) | 45.0–2.0 | |
| No. of reflections | 74,765 | |
| Rwork/Rfreec | 19.5/24.6 | |
| No. of atoms | ||
| Protein | 8139 | |
| Acetyl-CoA | 204 | |
| Water | 511 | |
| B-Factors | ||
| Protein | 35.4 | |
| Acetyl-CoA | 38.2 | |
| Water | 41.5 | |
| r.m.s.d.d | ||
| Bond lengths (Å) | 0.010 | |
| Bond angles | 1.11° | |
a Highest resolution shell is shown in parentheses.
b Figure of merit (defined as ∫φP(φ)eiφdφ, where P(φ) is the experimental phase probability distribution) is before (FOM) and after (FOM DM) density modification.
c Rwork = Σ(|Fobs| − k|Fcalc|)/Σ|Fobs|, and Rfree is the R-value for a test set of reflections consisting of a random 5% of the diffraction data not used in the refinement.
d r.m.s.d., root mean square deviation.
Kinetic Activity Assays
All activity assays were performed with N-hexahistidine-tagged FrbF in 50 mm HEPES (pH 7.25) at 30 °C. For determination of kinetic parameters, the concentration of one substrate was maintained at 400 μm, whereas the concentration of the other substrate was varied from 0 to 1000 μm. Samples were collected over a 7-min time course for product quantitation and determination of the initial rate. Reactions were initiated by the addition of 1.3 μm FrbF and quenched with 1% trifluoroacetic acid. All assays were performed in triplicate. Kinetic parameters were fit to the data using nonlinear least-squares regression in OriginPro 8 (OriginLab Corp.). Relative activity assays with in situ CMP-5′-H3APn generation were performed with 1.3 μm FrbF, 13 μm FrbG, 400 μm CMP-5′-3-aminopropylphosphonate (3APn), and 200 μm acetyl-CoA over a 30-min time course, and the rates obtained were compared with those measured under identical conditions in the absence of FrbG. Relative activity assays with alternative CoA substrates were performed with 400 μm CMP-5′-3APn and 200 μm CoA compound. Reactions were initiated with 15 μm FrbF, and samples were collected over a 60-min time course. For all assays, products were detected by UV absorbance at 254 nm on an Agilent 1100 Series HPLC system, with retention times confirmed using an Agilent XCT MSD ion-trap mass spectrometer at the Roy J. Carver Metabolomics Center (University of Illinois at Urbana-Champaign).
Site-directed Mutagenesis
All FrbF active site mutants were prepared using the megaprimer PCR method. Mutant frbF genes were digested with NdeI and HindIII, ligated into pET-28a, and sequenced to confirm inclusion of only the desired mutation. Relative activity assays with purified N-hexahistidine-tagged FrbF mutants were performed with 400 μm CMP-5′-3APn and 200 μm acetyl-CoA over a 7-min time course. For those mutants that exhibited significantly reduced activity, the assay time was extended up to 60 min. Samples were collected and analyzed as described above.
RESULTS
Kinetic Characterization of FrbF
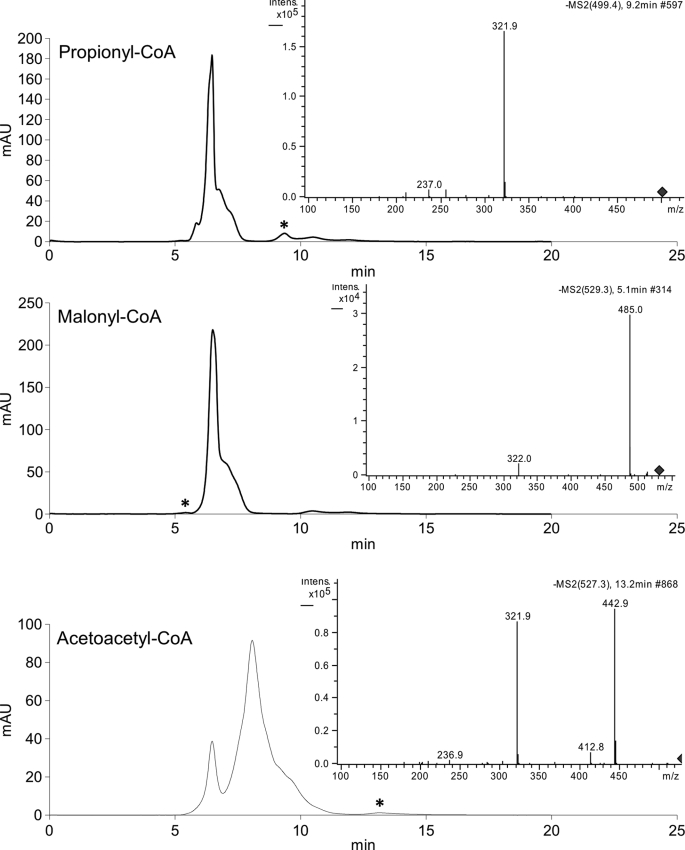
Kinetic parameters for the native FrbF substrate acetyl-CoA were determined by HPLC analysis, yielding the parameters given in Table 1. Previous analysis of the entire FR-900098 biosynthetic pathway demonstrated accumulation of a side product, CMP-5′-N-acetyl-3-aminopropylphosphonate (13), indicating that FrbF can accept both the native CMP-5′-H3APn substrate and the non-hydroxylated CMP-5′-3APn substrate, as recapitulated here. Kinetic parameters were determined with CMP-5′-3APn (Table 1). Because of the inherent instability of the native substrate CMP-5′-H3APn to non-enzymatic oxidation of the hydroxylamine moiety, kinetic parameters for this substrate could not be determined. Nevertheless, relative activity assays carried out with in situ CMP-5′-H3APn generation using 10-fold excess purified FrbG protein showed a 6.7-fold increase in FrbF activity, demonstrating that the native substrate for FR-900098 production is the preferred phosphonate substrate for FrbF. To determine the promiscuity of FrbF toward its CoA substrate, activity assays were carried out with several alternative acyl-CoAs. Of those compounds tested, propionyl-CoA, malonyl-CoA, and acetoacetyl-CoA yielded quantifiable amounts of product (Fig. 2). The activities of FrbF for these substrates, relative to its activity for the preferred acetyl-CoA substrate, are listed in Table 1. Two CoA compounds, isobutyryl-CoA and succinyl-CoA, yielded the expected reaction product as confirmed by LC-MS analysis but fell below the UV detection limit for quantitation of relative activity. The expected products with glutaryl-CoA and β-hydroxybutyryl-CoA were not detected by UV absorbance or LC-MS.
TABLE 1.
FrbF kinetic parameters (mean ± S.D. of three independent experiments)
| Substrate | Km | kcat | Relative activity |
|---|---|---|---|
| μm | min−1 | ||
| CMP-5′-3APn | 391 ± 61 | 2.04 ± 0.14 | 1.0 |
| CMP-5′-H3APn | NDa | ND | 6.7b |
| Acetyl-CoA | 19.5 ± 0.4 | 1.12 ± 0.03c | 1.0 |
| Propionyl-CoA | 23.6 ± 4.2 | 0.089 ± 0.024c | 0.080 ± 0.022 |
| Malonyl-CoA | ND | ND | 0.027 ± 0.006 |
| Acetoacetyl-CoA | ND | ND | 0.0044 ± 0.0013 |
| Isobutyryl-CoA | ND | ND | MS |
| Succinyl-CoA | ND | ND | MS |
a ND, not determined; MS, below UV detection limit (product observed by MS/MS only).
b Substrate was generated in situ with 10-fold excess FrbG.
c kcat was measured at 400 μm CMP-5′-3APn.
FIGURE 2.
HPLC traces following FrbF reactions with alternative acyl-CoAs. Insets, the MS/MS traces show fragmentation of the expected parent ion for each compound to the characteristic m/z = 322− (CMP) daughter ion.
Overall Structure and Non-crystallographic Symmetry
Crystals of FrbF in complex with acetyl-CoA diffract to a Bragg limit of at least 2.0 Å at an insertion device synchrotron source. Initial crystallographic phases were determined by single wavelength anomalous diffraction data collected on crystals grown using selenomethionine-labeled protein. 4-Fold symmetry averaging of the resultant phases, followed by cycle model building and refinement, yielded the current model that has been refined against data to a Bragg limit of 2.0 Å to a free R-factor of 24.8%. Of the 286 amino acids in the full-length polypeptide, the final model comprises residues 19–281 in chain A, residues 14–281 in chain B, residues 19–280 in chain C, and resides 19–282 in chain D. In addition, the final model contains four molecules of acetyl-CoA and 551 solvent molecules. Ramachandran analysis of the final model showed that 92.4% of the residues fall within the most energetically favored regions, and the remaining 7.5% fall in additionally allowed regions. Relevant data collection and refinement statistics are given in Table 2.
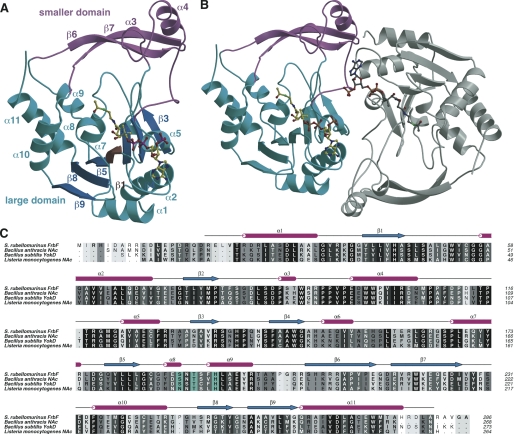
The overall structure of FrbF is distinct from that of members of the canonical GNAT superfamily of enzymes (14) and represents one of a family of four structurally characterized proteins with this new architecture. The structure is composed of two domains; the larger domain consists of seven β-strands flanked on either side by three α-helices and a smaller domain consisting largely of two long β-strands adjoined by two short helices (Fig. 3A). Strands β2, β3, and β4 form the first set, and strands β5, β8, and β9 form the second set. The arrangement of β-strands is somewhat unusual in that the two sets of outer three strands are antiparallel, but the middle strand (β1) that links the two sets is parallel to its neighbors. This unique arrangement is a result of the fact that only two of the strands are directly linked to each other, and the other six strands have several interweaving secondary structural elements in between. The strands are flanked by two sets (“upper” and “lower”) of α-helices, with the upper set composed of four helices (α8, α9, α10, and α11) and the lower set composed of three helices (α1, α2, and α5). Although these helices are composed from noncontiguous regions of the polypeptide, each set engages in interactions representative of a helical bundle. The smaller domain is composed of two large β-strands (strand β1 formed by residues Arg-205–Glu-215 and strand β2 formed by Arg-218–Ile-227) that form a turn structure. Two short α-helices, composed of Pro-87–Thr-90 (α4) and Glu-97–Gln-106 (α5), are located near the base of the β-turn.
FIGURE 3.
Overall structure and multiple sequence alignment. A, ribbon diagram showing the overall structure of the FrbF monomer in complex with acetyl-CoA (in yellow ball-and-stick). The acetyl-CoA-binding large domain is shown in cyan, and the smaller domain is shown in pink. B, ribbon diagram showing the structure of the FrbF dimer with the second molecule colored in gray. C, structure-based sequence alignment of FrbF along with the structural homologs B. anthracis N-acetyltransferase (NAc), B. subtilis YokD, and the aminoglycoside N-acetyltransferase from L. monocyotogenes. Sequence conservation is shaded such that the residues that show the highest conservation are darker. Residues colored in cyan are active site general acid/base catalysts.
The four molecules in the crystallographic asymmetric unit pack as two pairs of dimers (Fig. 3B), consistent with the behavior of the protein in solution as judged by gel filtration chromatography (data not shown). Formation of each dimer results in a burial of 1598 Å2 of surface area. Despite the fact that non-crystallographic restraints were not used during the course of refinement, the four molecules are essentially identical, with root mean square deviations for 260 equivalent main chain atoms ranging between 0.19 and 0.47 Å. The equivalency is highest between the polypeptides that form dimers (0.19–0.23 Å) than between polypeptides from one dimer to the other (0.43–0.47 Å). In these comparisons, the most significant deviations occur at the smaller domain, which undergoes a slight rigid body movement relative to the larger domain, in comparison of the polypeptides from different dimers.
Structural Homology
A lack of structural homology between FrbF and other members of the GNAT superfamily members is expected given the lack of appreciable similarities at the sequence level. A Dali search (15) against coordinates deposited in the Protein Data Bank (28) identified the closest homologs as uncharacterized enzymes whose crystal structures have been determined by various structural genomics consortia and whose function is annotated based solely on sequence similarities (Fig. 3C). Close structural homologs include the yokD gene product from B. subtilis (Protein Data Bank code 2NYG; 43% sequence identity; root mean square deviation of 1.6 Å over 258 aligned Cα atoms), and a putative aminoglycoside N-acetyltransferase from Bacillus anthracis (Protein Data Bank codes 3NOS and 3E4F; 43% sequence identity; root mean square deviation of 1.5 Å over 258 aligned Cα atoms). These two polypeptides likely represent homologs of the same gene, but their biological function has yet to be established. Searches against the GenBankTM Sequence Database identified numerous genes in Bacillus that share >45% sequence identity with FrbF, which have been annotated as aminoglycoside N-acetyltransferase, but their functions have not been experimentally verified (Fig. 3C). It is likely that these annotations are not all correct, as the corresponding genes product are equally likely to be involved in the biosynthesis of various metabolites in these organisms, similar to the role of FrbF in FR-900098 biosynthesis. The closest homolog for which a biological function has been experimentally verified is the resistance-associated N-acetyltransferase from a clinical isolate of Listeria monocytogenes (sequence identity of 39%) (29).
Active Site
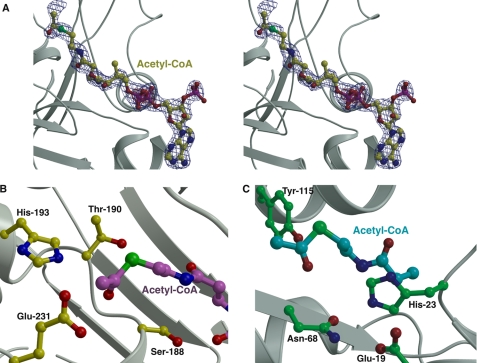
Clear and continuous electron density corresponding to the acetyl-CoA ligand can be observed within the larger domain (Fig. 4A). The phosphopantetheine moiety is oriented roughly perpendicular to the central β-strand core. As a result, the 3′,5′-phosphoadenosine is engaged by the lower helical bundle flanking the seven-β-strand core, whereas the acetyl group is situated near the upper helical bundle located on the opposite side of the β-strands. The adenosine ring of acetyl-CoA packs into a hydrophobic cleft defined by Leu-48, Val-54, and Trp-53, with favorable hydrogen bonding interactions with the adenine provided by Arg-19 and the backbone carbonyls of Gly-52 and Val-54. The CoA tail and the acetyl group are located at the interface between the β-strands' upper helical bundle.
FIGURE 4.
Active site of FrbF in complex with CoA. A, stereo view of the active site pocket with the acetyl-CoA shown in yellow ball-and-stick. Superimposed is a difference Fourier electron density map (contoured at 3σ over the background in blue and at 8σ over the background in red) calculated with coefficients |Fobs| − |Fcalc| and phases from the final refined model with the coordinates of acetyl-CoA deleted prior to one round of refinement. B, close-up view of the FrbF active site in the vicinity of the acyl moiety of the bound acetyl-CoA (shown in magenta ball-and-stick). Residues that participate in acid/base catalysis are shown in yellow ball-and-stick. C, the active site of the structurally unrelated GNAT superfamily acetyltransferase RimI (Protein Data Bank code 2CNS) is shown for comparison. The acetyl-CoA is shown in green, and active site residues are shown in cyan ball-and-stick.
In the co-crystal structure, the acetyl-CoA is bound with an anti-glycosidic torsion angle and an extended pantetheine arm, similar to the conformation observed for acetyl-CoA bound to other proteins. However, the ribose is in a 3′-endo-conformation rather than in the more typical 2′-endo-conformation observed in other acetyltransferase co-crystal structures. The pyrophosphate moiety of the 3′,5′-phosphoadenosine is engaged by interactions with main chain atoms of the flanking β-strands, as well as a spine of water molecules located in the acetyl-CoA-binding site. In addition, the adenine ring is engaged through an off-set π-π interaction with the guanidinium group of Arg-19 and the indole ring of Trp-53. There are limited interactions between the protein and the pantetheine arm. The extended conformation of the arm is maintained by hydrogen bonding interactions between one of its carbonyl groups and Ser-188 (Oγ-O distance of 2.7 Å) and between the adjacent amide nitrogen and the backbone carbonyl of Ala-46 (N-O distance of 2.65 Å). The thioester is located in the vicinity of polar residues Ser-188, Thr-190, and His-193, all of which are oriented within 6 Å of the acetyl group (Fig. 4B), suggesting that some of these residues might engage in acid/base chemistry during acetyl transfer. Notably, the constellation of active site residues that flank the acetyl group is different from those typically found in GNAT family members (Fig. 4C), suggesting that FrbF uses a different set of active site residues to facilitate acetyl transfer.
Catalytic Mechanism of FrbF
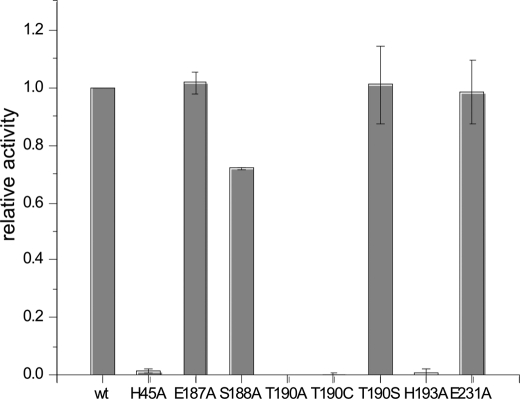
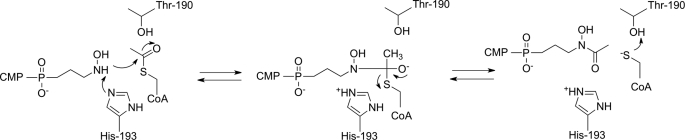
To elucidate the catalytic mechanism of FrbF, the residues located within 6 Å of the acyl moiety of the bound acetyl-CoA were analyzed (Fig. 4B). In principle, one could expect the reaction mechanism to proceed either via direct attack of the amine or hydroxylamine substrate on the carbonyl group of acetyl-CoA, as has been observed for the majority of GNAT superfamily members, or via an acyl-enzyme intermediate, as demonstrated in the yeast histone acetyltransferase Esa1 and in multiple mycobacteria (14, 30). Of the residues present in the active site, Glu-187, Ser-188, Thr-190, and Glu-231 were identified as possible general acids, whereas only His-193 was identified as a potential general base. Alanine mutants at each of these five positions were created, and their relative activities were measured in comparison with the wild-type enzyme (Fig. 5). The results indicate that the E187A and E231A mutants demonstrated activities similar to the wild-type enzyme, confirming that these two residues do not play a role in catalysis and further ruling out mechanisms that require formation of a covalent acyl intermediate with an active site carboxylate. The S188A mutant exhibited a modest decrease in activity, as anticipated based on its interaction with the phosphopantetheine arm of the acetyl-CoA substrate. In contrast, the activity of the H193A mutant was <1% of that of the wild-type enzyme, and no activity could be observed for the T190A mutant. These results strongly suggest the involvement of Thr-190 and His-193 in acid/base chemistry during acetyl group transfer. These results are consistent with the mechanistic route proposed in Fig. 6. Following the binding of the substrate conjugate CMP-5′-3APn and acetyl donor acetyl-CoA at the active site, His-193 functions as a general base to facilitate the attack of the amine or hydroxylamine on the electrophilic carbonyl of the acetyl group. Protonation of the resultant tetrahedral intermediate results in its collapse to yield the hydroxamate and CoA. Alternatively, His-193 could serve to stabilize the tetrahedral intermediate, as the N-O distance varies from 2.9 to 4.2 Å between the two pairs of FrbF dimers.
FIGURE 5.
Relative activity levels of each of the FrbF active site mutants. Error bars represent S.D. of three independent experiments.
FIGURE 6.
Proposed mechanism for N-acetylation of CMP-5′-H3APn based upon the structural and biochemical data presented in this work.
Further analysis was carried out at Thr-190 through mutations to both Ser and Cys, as well as mutation of His-45 to Ala. His-45 is the nearest residue to Thr-190, and so we speculated that this residue might serve to activate Thr-190 for a novel reaction mechanism involving attack of Thr-190 on the carbonyl of the acetyl group to form an acyl-enzyme intermediate. The H45A mutation reduced the relative activity of FrbF to 1.5% of the wild-type enzyme, supporting the catalytic relevance of this residue. However, high resolution mass spectrometry provided no evidence of an acyl-enzyme intermediate (data not shown). Further analysis of the crystal structure suggests that His-45 can participate in hydrogen bonding interactions both with a carbonyl group of the acetyl-CoA phosphopantetheine arm and with the backbone amide nitrogen of either Thr-190 or Ser-191, suggesting that this residue is critical for the positioning of acetyl-CoA and the catalytically requisite Thr-190 residue. Furthermore, the T190C mutation, which would more closely reflect the active site of GNAT enzymes that utilize an acyl-enzyme intermediate, lowered the activity dramatically, discrediting the prospect of a novel Thr-mediated ping-pong mechanism. Finally, the T190S mutation had no detectable effect on the relative activity.
DISCUSSION
The phosphonic acid fosmidomycin and its methylated derivative FR-900098 represent promising therapeutics against malarial parasites, especially when used in combination therapy with lincosamide antibiotics (31, 32). Although significant research efforts have focused on identifying synthetic derivatives of these compounds with improved efficacy, such compounds are cost-prohibitive (11, 33, 34). We have recently characterized the biosynthetic pathway for FR-900098 and have reconstituted the pathway through heterologous expression of the biosynthetic genes in E. coli, opening new avenues for the production of derivative compounds by genetic manipulation (12, 13). Such efforts will be aided by detailed biochemical and structural studies of the constituent enzymes in this pathway.
Here, we present the co-crystal structure of FrbF, revealing a new acetyltransferase fold that is distinct from that of canonical members of the GNAT superfamily. A structure-based sequence search revealed that this class of acetyltransferases encompasses a distinct superfamily of enzymes that have been annotated as resistance-associated N-acetyltransferase based upon sequence similarity to a gene encoded in a clinical isolate of L. monocytogenes (29). Our structural and functional analyses of FrbF extend the range of functions of such enzymes toward the biosynthesis of natural products, suggesting that other members of this superfamily may also be involved in production of antibiotics or other metabolites.
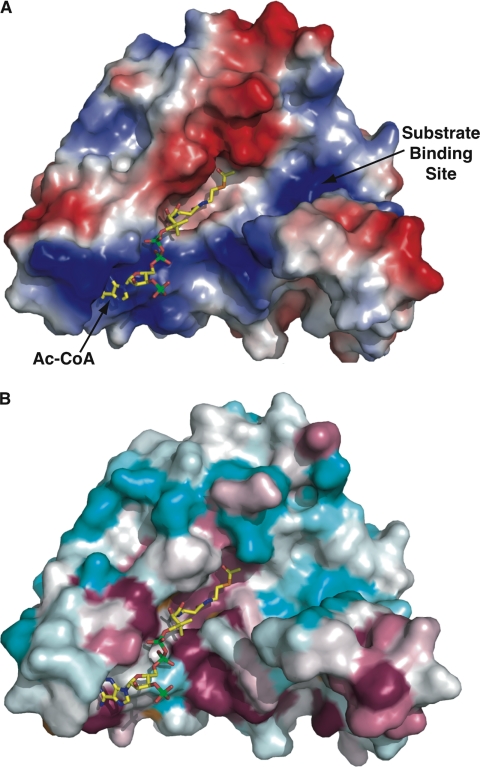
The co-crystal structure of FrbF bound to acetyl-CoA reveals that the phosphopantetheine moiety is oriented roughly perpendicular to the central β-strand core, resulting in the placement of the acetyl group proximal to residues that may serve in acid/base catalysis, and their functional roles have been confirmed by mutational analysis. Analysis of the molecular surface of FrbF reveals that the ligand is positioned in a pocket that is lined with basic residues that can stabilize the 3′,5′-phosphoadenosine moiety (Fig. 7A). Interestingly, there is a second channel located proximal to the acetyl-CoA-binding site that may serve as the substrate-binding cleft, and this cleft is similarly lined with basic residues that may stabilize interactions with the phosphate group of the CMP-5′-3APn substrate. Mapping the sequence conservation among other related N-acetyltransferases onto the solvent-accessible surface of FrbF demonstrated that the acetyl-CoA-binding site is highly conserved (Fig. 7B). However, the second presumptive substrate-binding channel is highly divergent, as would be expected given the divergent chemical structures of the substrates (aminoglycosides, phosphonates, etc.) utilized by these enzymes.
FIGURE 7.
FrbF pockets define the substrate- and acyl donor-binding sites. A, surface representation of the FrbF monomer with a vacuum electrostatic potential superimposed. The coordinates of acetyl-CoA are shown as yellow sticks. The acyl donor binds in a deep groove running along the width of the enzyme. A second pocket is located adjacent to the acyl group, and basic residues in this pocket may help stabilize the anionic diphosphates in the substrate conjugate. B, the same view of the FrbF surface but superimposed is a colored-coded representation of the sequence conservation shown as a gradient from cyan (least conserved) to pink (most conserved). Residues that line the acetyl-CoA-binding site and active site are conserved, as would be expected. Notice that residues in the putative substrate-binding pocket are less conserved, consistent with the range of substrates (such as aminoglycosides) that are accommodated by other members of this enzyme family.
In conclusion, the structure of the N-acetyltransferase involved in FR-900098 biosynthesis reveals a fold distinct from those of canonical members of the GNAT superfamily. The characterization of FrbF presented here expands the substrate scope of this family of enzymes to small molecules other than aminoglycosides and suggests a functional role for related enzymes that extends beyond antibiotic resistance. The structure allows for the identification of an extended cleft located adjacent to the acetyl donor that may serve as the binding site for substrates. Identification of the substrate binding determinants will require additional further biochemical and structural biological studies of site-specific mutants. Finally, the availability of the co-crystal structure should also facilitate engineering experiments directed at altering the donor specificity of this enzyme, which will allow the production of novel N-acylated derivatives of FR-900098.
Acknowledgments
We thank Wilfred van der Donk and William Metcalf for comments and continued support. We also thank Drs. Keith Brister, Spencer Anderson, and Joseph Brunzelle of the Life Sciences Collaborative Access Team (beamline 23-ID at the Advanced Photon Source, Argonne National Laboratory) for facilitating crystallographic data collection.
This work was supported by National Institutes of Health Grant P01 GM077596 (to H. Z. and S. K. N.) and National Institutes of Health Ruth L. Kirschstein National Research Award 5 T32 GM070421 from NIGMS (to R. E. C. and M. A. D.).
The atomic coordinates and structure factors (codes 3SMA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- H3APn
- N-hydroxy-3-aminopropylphosphonate
- GNAT
- GCN5-like N-acetyltransferase
- 3APn
- 3-aminopropylphosphonate.
REFERENCES
- 1. Feachem R. G., Phillips A. A., Hwang J., Cotter C., Wielgosz B., Greenwood B. M., Sabot O., Rodriguez M. H., Abeyasinghe R. R., Ghebreyesus T. A., Snow R. W. (2010) Lancet 376, 1566–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dondorp A. M., Yeung S., White L., Nguon C., Day N. P., Socheat D., von Seidlein L. (2010) Nat. Rev. Microbiol. 8, 272–280 [DOI] [PubMed] [Google Scholar]
- 3. Griffith K. S., Lewis L. S., Mali S., Parise M. E. (2007) JAMA 297, 2264–2277 [DOI] [PubMed] [Google Scholar]
- 4. Hastings I. (2011) Trends Parasitol. 27, 67–72 [DOI] [PubMed] [Google Scholar]
- 5. Burrows J. N., Chibale K., Wells T. N. (2011) Curr. Top. Med. Chem. 11, 1226–1254 [DOI] [PubMed] [Google Scholar]
- 6. Kuzuyama T., Shimizu T., Takahashi S., Seto H. (1998) Tetrahedron Lett. 39, 7913–7916 [Google Scholar]
- 7. Okuhara M., Kuroda Y., Goto T., Okamoto M., Terano H., Kohsaka M., Aoki H., Imanaka H. (1980) J. Antibiot. 33, 13–17 [DOI] [PubMed] [Google Scholar]
- 8. Proteau P. J. (2004) Bioorg. Chem. 32, 483–493 [DOI] [PubMed] [Google Scholar]
- 9. Jomaa H., Wiesner J., Sanderbrand S., Altincicek B., Weidemeyer C., Hintz M., Türbachova I., Eberl M., Zeidler J., Lichtenthaler H. K., Soldati D., Beck E. (1999) Science 285, 1573–1576 [DOI] [PubMed] [Google Scholar]
- 10. Kurz T., Schlüter K., Kaula U., Bergmann B., Walter R. D., Geffken D. (2006) Bioorg. Med. Chem. 14, 5121–5135 [DOI] [PubMed] [Google Scholar]
- 11. Schlüter K., Walter R. D., Bergmann B., Kurz T. (2006) Eur. J. Med. Chem. 41, 1385–1397 [DOI] [PubMed] [Google Scholar]
- 12. Eliot A. C., Griffin B. M., Thomas P. M., Johannes T. W., Kelleher N. L., Zhao H., Metcalf W. W. (2008) Chem. Biol. 15, 765–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johannes T. W., DeSieno M. A., Griffin B. M., Thomas P. M., Kelleher N. L., Metcalf W. W., Zhao H. (2010) Chem. Biol. 17, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vetting M. W., de Carvalho L. P., Yu M., Hegde S. S., Magnet S., Roderick S. L., Blanchard J. S. (2005) Arch. Biochem. Biophys. 433, 212–226 [DOI] [PubMed] [Google Scholar]
- 15. Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 16. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 18. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cowtan K., Main P. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 487–493 [DOI] [PubMed] [Google Scholar]
- 20. Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 25. Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 26. Kleywegt G. J., Brünger A. T. (1996) Structure 4, 897–904 [DOI] [PubMed] [Google Scholar]
- 27. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 28. Berman H. M., Battistuz T., Bhat T. N., Bluhm W. F., Bourne P. E., Burkhardt K., Feng Z., Gilliland G. L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J. D., Zardecki C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 899–907 [DOI] [PubMed] [Google Scholar]
- 29. Gilmour M. W., Graham M., Van Domselaar G., Tyler S., Kent H., Trout-Yakel K. M., Larios O., Allen V., Lee B., Nadon C. (2010) BMC Genomics 11, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sim E., Sandy J., Evangelopoulos D., Fullam E., Bhakta S., Westwood I., Krylova A., Lack N., Noble M. (2008) Curr. Drug Metab. 9, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wiesner J., Borrmann S., Jomaa H. (2003) Parasitol. Res. 90, S71–S76 [DOI] [PubMed] [Google Scholar]
- 32. Wiesner J., Henschker D., Hutchinson D. B., Beck E., Jomaa H. (2002) Antimicrob. Agents Chemother. 46, 2889–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ortmann R., Wiesner J., Reichenberg A., Henschker D., Beck E., Jomaa H., Schlitzer M. (2005) Arch. Pharm. 338, 305–314 [DOI] [PubMed] [Google Scholar]
- 34. Reichenberg A., Wiesner J., Weidemeyer C., Dreiseidler E., Sanderbrand S., Altincicek B., Beck E., Schlitzer M., Jomaa H. (2001) Bioorg. Med. Chem. Lett. 11, 833–835 [DOI] [PubMed] [Google Scholar]