Background: Acid ceramidase is an enzyme of sphingolipid metabolism whose cellular activity is carefully regulated.
Results: Cystatin SA inhibits acid ceramidase activity without affecting precursor processing. Small peptides derived from cystatin SA also are inhibitory.
Conclusion: Cystatin SA inhibits acid ceramidase activity and contributes to the regulation of this enzyme function.
Significance: The first physiological inhibitor of acid ceramidase has been described.
Keywords: Enzyme inhibitors, Enzyme Mechanisms, Peptides, Sphingolipid, Storage Diseases, Ceramidase, Ceramide, Cystatins
Abstract
Autoproteolytic cleavage of the inactive acid ceramidase (AC) precursor into the active heterodimer exposes a free cysteine residue, leading us to study whether AC could be regulated by one or more members of the cystatin family. Co-expression of the full-length AC and cystatin SA (cysSA) cDNAs led to significant reduction of AC activity in the transfected cells. Expression of cysSA also inhibited endogenous AC activity in cells and increased ceramide. Conversely, cysSA siRNA expression led to elevated AC activity and reduction in ceramide. The effects of cysSA siRNA expression could be reversed by the addition of recombinant cysSA into the culture media. These results were consistent with detection of a physical interaction between AC and cysSA, assessed by co-immunoprecipitation and nickel-nitrilotriacetic acid affinity chromatography, and further supported by co-localization of the endogenous proteins using confocal microscopy. In vitro kinetic analysis of purified, recombinant AC and cysSA confirmed the transfection results and suggested a non-competitive type of inhibition with a Ki in the low micromolar range. Processing of the AC precursor into the active form was not affected by cysSA expression, suggesting that it likely inhibits AC by allosteric interference. Computer modeling and expression studies identified several potential inhibitory domains in cysSA, including a small “AC-like” domain (identical to the AC cleavage site, TICT). Small peptides, synthesized with combinations of this and a “cystatin-like” domain (QXVXG), exhibited significant AC inhibition as well. Such peptide-based AC inhibitors could potentially be used to regulate AC activity in cancer cells that are known to overexpress this enzyme alone and in combination with conventional anti-cancer drugs.
Introduction
During the past decade sphingolipids have been recognized as important bioactive signaling molecules involved in the regulation of cell differentiation, proliferation, and death (1, 2). Acid ceramidase (AC3; N-acylsphingosine deacylase; EC 3.5.1.23) is a key enzyme in this pathway required to maintain the proper balance between the pro-apoptotic lipid, ceramide, and the anti-apoptotic lipid, sphingosine-1-phosphate. Mutations in the AC gene (Asah1) cause the severe lipid storage disorder, Farber lipogranulomatosis (i.e. Farber disease), associated with the accumulation of ceramide in various tissues (3, 4). Farber disease is an extremely rare disorder and has been associated with embryonic lethality (5). Complete deletion of the AC gene in mice also leads to embryonic lethality (6), highlighting the importance of this enzyme in mammalian development.
We have recently shown that the inactive AC precursor undergoes self-cleavage to form a mature, active enzyme, and that the mechanism of this transformation is similar to other members of the N-terminal nucleophile hydrolase superfamily (7). Typically, the activity of one N-terminal nucleophile hydrolase subfamily member, the cysteine protease, is inhibited by small proteins known as cystatins (8). Cystatins are evolutionary related proteins, all of which are composed of at least one cystatin-like domain (CLD) with conserved sequence motifs. Aberrant regulation of cystatins occurs in a number of human diseases, including certain neurodegenerative disorders and cancer (9). For example, the cystatin A (Stefin A) gene is differentially expressed in primary and metastatic mammary tumors (10). Cystatin B also is elevated in tissues and the urine of bladder cancer patients, and its levels in urine are positively correlated with tumor grade, stage, and a shorter time to disease recurrence and progression (11). Decreased levels of cystatin C were found in the plasma of mice with Lewis lung adenocarcinoma (12), and cystatin E/M is a suppressor gene of cervical and breast cancer (13, 14). One member of the salivary cystatins (cystatin SN) also was found to be differentially regulated (activated or suppressed) in cancerous lesions of gastric cancer patient tissues (15).
Based on the AC self-cleavage and activation mechanism (7), which exposes a free cysteine residue, we hypothesized that one or more cystatins might also inhibit AC activity. We, therefore, assessed the effect of five candidate cystatins (A, B, C, E/M, and SA) on AC cleavage and/or activity. In the current study we identified cystatin SA (cysSA) as a potential physiological inhibitor of AC that affects activity of the enzyme without affecting precursor cleavage. We also carried out computer modeling to predict the interaction of cysSA and AC and identified an AC-like domain (ACLD) within cysSA that might facilitate this interaction. Two short peptides were synthesized containing the ACLD and CLD, and also were shown to inhibit AC activity in vitro. The interaction of AC and cysSA in cells was further demonstrated by co-immunoprecipitation and co-localization studies using confocal microscopy and by showing that elevated AC activity resulted from the expression of cysSA siRNA in cancer cells. Based on these results, we propose that cysSA affects AC activity by binding of its ACLD to an area adjacent to the active site of the enzyme. This potentially alters the affinity of the enzyme to the substrate and/or the product dissociation rate. This is the first report of a physiological inhibitor of AC and of a novel function for cysSA in the regulation of ceramide metabolism. In the future cysSA-based peptides might be used to treat various types of cancer in which AC is overexpressed alone or in combination with other cancer drugs.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibody reagents were used from Santa Cruz Biotechnology: anti-AC goat polyclonal IgG, catalog # sc-28486; anti-cysSA monoclonal IgG2b, catalog # sc-73884; donkey anti-goat IgG-horseradish peroxidase (HRP) conjugated, catalog # sc-2020; goat anti-mouse IgG-HRP conjugated, catalog # sc-2005. Anti-AC mouse monoclonal IgM also was obtained from BD Transduction Laboratories, catalog # 612302; Hoechst was purchased from Sigma, catalog # 33342. The Lipofectamine transfection reagent was purchased from Invitrogen, catalog # 11668019. Full-length cDNAs encoding cystatins A, B, C, E/M, and SA in the pCMV vector were purchased from OriGene Technologies. Sense and antisense oligonucleotides for cysSA were purchased from Sigma. Ribojuice was purchased from EMD Medical, catalog # 71115. Recombinant cysSA was purchased from R & D Systems, catalog #1201PI.
Oligonucleotide Generation and cDNA Cloning
The full-length human AC cDNA was subcloned in-frame into the pCMV vector (Sigma). Commercial cDNAs of cystatins A, B, C, E/M, and SA in the pCMV vector also were used. Partial cysSA cDNA fragments were generated by annealing of single-stranded, synthetic sense (see below) and antisense oligonucleotides flanked with HindIII and BamHI restriction sites using graduated temperature decrease. Cloning sites were then created by restriction digest using HindIII and BamHI and subcloned into the pCMV vector, digested by the same enzymes. The newly synthesized cDNA constructs were transfected into Top10 competent bacterial cells, and the integrity of the constructs was confirmed by sequencing. The sequences of the sense primers used for the fragment constructs were: Fragment 1, AATTAAGCTTTGGAGCCCCCAGGAGGAGGACAGGATAATCGAGGGTGGCATCTATGATGCAGACCTCAATGATGAGCGGGTACAGCGTGCCCTTCACTTTGTCATAGGGATCC; Fragment 2, AATTAAGCTTAGACGCCTGCTGCGGGTGCTACGAGCCAGGGAGCAGATCGTGGGCGGGGTGAATTACTTCTTCGACATAGAGGTGGGCCGAACCATATGTACCTAGGGATCC; Fragment 3, AATTAAGCTTCAGATCGTGGGCGGGGTGAATTACTTCTTCGACATAGAGGTGGGCCGAACCATATGTACCAAGTCCCAGCCCAACTTGGACACCTGTGCCTTCTAGGGATCC; Fragment 4, AATTAAGCTTTGGAGCCCCCAGGAGGAGGACAGGATAATCGAGGGTGGCATCACCATATGTACCAAGTCCCAGCCCAACTTGGACACCTGTGCCTTCCATTAGGGATCC; Fragment 5, AATTAAGCTTTGGAGCCCCCAGGAGGAGGACAGGATAATCGAGGGTGGCATCCGAGCCAGGGAGCAGATCGTGGGCGGGGTGAATTACTTCTTCGACTAGGGATCC.
Transient Transfection of HEK 293T17 Cells
For transient expression of the AC and cystatin cDNAs into HEK 293T17 cells, cDNA constructs were preincubated with the Lipofectamine-2000 transfection reagent in media according to the commercial instructions (Opti-MEM; Invitrogen). DNA-Lipofectamine-2000 complexes were then added to cells cultured overnight in 0.5 ml of antibiotic free DMEM media. After 24 or 48 h, the treated cells were harvested, centrifuged at 800 × g for 5 min at 4 °C, and kept at −20 °C. To prepare cell lysates, the cell pellets were lysed with the celLytic reagent (Sigma) and centrifuged at 10,000 × g.
Inhibition of Cystatin SA with siRNA
200 pmol of cysSA-specific (Santa Cruz Biotechnologies, sc-44521) or control siRNA (Dharmacon, D-001210-02-20) was transfected into SK-melanoma (SK-MEL) cells at 30% confluency in 6-well plates using the RiboJuiceTM siRNA Transfection Reagent (EMD Biosciences). Cell lysates were prepared 72 h after transfection and subjected to further analysis. In some (rescue) experiments recombinant cysSA (R&D Systems) was added to the culture media 48 h after transfection (0.4 μm). The treated cells were harvested 24 h later for the analysis of AC activity and ceramide levels.
Quantitative RT-PCR for Cystatin SA
Total RNA was extracted from SK-MEL cells (transfected with or without cysSA siRNA) using Cell Direct reagents (Invitrogen, catalog # 11739-010), as recommended by the manufacturer. An equal amount of RNA was reverse-transcribed using the Superscript III first strand kit (Invitrogen, catalog # 18080-051). mRNA levels were analyzed by quantitative PCR using Taq polymerase and SYBR Green fluorescent DNA binding dye and forward and reverse primers specific for cysSA and RPS18 (see the sequences below). The PCR products were analyzed using an ABI PRISM 7900 HT sequence detection system (Applied Biosystems). Fluorescence signals were analyzed during each of 40 cycles (denaturation, 30 s at 94 °C; annealing, 30 s at 55 °C; extension, 30 s at 72 °C). Relative quantification was calculated using the comparative threshold cycle (CT) method, as described in User Bulletin #2, ABI PRISM 7900HT Sequence Detection System. Median CT of triplicate measurements was used to calculate DCT as the difference in CT for the target gene (cysSA) and reference (RPS18). ΔCT values for each sample were compared with the corresponding mean control ΔCT and expressed as ΔΔCT. Relative quantification was expressed as -fold change of the gene of interest compared with the control according to the formula 2−ΔΔCT. Target gene expression between experimental and control groups were then compared using mean -fold changes. Sequences were: CysSA forward, AGCGTGCCCTTCACTTTGTCAT; CysSA reverse, TACATATGGTTCGGCCCACCTCT; RPS18 forward, TTCGGAATCGAGGCCATGAT; RPS18 reverse, TTTCGCTCTGGTCCGTCTTG.
Enzymatic Activity Assay
AC activity assays were performed using two types of synthetic substrates, BODIPY or NBD-conjugated C12-ceramide. The protocol for using the BODIPY-conjugated substrate was previously described (16). Briefly, cell lysates were incubated for 22 h at 37 °C with 0.2 mm BODIPY-conjugated C12-ceramide (a gift from Dr. Arie Dagan, Hadassah, Hebrew University, Israel) in 0.1 m citrate/phosphate buffer, pH 4.5, 150 mm NaCl, 0.05% BSA, and 0.1% Igepal CA-630. The reactions were stopped by ethanol and centrifuged, and the supernatants were analyzed using an HPLC separation system (Waters). Fluorescence was quantified using a Waters 474 fluorescence detector set to excitation and emission wavelengths of 505 and 540 nm for the product (i.e. BODIPY-conjugated C12-fatty acid) and substrate, respectively. The amount of product was calculated using a regression equation that was established from a standard curve using BODIPY-conjugated C12 fatty acid. The activity of neutral ceramidase in the cell lysates also was determined by the protocol described above, except that the buffer was adjusted to pH 7.
NBD-conjugated C12-ceramide was purchased from Cayman Chemical (catalog # 10007958), and the assay method was adopted from a previously described protocol (17). Briefly, recombinant proteins (AC with or without cysSA or cysSA-derived peptides) were incubated at 37 °C with 0.2 mm NBD substrate in 0.1 m citrate/phosphate buffer, pH 4.5, 150 mm NaCl, 0.05% BSA, and 0.1% Igepal CA-630. The reactions were stopped by ethanol and centrifuged, and the supernatants were analyzed using an ACQUITY UPLC separation system (Waters). Fluorescence was quantified using an ACQUITY UPLC fluorescence detector set to excitation and emission wavelengths of 435 and 525 nm for the product (i.e. NBD-conjugated C12-fatty acid) and substrate, respectively. The amount of product was calculated using a regression equation that was established from a standard curve using NBD-conjugated C12-fatty acid.
Ceramide Quantification
SK-Melanoma cells cultured to 40% confluency were transiently transfected with the cysSA cDNA or cysSA siRNA (or control siRNA, see above), and after 72 h cell extracts were prepared by three cycles of freeze/thaw. Lipids were extracted by mixing 150 μl with chloroform:methanol (1:2, v/v) followed by sonication for 5 min. After sonication, 100 μl of 1 m NaCl and 10 μl of concentrated HCl were added, vortexed, and centrifuged at 10,000 × g for 2 min. The lower organic phase was transferred to a new tube, dried with a SpeedVac concentrator, and resuspended in 10 μl of ethanol. Ceramide was quantified from the lipid extracts using the diacylglycerol kinase method (18).
Western Blot Analysis
Samples were separated by SDS-PAGE using 12% precast NuPAGE Bis-Tris gels under reducing conditions and MES running buffer (Invitrogen), and then transferred onto nitrocellulose membranes (Amersham Biosciences) using a semidry transfer apparatus (Bio-Rad) and NuPAGE-MOPS transfer buffer. For immunoblot analysis, membranes were blocked with TBS/Tween containing 5% dry milk and then incubated with specific antibodies that were recognized by secondary antibodies conjugated to HRP. Detection was achieved using an enhanced chemiluminescence detection reagent (Amersham Biosciences). Approximate molecular masses were determined by comparison with the migration of prestained protein standards (Bio-Rad). For quantitative analysis, the total intensity of the bands was assessed using the ImageJ software. Briefly, quantification was calculated by multiplying the area of the band by the average intensity (average number of pixels/area).
In Vitro, Auto-Proteolytic Cleavage Analysis
Cell lysates were prepared as described above and incubated at 4 or 37 °C. At 2, 4, 8, 24, and 48 h, an aliquot was withdrawn and subjected to SDS-PAGE and immunoblotting as described above.
Immunohistochemistry
For immunostaining of gingival fibroblasts, cells were cultured in minimum essential media on chamber slides. Slides were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton, and labeled with anti-AC and cysSA antibodies. Localization of the primary antibodies was visualized using a fluorescent second antibody (Cy-3/2) and laser-scanning confocal microscopy.
Immunoprecipitation
For co-immunoprecipitation of AC and cysSA, HEK 293T17 cells were lysed 24 h after transfection with the full-length cDNAs and incubated overnight with polyclonal anti-AC serum followed by magnetic beads precipitation (Dynabeads, Invitrogen) according to the manufacturer's protocol. The eluted proteins were separated using SDS-PAGE and detected by Western blotting using specific antibodies for AC and cysSA. For pulldown assays, recombinant AC and His-tagged cysSA were incubated at 4 °C overnight, and a pulldown assay was performed using Ni-NTA resin (Novagen, catalog # 70691) and the Ni-NTA buffer kit (Novagen, catalog # 70899-3).
Generation of Cystatin SA-based Peptides
Peptides P1 and P2 (see below) containing the AC-like (italics) and cystatin-like (bold) domains were synthesized by the Proteomics Resource Center in The Rockefeller University. The peptides were synthesized with an Applied Biosystems Model 430A instrument and purified using reversed phase HPLC. Purity of 96% was obtained for peptide P1 and 90% for P2. Peptide P2 was circular. Lyophilized peptides were reconstituted in water and stored at −80 °C. Peptides were: P1, QIVGGTICT; P2, CREQIVGGTICT.
Statistical Analyses
All experiments were repeated at least three times. The combined data from triplicate experiments were subjected to t test analysis, and the p values are indicated in each figure.
RESULTS
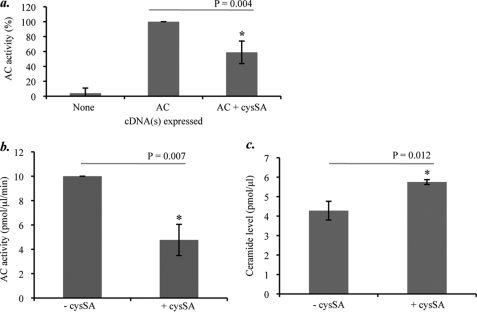
To initially assess the effect of different cystatins on AC activity, the full-length cDNAs encoding cystatins A, B, C, E/M, or SA were co-transfected with the AC cDNA into HEK 293T17 cells. AC activity was determined in cell extracts 24 h post-transfection, and the data were combined from three separate experiments. As shown in Fig. 1a, cysSA exhibited significant, albeit partial, inhibition (p = 0.004). Expression of the other cystatins did not have a significant effect on the co-expressed AC activity (data not shown). Note that in the absence of transfection endogenous AC, expression was nearly non-detectable in HEK 293T17 cells.
FIGURE 1.
The effects of cysSA expression on AC activity and ceramide in cells. a, AC activity was measured in HEK 293T17 cell lysates 24 h after transient expression of the full-length AC cDNA alone or co-transfection of the AC and the cysSA cDNAs. Note that AC activity was significantly lower after co-expression of AC and cysSA as compared with AC only (the asterisk indicates statistical significance, t test, p = 0.004). Expression of the cystatins A, B, C, and E/M cDNAs did not affect AC activity (data not shown). b, after expression of the full-length cysSA cDNA in SK-MEL cells, endogenous AC activity was significantly reduced in the cell lysates 48 h later (p = 0.007). c, ceramide levels also were significantly elevated in the transfected SK-MEL cells at 48 h (p = 0.012). Data represent the mean ± S.E., n = 3 independent experiments.
To further examine the effects of cysSA on AC, we therefore transfected the cysSA cDNA into SK-MEL cells, which express high endogenous levels of this enzyme. As shown in Fig. 1, b and c, expression of cysSA led to reduction of endogenous AC activity in the SK-MEL cells and elevation of ceramide (p = 0.007 and p = 0.012, respectively).
To investigate this interaction further, the effect of expressing cysSA siRNA on AC activity in SK-MEL cells was examined. As shown in Fig. 2a, expression of cysSA siRNA led to a significant increase (p = 0.003) in AC activity as compared with control siRNA. The addition of recombinant cysSA (0.4 μm) into the culture media 48 h after transfection of the cysSA siRNA led to partial reduction of AC activity (p = 0.044), revealing that the recombinant protein could partially rescue the effect of cysSA siRNA expression.
FIGURE 2.
AC and ceramide levels in SK-MEL cells after cysSA siRNA expression. a, AC activity was determined in SK-MEL cell lysates 72 h after transfection with control siRNA (con siRNA), cysSA siRNA, or cysSA siRNA exposed to recombinant cysSA in the culture media (cysSA siRNA + SA). In the latter experiments recombinant cysSA was added to the culture media 48 h after transfection (i.e. cells were exposed to recombinant cysSA for 24 h before being analyzed). Note that AC activity was significantly elevated after cysSA siRNA expression and that this effect was partially (but significantly) mitigated by the addition of recombinant cysSA into the culture media (the asterisk indicates statistical significance, t test, p = 0.003 and p = 0.044, respectively). Data represent the mean ± S.E., n = 3 independent experiments. b, to examine the effect of cysSA siRNA expression on ceramide, the levels were measured 72 h after siRNA transfection ± the inclusion of recombinant cysSA in the media. Note that the amount of ceramide in the cell lysates was significantly decreased after cysSA siRNA transfection, consistent with the elevation of AC activity (p = 0.005). Similarly, the inclusion of recombinant cysSA led to a partial but significant increase in the ceramide level (p = 0.045). c, to confirm the effect of cysSA siRNA expression on the endogenous cysSA mRNA, the levels were quantified by SYBR Green quantitative PCR after transfection with cysSA siRNA or control (con) siRNA. The cysSA mRNA levels were normalized to RPS18 rRNA as an internal control. Bar heights represent the mean values (-fold difference comparing cysSA to SRP18 expression) from three independent experiments. These quantitative PCR findings demonstrated that expression of the cysSA siRNA led to an ∼5-fold reduction of the endogenous cysSA mRNA levels (p = 0.002). d, expression of cysSA protein in the transfected and control cells was detected by Western blotting using a monoclonal antibody specific to cysSA. GAPDH was used as a loading control. Quantification was performed by densitometric scans of the Western blot images. The black bars indicate cysSA; gray bars indicate GAPDH. These results confirmed that in addition to reduction of cysSA mRNA, expression of cysSA siRNA led to reduction of cysSA protein.
Ceramide levels also were reduced by cysSA siRNA expression (p = 0.005) (Fig. 2b), consistent with the increase in AC activity, and were significantly increased after reconstitution by recombinant cysSA (p = 0.045). Quantitative RT-PCR and Western blot analysis were performed to confirm the reduction of endogenous cysSA mRNA and protein after cysSA siRNA transfection. As shown in Fig. 2c, cysSA mRNA levels were reduced ∼5-fold after expression of cysSA siRNA compared with a control siRNA (p = 0.002). The level of RPS18, used as a reference mRNA, remained unchanged (data not shown). Importantly, cysSA protein expression also was reduced ∼65% after cysSA siRNA transfection compared with the control (Fig. 2d).
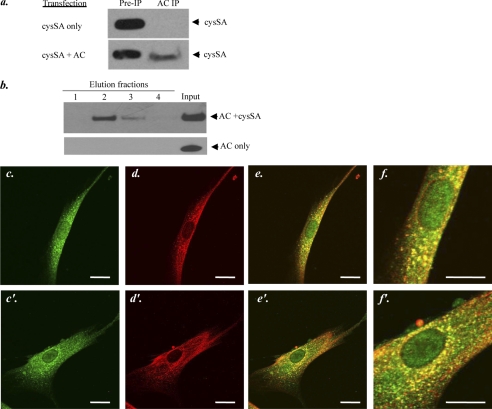
The interaction between AC and cysSA was further examined by immunoprecipitation studies after co-transfection of the AC and cysSA cDNAs in HEK 293T17 cells (Fig. 3a). AC was immunoprecipitated using a polyclonal anti-AC antibody, and the presence of cysSA in the precipitate was detected by Western blot analysis. As shown in this figure, after immunoprecipitation with the anti-AC antibody, we could readily detect cysSA in cells that were co-transfected with both cDNAs, suggesting that AC and cysSA formed a complex in these cells after co-expression. Importantly, the AC antibody alone did not precipitate cysSA in the absence of AC co-expression (Fig. 3a, upper right lane), implying that the interaction required expression of both proteins and was not due to nonspecific binding of cysSA to the anti-AC antibody. In addition, this control experiment indicated that the AC antibody did not recognize any endogenous protein(s) in HEK 293T17 cells that might be responsible for the precipitation of cysSA.
FIGURE 3.
Interaction between AC and cysSA. a, shown is co-immunoprecipitation (IP) of AC and cysSA from cell lysates after transient transfection of HEK 293T17 cells with full-length cDNAs of cysSA only or cysSA and AC together. Immunoprecipitation was performed using a polyclonal anti-AC serum. Western blotting was then performed using a polyclonal anti-cysSA antibody. These experiments indicated that immunoprecipitation of cysSA only occurred if both cysSA and AC were co-expressed. b, shown is a pulldown assay of recombinant AC and recombinant His-tagged cysSA, incubated in vitro for 24 h, or AC alone. Pulldown was performed using Ni-NTA resin followed by Western blotting and detection of AC in the elution fractions using a monoclonal anti-AC antibody. The results indicate that only in the presence of cysSA, could AC be bound to the Ni-NTA resin and detected in the elution fractions. c and c′–f and f′, shown is localization of AC and cysSA within human gingival fibroblasts, detected by immunohistochemistry. The upper (c–f) and lower (c′–f′) panels show images for two different cells. Staining was performed using anti-cysSA (red, c and c′) and anti-AC (green, d and d′) antibodies for protein detection. The upper (c–f) and lower (c′–f′) panels show staining for two different cells. Localization of the primary antibodies was visualized using a fluorescent second antibody Cy-3/2 and laser-scanning confocal microscopy. e and e′ are merged images showing co-localization of the two proteins (yellow). Scale bar = 20 μm. f and f′ are enlarged images of e and e′ to better illustrate the staining patterns. The images are representative of more than 50 cells analyzed from three independent immunostaining experiments.
To obtain additional evidence for this protein-protein interaction, we performed an in vitro binding assay using recombinant human AC and His-tagged cysSA. Both proteins or AC alone were incubated in vitro followed by pulldown using Ni-NTA resin and spin columns as described under “Experimental Procedures.” Proteins were eluted using a denaturing elution buffer, and AC was detected in the elution fractions after incubation with cysSA by Western blot analysis using a monoclonal anti-AC antibody (Fig. 3b). These results showed that AC eluted from the beads in a gradient manner, peaking in elution fraction 2. The fact that there was no detectable AC in fraction 1 indicated that the interaction and elution from the Ni-NTA columns was specific and not due to “spill-over” from the input. Moreover, in the absence of cysSA, AC did not bind to the Ni-NTA beads and was not observed in the elution fractions (Fig. 3b, bottom lanes). These findings were further supported by immunostaining of non-transfected gingival fibroblasts, which are known to express endogenous cysSA (Fig. 3, c–f). Co-localization (yellow, panels e, e′, f, and f′) of AC and cysSA was observed in these cells.
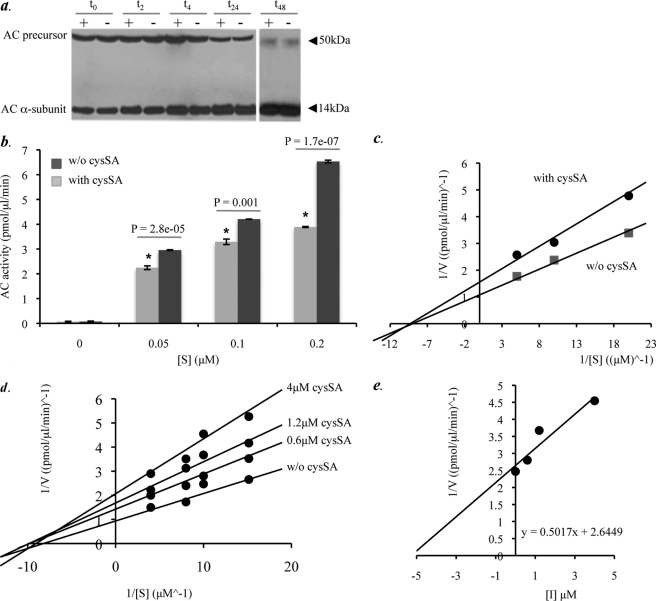
We next investigated the mechanism of AC inhibition by cysSA. For this purpose we assessed whether expression of cysSA blocked transition of the inactive AC precursor into the active heterodimer. HEK 293T17 cells were transiently transfected with cDNAs encoding AC alone or both AC and cysSA. Cell lysates were prepared 24 h post-transfection and then incubated at 37 °C for 2, 4, 8, 24, and 48 h. After incubation the protein lysates were analyzed by Western blotting using a monoclonal anti-AC antibody that recognized the AC α-subunit (14 kDa). This allowed detection of both the inactive 50-kDa precursor and heterodimeric active form (Fig. 4a). The results revealed that the conversion of AC from the precursor to active form was the same regardless of cysSA co-transfection. Thus, cysSA did not interfere with AC processing despite inhibiting activity.
FIGURE 4.
Characterization of the AC inhibitory mechanism by cysSA. a, protein lysates were prepared from HEK 293T17 cells transiently transfected with the AC cDNA, alone (−), or in combination with cysSA (+). Cleavage of the AC precursor into the active heterodimer was then assessed by incubation of the lysates at 37 °C at several time points (2, 4, 8, 24, and 48 h). Note that AC precursor cleavage was not affected by co-expression of cysSA at any of the time points. b, AC activity in the transfected HEK 293T17 cell lysates was determined at three different concentrations of substrate (BODIPY C12-ceramide). AC activity at each substrate concentration was significantly reduced (p = 2.8e−05, p = 0.001, p = 1.7e−07, respectively (indicated by the asterisk)) in the presence of cysSA co-transfection. For these experiments equal amounts of cell lysate protein (400 μg) from the transfected cells were incubated in a 100-μl reaction mixture containing BODIPY C12-ceramide. Data represent the mean ± S.E., n = 3 independent experiments. c, shown is a Lineweaver-Burk plot for the data of b, represented by plots y = 0.1509x + 1.7023 (cysSA co-transfection) and y = 0.1069x + 1.2654 (without (w/o) cysSA co-transfection). Interception of the x axes represents −1/Km. The predicted Km values were 0.88 and 0.83 with and without cysSA, respectively. Data represent the mean ± S.E., n = 3 independent experiments. d, shown is a Lineweaver-Burk plot depicting the activity of pure, recombinant AC in the presence of three different concentrations of pure, recombinant cysSA (0.6, 1.2, and 4 μm) at four different concentrations of substrate (NBD C12-ceramide) concentrations (0.067, 0.1, 0.125, and 0.25 μm). e, shown is a Dixon plot (1/V versus [I]), used to predict the Ki through the equation y = 0.5017x + 2.6449. Data represent three independent experiments. AC inhibition was statistically significant for all of the inhibitor concentrations with p values of 0.0006, 1.24e−05, and 2.9e−07 for 0.6, 1.2, and 4 μm cysSA, respectively.
To characterize the inhibitory effect further, we measured AC activity in the transfected HEK 293T17 cell lysates at three different substrate (BODIPY-C12 ceramide) concentrations (Fig. 4, b and c). In the presence of cysSA, the inhibition of AC exhibited the characteristics of a noncompetitive inhibitor, as enzyme activity (velocity) was reduced at all substrate concentrations, whereas the Km remained unchanged. This observation implies that cysSA binds AC at a site other than the enzyme active site (allosteric site) and is in agreement with the model of endopeptidase inhibition by cystatins described by Bode and Huber (19). To confirm that the effect was specific to AC, the activity of neutral ceramidase in the cell lysates also was measured, and no differences between the transfection and control groups were observed (data not shown).
The activity of pure, recombinant AC was then measured in the presence of three different concentrations of pure, recombinant cysSA using NBD-C12 ceramide as the substrate. NBD-C12 ceramide was used for these studies (as opposed to BODIPY-C12 ceramide) because it is commercially available and the in vitro kinetics for this substrate are well established (17). As illustrated by the Lineweaver-Burk plot (Fig. 4d), in the presence of cysSA (0.6, 1.2, and 4 μm) the AC activity was reduced for all concentrations of substrate (0.06, 0.1, 0.125, and 0.25 μm). AC inhibition was statistically significant for all of the inhibitor concentrations with p values of 0.0006, 1.24e−05, and 2.9e−07 for 0.6, 1.2, and 4 μm cysSA, respectively. As was observed with the in situ (co-transfection) analysis described above, the Lineweaver-Burk plot also indicated a noncompetitive type of inhibition. The Ki value for cysSA as calculated from a 1/V versus [I] plot equation (y = 0.5017x + 2.6449) was 5 μm (Fig. 4e).
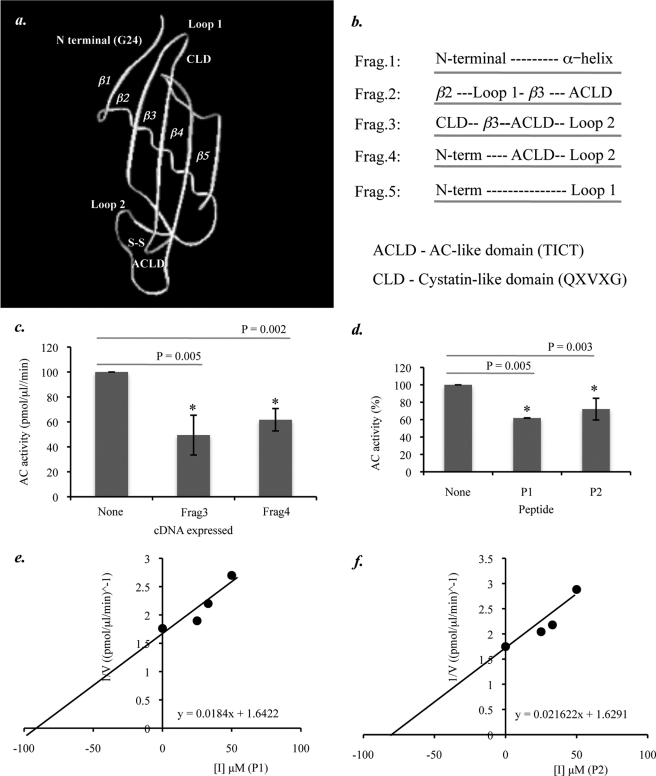
The three-dimensional structure of cysSA has not yet been resolved (20). Thus, we modeled the protein's secondary structure based on sequence homology to chicken cystatin C (Protein Data Bank accession code 1CEW) (Fig. 5a). Visualization of the predicted cysSA structure together with data obtained from published sources (21, 22) and analysis of the cysSA amino acid sequence revealed several structural components that we hypothesized might be involved in the AC inhibitory process. These regions included the N-terminal segment, containing a conserved Gly at residue 24, a conserved hairpin loop (loop 1) containing the CLD (QXVXG), and a second hairpin loop (loop 2) containing an ACLD (TICT). The latter AC-like domain was defined by a region within cysSA (residues 92–95) that was homologous to a region in human AC (residues 141–145) and formed by a disulfide bridge between residues Cys-94 and Cys-104. Of note, the corresponding sequence in AC (TICT) comprised the precursor cleavage site (i.e. the cysteine residue is the N terminus of the β-subunit).
FIGURE 5.
Computer modeling and construction of cysSA-based peptides. a, shown is a proposed model of cysSA, generated by 3D-JIGSAW protein homology modeling based on PDB entry 1rn7, showing the location of potential functional domains (i.e. CLD; QXVXG), the N-terminal region, Loop 1, Loop 2, and ACLD (TICT). b, shown is the design of partial cDNA fragments encoding the potential functional domains of cysSA. c, AC activity was measured in HEK 293T17 cell extracts after transient expression of the AC cDNA and co-expression of Fragments 1–5, subcloned into the pCMV vector. AC activity in the presence of Fragments 2 and 3, both of which contain the AC-like domain, was significantly lower in comparison to AC only (t test, p = 0.005 and 0.002, respectively). Other peptides did not have a significant effect on AC activity (data not shown). Data represent the mean ± S.E., n = 3 independent experiments. d, the activity of pure, recombinant AC was determined in the presence of three different amounts (25, 33, and 50 μm) of peptides P1 and P2 as well as a nonspecific control peptide (Pc) using 0.1 μm NBD-C12 ceramide. The relative AC activity, averaged for all peptides concentrations, was significantly reduced for both of the cysSA-based peptides (p = 0.005 and 0.003, respectively) but not for the control peptide. e and f, Dixon plots (1/V versus [I]) were used to estimate the Ki values for P1 (e) and P2 (f) though equations (y = 0.0184x + 1.6422 and y = 0.021622x + 1.6291, respectively). Data represent the mean ± S.E., n = 3 independent experiments.
Five partial cysSA cDNA fragments containing these potential inhibitory domains in different combinations were synthesized, and their effect on AC activity was assessed in HEK 293T17 cells 24 h after co-transfection with the full-length AC cDNA (Fig. 5, b and c). This analysis revealed that Fragment 3, containing the CLD, β3 region, ACLD, and loop 2 regions, and Fragment 4, containing the N-terminal region, ACLD, and loop 2 regions, exhibited statistically significant inhibition (p = 0.005 and p = 0.002, respectively). Fragments containing the N-terminal α-helix domain or β2 regions alone had no effect on AC inhibition (data not shown).
These results suggested that the ACLD of cysSA was important, but not sufficient, for AC inhibition. Based on these results, we next synthesized two small peptides and tested their effects on AC inhibition. Peptide P1 was linear and consisted of contiguous cystatin-like and AC-like domain sequences (QIVGGTICT). Peptide P2 had an additional three amino acids at the N-terminal end to allow formation of a circular structure (CREQIVGGTICT). AC activity in the presence of the peptides (25, 33, and 50 μm) was assessed in vitro using 0.1 μm NBD-C12 ceramide. The combined data showed that AC activity was significantly reduced in the presence of both peptides (p = 0.003 and 0.005, respectively) (Fig. 5d). A control peptide (derived from β-galactosidase) did not effect AC activity (data not shown). The calculated Ki values for peptides P1 and P2 using Dixon plots equations (y = 0.0184x + 1.6422 and y = 0.021622x + 1.6291, respectively) were 90 and 75 μm, respectively (Fig. 5, e and f).
DISCUSSION
We have previously reported that the inactive AC precursor undergoes self-cleavage and activation, similar to other N-terminal nucleophile hydrolase family members (7). To the best of our knowledge this is the only mammalian ceramidase that is known to undergo this activation mechanism. Having cysteine residues at the cleavage site implied that AC is within the same subcategory of N-terminal nucleophile hydrolases as the cysteine proteases. In the current study we therefore investigated whether AC could be inhibited by the cysteine protease inhibitors, cystatins, particularly by cystatins A, B, C, E/M, and/or SA. Our initial approach relied on co-expression of AC and the cystatin cDNAs in HEK 293T17 cells to produce enough of the proteins for co-immunoprecipitation and other analyses. These studies suggested that cysSA had an inhibitory effect, and therefore, the remainder of our work was focused on this cystatin.
To evaluate a more physiological system, we next followed the activity of endogenous AC. A melanoma cell line (SK-MEL) was chosen for these studies because it expressed high levels of AC, allowing confident analysis of the activity changes. Overexpression of cysSA in these cells significantly reduced endogenous AC activity. Ceramide levels also were elevated, supporting the results obtained in the co-expression system. We also used siRNA to inhibit cysSA in the SK-MEL cells, resulting in elevated AC activity and reduced levels of ceramide. Expression of control siRNA did not elicit these changes. Moreover, supplementation of recombinant cysSA into the media recovered the effects of cysSA siRNA on AC activity and ceramide levels, supporting the conclusion that these changes occurred due to down-regulation of cysSA and were not due to nonspecific effects of cysSA siRNA expression on other mRNAs and proteins.
A physical interaction between AC and cysSA also was demonstrated after co-expression of the two proteins in HEK 293T17 cells by co-immunoprecipitation and purification. Confocal microscopy in gingival fibroblasts further revealed co-localization of endogenous AC and cysSA. While these experiments do not rule out the possibility of an indirect mechanism of cysSA action, together with the data described above they strongly suggest that cysSA affects AC activity through a physical interaction.
Published studies describing the mechanism of inhibition of cystatins on cysteine proteases are consistent with this hypothesis and suggest that cystatins bind to the enzymes and create allosteric interference with the active site domains (19). This would be consistent with our observation that expression of cysSA did not affect processing of the AC precursor into the active form heterodimer despite inhibiting activity.
To investigate the mechanism of cysSA on AC further and to definitively establish a direct interaction between the two proteins, we next performed a series of kinetic studies using pure, recombinant forms of cysSA and AC. At all substrate concentrations tested, significant inhibition of AC activity by cysSA was observed without changes in the Km, suggesting non-competitive inhibition and supporting the proposed model of inhibition for other cystatins on the endopeptidases (19). To identify potential regions of cysSA that might facilitate an interaction with AC, computer modeling was used and identified an ACLD that shared a short amino acid sequence (TICT) with the precursor cleavage site of AC. We hypothesized that this sequence may facilitate the binding of cysSA to an allosteric site on the enzyme and could influence the enzyme-substrate interaction.
To explore this hypothesis, we constructed partial cysSA cDNA fragments that included various combinations of known functional domains (e.g. the N-terminal and CLD domains) as well as new potential binding domains identified by protein modeling, including several α-helices, the ACLD, and loop 2. Although we cannot confirm that proper folding of these cysSA-based fragments occurred after cDNA expression, the results revealed that the inhibitory effect was most significant for fragments containing the ACLD, CLD, and loop 2 regions. This combination of common (e.g. CLD) and unique (e.g. ACLD) features could presumably lead to preferential inhibition of AC by cysSA, as compared with other enzymes that may also be affected by this protein (e.g. cathepsin C and L, 22).
These findings also led us to synthesize two small peptides that contained the CLD and ACLD sequences in order to confirm the results. The only difference between the two peptides was the presence of an additional three amino acids in peptide 2 that facilitated circularization. Both peptides were found to be effective AC inhibitors in vitro, confirming the observations from co-transfection of the partial cysSA cDNAs. Another short peptide (Pc) that did not contain these sequences was used as a control and did not exhibit any AC inhibition. Future studies are required to further analyze the properties of the peptides (e.g. specificity for AC, cytotoxicity etc.) in comparison to other existing AC inhibitors.
Overall, the results presented here describe a new inhibitor and inhibitory mechanism for AC and a unique form of post-translational regulation for this enzyme. In addition, based on these observations, two small peptides were synthesized with inhibitory activity. Clearly, the functional regulation of AC in vivo is likely to be very complex, particularly given its the critical role in sphingolipid metabolism and cell survival, and although we hypothesize that cysSA is an important part of this regulatory process, it is certainly not expected to be the only factor. For example, AC is known to exist in a complex with several other lipid hydrolyases (23), suggesting some type of coordinated, functional regulation of the enzymatic machinery needed for sphingolipid metabolism. In addition, it requires “activator” proteins (e.g. Sap D, Ref. 24) to facilitate its interaction with the very hydrophobic, membrane-embedded ceramide substrate, and the interaction of these and other regulatory molecules (e.g. cysSA) is likely to vary greatly based on cell type, the stage of cell cycle, stress, and other factors.
In conclusion, we reveal for the first time that cysSA is a physiological inhibitor of AC and that it contributes to the complex regulation of ceramide metabolism by this enzyme. We have also identified functional domains within cysSA that are responsible for the inhibitory effect and designed cysSA-based peptide inhibitors that could be used in the future as reagents to modulate AC activity and ceramide metabolism.
Acknowledgment
We acknowledge the Proteomics Resource Center at the Rockefeller University (New York) for synthesis of the peptides.
This work was supported, in whole or in part, by National Institutes of Health Grant 2 R01 DK54830. This work was also supported by Binational Science Foundation Grant 2009212. N. S., E. E., X. H., and E. H. S. are inventors on AC-related patent applications, which if licensed could generate royalties for them and the Mount Sinai School of Medicine.
- AC
- acid ceramidase
- CLD
- cystatin-like domain
- ACLD
- acid ceramidase-like domain
- cysSA
- cystatin SA
- SK-MEL
- SK melanoma cells
- Ni-NTA
- nickel-nitrilotriacetic acid
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Bartke N., Hannun Y. A. (2009) J. Lipid Res. 50, S91–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang Y., Li X., Becker K. A., Gulbins E. (2009) Biochim. Biophys. Acta 1788, 178–183 [DOI] [PubMed] [Google Scholar]
- 3. Sugita M., Dulaney J. T., Moser H. W. (1972) Science 178, 1100–1102 [DOI] [PubMed] [Google Scholar]
- 4. Moser H. W., Linke T., Fensom A. H., Levade T., Sandhoff K. (2001) (Scriver C. R., Beaudet A. L., Valle D., Sly W. S. eds) pp. 3573–3588, McGraw-Hill Inc., New York [Google Scholar]
- 5. van Lijnschoten G., Groener J. E., Maas S. M., Ben-Yoseph Y., Dingemans K. P., Offerhaus G. J. (2000) Pediatr. Dev. Pathol. 3, 597–602 [DOI] [PubMed] [Google Scholar]
- 6. Eliyahu E., Park J. H., Shtraizent N., He X., Schuchman E. H. (2007) FASEB J. 21, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 7. Shtraizent N., Eliyahu E., Park J. H., He X., Shalgi R., Schuchman E. H. (2008) J. Biol. Chem. 283, 11253–11259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rzychon M., Chmiel D., Stec-Niemczyk J. (2004) Acta Biochim. Pol. 51, 861–873 [PubMed] [Google Scholar]
- 9. Rivenbark A. G., Coleman W. B. (2009) Front. Biosci. 14, 453–462 [DOI] [PubMed] [Google Scholar]
- 10. Parker B. S., Ciocca D. R., Bidwell B. N., Gago F. E., Fanelli M. A., George J., Slavin J. L., Möller A., Steel R., Pouliot N., Eckhardt B. L., Henderson M. A., Anderson R. L. (2008) J. Pathol. 214, 337–346 [DOI] [PubMed] [Google Scholar]
- 11. Feldman A. S., Banyard J., Wu C. L., McDougal W. S., Zetter B. R. (2009) Clin. Cancer Res. 15, 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Usova T. A., Poteryaeva O. N., Zhanayeva S. Y., Yarygina E. S., Korolenko T. A. (2003) Bull. Exp. Biol. Med. 135, 81–84 [DOI] [PubMed] [Google Scholar]
- 13. Veena M. S., Lee G., Keppler D., Mendonca M. S., Redpath J. L., Stanbridge E. J., Wilczynski S. P., Srivatsan E. S. (2008) Genes Chromosomes Cancer 47, 740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang J., Shridhar R., Dai Q., Song J., Barlow S. C., Yin L., Sloane B. F., Miller F. R., Meschonat C., Li B. D., Abreo F., Keppler D. (2004) Cancer Res. 64, 6957–6964 [DOI] [PubMed] [Google Scholar]
- 15. Choi E. H., Kim J. T., Kim J. H., Kim S. Y., Song E. Y., Kim J. W., Kim S. Y., Yeom Y. I., Kim I. H., Lee H. G. (2009) Clin. Chim. Acta 406, 45–51 [DOI] [PubMed] [Google Scholar]
- 16. He X., Chen F., Gatt S., Schuchman E. H. (2001) Anal. Biochem. 293, 204–211 [DOI] [PubMed] [Google Scholar]
- 17. Tani M., Okino N., Mitsutake S., Ito M. (1999) J. Biochem. 125, 746–749 [DOI] [PubMed] [Google Scholar]
- 18. Bielawska A., Perry D. K., Hannun Y. A. (2001) Anal. Biochem. 298, 141–150 [DOI] [PubMed] [Google Scholar]
- 19. Bode W., Huber R. (2000) Biochim. Biophys. Acta 1477, 241–252 [DOI] [PubMed] [Google Scholar]
- 20. Vray B., Hartmann S., Hoebeke J. (2002) Cell. Mol. Life Sci. 59, 1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hall A., Dalbøge H., Grubb A., Abrahamson M. (1993) Biochem. J. 291, 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saitoh E., Minaguchi K., Ishibashi O. (1998) Arch. Biochem. Biophys. 352, 199–206 [DOI] [PubMed] [Google Scholar]
- 23. He X., Okino N., Dhami R., Dagan A., Gatt S., Schulze H., Sandhoff K., Schuchman E. H. (2003) J. Biol. Chem. 278, 32978–32986 [DOI] [PubMed] [Google Scholar]
- 24. Azuma N., O'Brien J. S., Moser H. W., Kishimoto Y. (1994) Arch. Biochem. Biophys. 311, 354–357 [DOI] [PubMed] [Google Scholar]