Background: The C-ring has a crucial role in bacterial type III secretion.
Results: Salmonella ssaQ produces two proteins by tandem translation: a short protein binds to its corresponding region within the larger putative C-ring protein and stabilizes it.
Conclusion: SsaQ function is optimized by a novel chaperone-like protein, produced by tandem translation from its own mRNA.
Significance: The data increase the understanding of type III secretion.
Keywords: Bacterial Pathogenesis, Chaperone Chaperonin, Microbial Pathogenesis, Protein Secretion, Type III Secretion System
Abstract
Type III secretion systems (T3SSs) of bacterial pathogens involve the assembly of a surface-localized needle complex, through which translocon proteins are secreted to form a pore in the eukaryotic cell membrane. This enables the transfer of effector proteins from the bacterial cytoplasm to the host cell. A structure known as the C-ring is thought to have a crucial role in secretion by acting as a cytoplasmic sorting platform at the base of the T3SS. Here, we studied SsaQ, an FliN-like putative C-ring protein of the Salmonella pathogenicity island 2 (SPI-2)-encoded T3SS. ssaQ produces two proteins by tandem translation: a long form (SsaQL) composed of 322 amino acids and a shorter protein (SsaQS) comprising the C-terminal 106 residues of SsaQL. SsaQL is essential for SPI-2 T3SS function. Loss of SsaQS impairs the function of the T3SS both ex vivo and in vivo. SsaQS binds to its corresponding region within SsaQL and stabilizes the larger protein. Therefore, SsaQL function is optimized by a novel chaperone-like protein, produced by tandem translation from its own mRNA species.
Introduction
Following uptake or invasion of mammalian host cells, Salmonella enterica serovar Typhimurium (sv. Typhimurium) replicates within membrane-bound compartments known as Salmonella-containing vacuoles. Acidification of lumen of the Salmonella-containing vacuole stimulates expression of genes involved in the assembly of a multiprotein structure that spans the bacterial cell envelope, called the Salmonella pathogenicity island 2 (SPI-2)2 type III secretion system (T3SS) (1–3). This T3SS secretes proteins that assemble a needle structure and a translocon pore in the vacuolar membrane (1). Sensing of the near-neutral pH of the host cell cytosol by unknown component(s) of the T3SS triggers the dissolution and loss in the bacterial cytoplasm of a T3SS-associated regulatory complex comprising three proteins: SsaL, SpiC, and SsaM (4). This relieves repression of secretion of ∼25 effector proteins that are translocated across the vacuolar membrane (4). The effectors have many different functions, affecting immune signaling (5), bacterial and host cell motility (6, 7), and intracellular replication of bacteria in a variety of host cell types, including epithelial cells and macrophages (8). As a result, SPI-2 T3SS null mutants are strongly attenuated in virulence in various hosts (9–12).
ssaQ is the last gene in the ssaMVNOPQ operon within SPI-2 (13, 14). The predicted product of ssaQ is a member of the FliN/YscQ/Spa33/HrcQ family of flagellum and T3SS proteins (15). These proteins belong to the conserved essential core of these structures and are thought to constitute a cytoplasmic platform (C-ring) connected to the base of the secretion system (16, 17). In this work, we studied the ssaQ gene of sv. Typhimurium. We found that it produces two proteins from one transcript: a protein of the expected size and a second translational product corresponding to the C-terminal 106 amino acids. The tandem translated small protein acts as a chaperone, binding to and stabilizing the larger protein, and is important for the overall efficiency of the secretion system.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The sv. Typhimurium strains used in this study are derivatives of wild-type strain 12023 and are listed in Table 1. Bacteria were grown in LB medium supplemented with carbenicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (15 μg/ml), or tetracycline (25 μg/ml) for strains resistant to these antibiotics (Ampr, Kanr, Camr, and Tetr, respectively). To induce SPI-2 gene expression and SPI-2-dependent secretion, bacteria were grown in magnesium minimal medium (MgM)/MES (3) at pH 5.0 with the corresponding antibiotics when appropriate.
TABLE 1.
sv. Typhimurium strains constructed and used in this study
| Name | Description | Source or Ref. |
|---|---|---|
| 12023 | Wild-type | NCTC |
| P3F4 | ssrA::mTn5 in 12023 (Kanr) | Ref. 9 |
| HH216 | sseJ-2HA::cat in 12023 (Camr) | Ref. 21 |
| HH225 | ΔssaQ18–311::Kan in 12023 (Kanr) | This study |
| HH226 | sseJ-2HA::cat in ΔssaQ18–311::Kan mutant | This study |
| HH227 | ΔssaQ104–223::cat in 12023 (Camr) | This study |
| HH228 | ssaQ replaced with ssaQM217L in 12023 | This study |
| HH229 | ssaQ replaced with ssaQY206oc in 12023 | This study |
| HH230 | sseJ-2HA::cat in HH229 | This study |
| HH231 | sseJ-2HA::cat in HH228 | This study |
| HH232 | ssaQ replaced with ssaQM217L-HA::Kan in 12023 | This study |
The λ Red recombination system (18) was used to construct ssaQ deletion mutants HH225 and HH227 using primers ssaQd1 and ssaQd2 or primers ssaQd3 and ssaQd4, respectively. Primers are listed in the supplemental table. Chromosomal allelic exchange was used to construct ssaQS point mutant HH228 or ssaQL point mutant HH229. Briefly, ssaQ was ligated into pCR2.1-TOPO (Invitrogen) using primers ssaQf and ssaQb to construct pCR-ssaQ, followed by site-directed mutagenesis to create pCR-ssaQM217L and pCR-ssaQY206oc using primers ssaQM217Lf and ssaQM217Lb or primers ssaQY206ocf and ssaQY206ocb, respectively. Mutant versions of ssaQ were subcloned into suicide vector pCVD442 for transferring mutations to HH227 by conjugation, and exconjugants were selected as described previously (19). Primers ssaQHAf and ssaQdb were used to fuse DNA segments encoding the HA epitope to the 3′ termini of the coding region of ssaQ or ssaQL in WT or HH228 strains as described previously (20).
To express sseJ-2HA from chromosomal DNA, sseJ-2HA::cat from HH216 (21) was transduced into different strains of sv. Typhimurium by phage P22 as described previously (22). When necessary, the FRT-flanked antibiotic resistance cassette was removed after transformation with pCP20 as described (18).
Plasmids
The spiC promoter was ligated into pWSK29 (23) using primers spiCn1 and spiCn2 to create pspiCpr. Plasmids pssaQ and pssaQL were constructed by subcloning ssaQ and ssaQM217L fragments from pCR-ssaQ and pCR-ssaQM217L into pspiCpr. The ssaQS gene was amplified using primers ssaQSf and ssaQb and ligated into pspiCpr to construct pssaQS. The HA-tagged ssaQ, ssaQM217L, and ssaQY206oc DNA fragments were amplified from pCR-ssaQ, pCR-ssaQM217L, and pCR-ssaQY206oc using primers ssaQf and ssaQHAb and ligated into pspiCpr to create plasmids pssaQ-HA, pssaQM217L-HA, and pssaQY206oc-HA. The ssaQLN216-HA fragment was amplified using primers ssaQf and ssaQLN216HAb and ligated into pspiCpr to construct pssaQLN216-HA. The ssaQL-T7 fragment was amplified from pssaQM217L-HA using primers ssaQf-SacI and ssaQT7b-SacI and ligated into pspiCpr, pssaQM217L-HA, and pssaQLN216-HA to create plasmids pssaQL-T7, pssaQL-HA/ssaQL-T7, and pssaQLN216-HA/ssaQL-T7, respectively. Plasmid pssaQS-T7 was constructed by replacing the cat gene of pACYC184 (24) with ssaQS-T7 following PCR amplification with primers ssaQSf and ssaQT7b. The expression of ssaQS-T7 is under the control of the constitutive promoter of cat.
The PCR products including the promoter region of ssaM to the start codon of ssaQS (using primers 102ssaM-KpnI and ssaQL217r-XbaI) or beginning from the start codon of ssaM to the start codon of ssaQS (using primers ssaM-KpnI and ssaQL217r-XbaI), amplified from genomic DNA of sv. Typhimurium strain 12023, were ligated into the KpnI and XbaI sites of pFPV25, a vector carrying promoterless gfpmut3A (25), to create p102ssaMQLN217 and pssaMQLN217, respectively. To express ssaQS with a His6 tag, the cleaved ssaQS PCR product (using primers ssaQS-EcoRIf and ssaQS-XhoIb) was ligated into pET-22b (Novagen) to construct pET-ssaQS. Plasmids constructed as part of this study were verified by DNA sequencing and are listed in Table 2. Plasmid psteC-2HA was described previously (26).
TABLE 2.
Plasmids used in this study
| Name | Description | Source or Ref. |
|---|---|---|
| pWSK29 | pSC101 ori, low copy number vector, Ampr | Ref. 23 |
| pFPV25 | Carrying promoterless gfpmut3A, Ampr | Ref. 25 |
| pCVD442 | Suicide vector for DNA allelic exchange, Ampr | Ref. 19 |
| pACYC184 | p15A ori, medium copy number vector, Tetr, Camr | Ref. 24 |
| pspiCpr | spiC promoter on pWSK29, Ampr | This study |
| pssaQ | ssaQ gene on pspiCpr, Ampr | This study |
| pssaQ-HA | ssaQ-HA gene on pspiCpr, Ampr | This study |
| pssaQM217L-HA | ssaQM217L-HA gene on pspiCpr, Ampr | This study |
| pssaQY206oc-HA | ssaQY206oc-HA gene on pspiCpr, Ampr | This study |
| p102ssaMQLN217 | Sequence composed of promoter region of ssaM to start codon of SsaQS on pFPV25, Ampr | This study |
| pssaMQLN217 | Sequence composed of start codon of ssaM to start codon of SsaQS on pFPV25, Ampr | This study |
| pssaQL | ssaQM217L gene on pspiCpr, Ampr | This study |
| pssaQS | ssaQS gene on pspiCpr, Ampr | This study |
| pssaQS-T7 | ssaQS-T7 gene on pACYC184, Tetr | This study |
| pssaQLN216-HA | ssaQLN216-HA gene on pspiCpr, Ampr | This study |
| pET-ssaQS | ssaQS gene on pET-22b, Ampr | This study |
| pssaQL-T7 | ssaQL-T7 gene on pspiCpr, Ampr | This study |
| pssaQL-HA/ssaQL-T7 | ssaQL-T7 gene on pssaQM217L-HA, Ampr | This study |
| pssaQLN216-HA/ssaQL-T7 | ssaQL-T7 gene on pssaQLN216-HA, Ampr | This study |
| psteC-2HA | steC-2HA gene with steC promoter on pWSK29, Ampr | Ref. 26 |
Antibodies
The following primary antibodies were used for immunoprecipitation, immunofluorescence staining, and immunoblot analysis: rat anti-HA antibody (3F10, Roche Applied Science); mouse anti-T7 antibody (Novagen); mouse anti-HA monoclonal antibody HA.11 (MMS-101P, Covance); mouse anti-DnaK antibody (Assay Designs); goat anti-Salmonella polyclonal antibody CSA-1 (Kirkegaard & Perry Laboratories); and rabbit anti-SseB, anti-SseC, and anti-SseD polyclonal antibodies (21). Texas Red sulfonyl chloride-conjugated donkey anti-goat antibody (Jackson ImmunoResearch Laboratories) was used for immunofluorescence microscopy. Alexa Fluor 488-conjugated donkey anti-goat antibody and Alexa Fluor 647-conjugated donkey anti-mouse antibody (Invitrogen) were used for flow cytometric analysis. The following HRP-conjugated secondary antibodies were used for immunoblot analysis: donkey anti-rabbit and sheep anti-mouse antibodies (Amersham Biosciences) and rabbit anti-rat antibody (Dako).
Preparation of Protein Fractions from Bacteria Grown in Vitro
Bacterial strains were grown in MgM/MES for secretion assays as described previously (4, 21). To ensure that protein from equal numbers of cells was analyzed, in all experiments, protein samples were adjusted to A600 values such that a volume corresponding to 10 ml of a culture with A600 = 0.6 was taken up in 100 μl of protein denaturing buffer for secreted and bacterial surface-associated fractions and in 600 μl of protein denaturing buffer for the total bacterial fraction.
Immunoprecipitation Assays
Volumes corresponding to 20 ml of a bacterial culture with A600 = 0.6 after 5 h of growth of sv. Typhimurium in MgM/MES at pH 5.0 were used to perform immunoprecipitation assays. Bacteria were collected by centrifugation and resuspended in a solution comprising 1 ml of 50 mm glucose, 10 mm EDTA, 4 mg/ml lysozyme, and 25 mm Tris-Cl (pH 8.0) and incubated for 5 min at room temperature to generate spheroplasts. The spheroplasts were resuspended into 1.5 ml of lysis buffer (50 mm Tris-Cl (pH 8.0, 100 mm NaCl, 1% Triton X-100, and 1 mm PMSF) and incubated on ice for 30 min with occasional mixing. The lysate was centrifuged for 10 min at 10,000 × g, and the supernatant was transferred into a fresh tube and incubated with 50 μl of protein G-immobilized agarose (Pierce) for 1 h to preclear the lysate. The precleared supernatant was incubated with 10 μl of rat anti-HA antibody or mouse anti-T7 antibody for 2 h at 4 °C to form antibody-antigen complexes, and 50 μl of protein G-immobilized agarose was added to the reaction and incubated for 2 h at 4 °C. The beads were collected by centrifugation at 500 × g for 4 min and washed four times with 700 μl of lysis buffer prior to boiling in 50 μl of SDS-PAGE sample buffer.
N-terminal Sequencing of SsaQS
The bacterial lysate of ssaQ pssaQ-HA culture grown in MgM/MES was subjected to immunoprecipitation with rat anti-HA antibody. SsaQS-HA was recovered from the PVDF membrane and sequenced by Edman degradation (Protein & Nucleic Acid Chemistry Facility, University of Cambridge). The first six amino acids obtained for SsaQS were MKFDEL.
Cross-linking and Protein Stability Assays
Escherichia coli BL21(DE3) cells containing plasmid pET-ssaQS were cultured in LB medium and induced for 2 h at 30 °C with 1 mm isopropyl β-d-thiogalactopyranoside to express SsaQS-His6. The bacteria were treated with DMSO (Sigma) or the cross-linker disuccinimidyl suberate (1 mm; Pierce) as recommended by the manufacturer and subjected to Western blotting with HRP-conjugated anti-His antibody (ab1187, Abcam).
For the SsaQL-HA stability assay, bacterial strains were cultured in MgM/MES at pH 5.0 for 4 h, and tetracycline was then added to a final concentration 20 μg/ml to stop protein synthesis. Samples were removed at different time points for immunoblotting.
PAGE and Immunoblot Analysis of Proteins
Protein fractions were dissolved in the appropriate volume of protein denaturing buffer and held at 100 °C for 10 min. Proteins were immediately separated on 12% SDS-polyacrylamide gels. For immunoblot analysis, proteins were transferred to Immobilon-P (PVDF) membranes (Millipore) and examined using the ECL detection system (Amersham Biosciences) under conditions recommended by the manufacturer. Incubation of membranes with primary antibodies was followed by incubation with HRP-conjugated secondary antibodies.
Cell Culture, Immunofluorescence, and Flow Cytometric Analysis
HeLa epithelial cells (clone HtTA1) were grown in DMEM (Invitrogen) supplemented with 10% FCS and 2 mm glutamine at 37 °C in 5% CO2 and infected with exponential phase sv. Typhimurium as described previously (27).
To visualize GFP expression, HeLa cells were fixed with 3% paraformaldehyde at 5 h post-invasion. Bacteria were labeled first with antibody CSA-1 and then with Texas Red sulfonyl chloride-conjugated donkey anti-goat antibody. Images were taken on a Zeiss LSM 510 confocal laser scanning microscope.
To detect translocation of SseJ-2HA, HeLa cells infected with Salmonella strains for 13 h were trypsinized and then fixed with 3% paraformaldehyde. Antibodies CSA-1 and HA.11 were used to label bacteria and translocated SseJ-2HA in the presence of 0.1% saponin (Sigma), followed by labeling with secondary antibodies conjugated to Alexa Fluor 488 or 647. Approximately 1 × 105 infected cells were analyzed on a FACSCalibur cytometer (BD Biosciences), and bacteria and SseJ-2HA were detected at 525 nm in the FL1 channel and at 633 nm in the FL4 channel. Data were analyzed with FlowJo 8.8.6 software.
Bioinformatic Analysis and Homology Modeling
Homology models of SsaQS were calculated using the Phyre protein structure prediction server (28). The homology model of SsaQS was calculated from the primary amino acid sequence of the flagellar rotor protein FliN from Thermotoga maritima (29). The SsaQS sequence was aligned, submitted to multiple secondary structure prediction, and compared against the Phyre fold library. The top 10 alignments were used to produce an ensemble of three-dimensional models. The top-scoring Phyre homology model was used as the representative monomer structure, and a final model for the dimer was assembled by superposition on the dimer of FliN. ClustalW2 was used to align the two protein sequences.
Mouse Mixed Infections
Female BALB/c mice (20–25 g) were used for competitive index (CI) studies. Four mice were inoculated intraperitoneally with a mixture of two strains comprising 500 colony-forming units of each strain in physiological saline, and the CIs were determined from spleen homogenates 96 h post-inoculation as described previously (30). The p value was determined by two-tailed t test.
RESULTS
Products of SsaQ
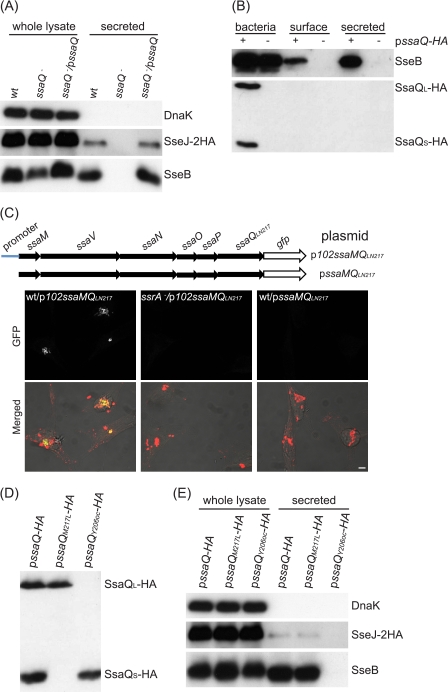
SsaQ is encoded by SPI-2 and shares weak overall similarity (14.6% identity and 38.5% similarity) with Spa33, the C-ring protein of the Shigella T3SS (16). The C-ring proteins show greater conservation in the C-terminal regions, and an alignment of this region revealed several conserved residues that are also found in SsaQ (15). To investigate the function of SsaQ in SPI-2 type III secretion, an ssaQ deletion mutant with the chromosomally encoded epitope-tagged effector SseJ-2HA was constructed and analyzed. In minimal medium at pH 5.0, the SPI-2 T3SS is activated and secretes high levels of translocon proteins and very low levels of effectors (3, 4). Under these conditions, secretion of the translocon protein SseB and SseJ-2HA from the mutant strain was undetectable, and this phenotype was complemented by introduction of a plasmid expressing wild-type SsaQ (Fig. 1A).
FIGURE 1.
Analysis of ssaQ gene products and their function. A, essential role of SsaQ in secretion of SseB and SseJ. Bacterial strains expressing SseJ-2HA from the chromosome were grown in MgM/MES at pH 5.0 for 5 h, and whole bacterial cell lysates and secreted fractions were subjected to immunoblot analysis. The intrabacterial protein DnaK was used as a control. B, neither SsaQL-HA nor SsaQS-HA was secreted or presented on the bacterial surface. Plasmid pssaQ-HA was transformed into the ssaQ mutant (HH225) for analysis. Bacterial surface proteins were extracted with n-hexadecane. C, the promoter upstream of ssaM is the only one driving expression of the ssaMVNOPQ operon. The schematic illustrates the plasmids used in the transcriptional assay. The confocal micrographs show HeLa cells infected with the indicated bacterial strains for 5 h. Green, GFP; red, CSA-1-labeled bacteria. Scale bar = 5 μm. D, SsaQS-HA is a translated product rather than a cleavage product of SsaQL-HA. The ssaQ mutant (HH225) was transformed with different plasmids and grown in MgM/MES at pH 5.0 for 5 h, and whole bacterial cell lysates were subjected to immunoblot analysis. E, SsaQL, but not SsaQS, is essential for secretion of SseB and SseJ. The ssaQ mutant expressing SseJ-2HA from chromosomal DNA (HH226) was transformed with the indicated plasmids and used for analysis.
As well as being a component of the C-ring, Spa33 has been proposed to be a secreted substrate of the T3SS (31). To determine whether SsaQ is secreted, a plasmid expressing C-terminally HA-tagged SsaQ (SsaQ-HA) was introduced into the ssaQ deletion mutant. SsaQ-HA was functional, as shown by its ability to restore SseB secretion in minimal medium at pH 5.0 (Fig. 1B). However, under these conditions, SsaQ-HA was not detected in culture supernatants or on the bacterial cell surface (Fig. 1B). Interestingly, two proteins were detected in the bacterial lysate with an anti-HA antibody. One corresponds to the expected mass of SsaQ (36 kDa), and the other is ∼12 kDa. The small protein was not due to the overexpression of SsaQ-HA from the plasmid, as it was also detected when SsaQ-HA was expressed from chromosomal DNA (data not shown). To establish the identity of the small protein, it was gel-purified and subjected to N-terminal sequencing. This revealed that it comprises the C-terminal 106 amino acids of SsaQ, with Met-217 of SsaQ at its N terminus. The full-length protein was designated SsaQL, and the smaller protein was designated SsaQS.
The expression of ssaQ is controlled by a promoter of the 5.4-kb ssaMVNOPQ operon (13, 14). To determine whether SsaQS is derived from an independent transcript, DNA fragments either including the 102-bp promoter sequence upstream of ssaM or beginning from the start codon of ssaM to the putative start codon of ssaQS were ligated into a gfp reporter plasmid to generate plasmids p102ssaMQLN217 and pssaMQLN217 respectively (Fig. 1C). The plasmids were transformed into sv. Typhimurium strains, which were then used to infect HeLa cells for 5 h. There was no detectable GFP fluorescence in bacterial cells carrying the plasmid lacking the 102-bp promoter sequence. GFP production was observed in >90% of bacterial cells containing the 102-bp promoter sequence and was completely dependent on the SPI-2 two-component regulatory system SsrA-B (Fig. 1C) (32). These results indicate that both SsaQL and SsaQS are translated from the ssaMVNOPQ transcript.
To determine whether SsaQS is a cleavage product of SsaQL or is translated separately, ssaQ-HA was subjected to site-directed mutagenesis, and the effects were examined following production of mutant proteins from a plasmid in an sv. Typhimurium ssaQ null mutant strain grown in minimal medium at pH 5.0. If SsaQS is the product of cleavage of SsaQL, then it should not be produced if SsaQL is truncated before Met-217. Therefore, the codon for Tyr-206 of SsaQL-HA was replaced with a stop codon (ochre mutation, TAA). As expected, SsaQL-HA was not detected upon expression of SsaQY206oc-HA, but SsaQS-HA was still produced by this mutant (Fig. 1D). Substituting the codon for Met-217 (the putative start codon for SsaQS) with a leucine codon (SsaQM217L) resulted in an absence of SsaQS-HA but not SsaQL-HA (Fig. 1D). These results show that SsaQS is not a product of cleavage of SsaQL and that SsaQL and SsaQS are translated independently from the same mRNA species. Consistent with this, there is a purine-rich sequence upstream of the start codon of ssaQS (AGAGGATAACACGATG), which is likely to be the Shine-Dalgarno sequence for translational initiation (33).
We next used these strains to examine the involvement of SsaQL and SsaQS in SPI-2 T3SS function. In the absence of SsaQL (the ssaQ null mutant strain expressing SsaQY206oc-HA from a plasmid), there was no detectable secretion in minimal medium at pH 5.0 of either the translocon protein SseB or the effector SseJ (Fig. 1E). However, the absence of SsaQS (the ssaQ null mutant strain expressing SsaQM217L-HA from a plasmid) did not noticeably affect secretion of SseB or SseJ at pH 5.0 (Fig. 1E). This result indicates that SsaQM217L-HA is functional and that SsaQL is essential for the SPI-2 T3SS, whereas SsaQS is not.
SsaQS Is Required for Efficient Secretion of Translocon and Effector Proteins
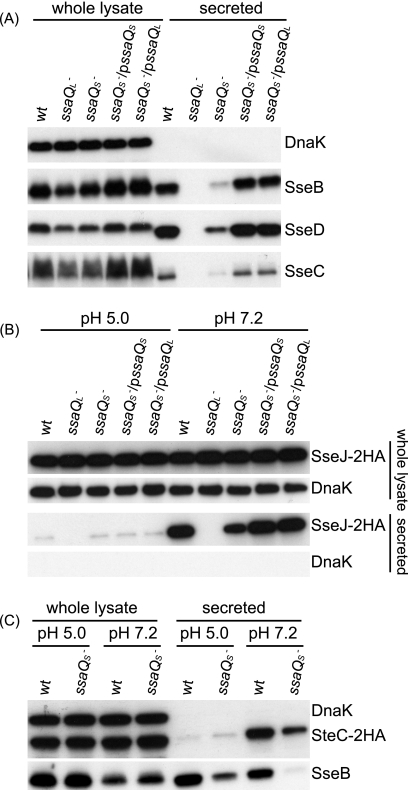
To further investigate the possible function of SsaQS, the point mutation for M217L was introduced into the chromosomal ssaQ gene to create a mutant in which SsaQL is produced as a result of expression from chromosomal DNA but in which SsaQS is lacking. In minimal medium at pH 5.0, the mutant displayed highly reduced secretion of all three translocon proteins: SseB, SseC, and SseD (Fig. 2A). The decreased secretion of these proteins in the ssaQS mutant was restored to wild-type levels by introduction of a plasmid expressing either ssaQS or ssaQL (Fig. 2A). The ability of overexpressed SsaQL to compensate for loss of SsaQS was also evident when ssaQL was overexpressed in the ssaQ mutant (Fig. 1E). These results indicate that SsaQS is required for the efficient secretion of translocon proteins and that overexpression of SsaQL can compensate for the loss of SsaQS.
FIGURE 2.
Role of SsaQS in secretion of translocon and effector proteins. The ssaQ gene of the wild-type strain of sv. Typhimurium was replaced with ssaQY206oc or ssaQM217L to create the ssaQL (HH229) or ssaQS (HH228) mutant, respectively, and used for secretion assays. A, secretion of translocon proteins. Bacterial strains were grown for 5 h in MgM/MES at pH 5.0 for analysis. B, secretion of effector SseJ-2HA upon pH shift. Strains expressing SseJ-2HA from chromosomal DNA were grown for 4 h in MgM/MES at pH 5.0 to activate SPI-2 T3SS and then changed to MgM/MES at the indicated pH and incubated for another 1.5 h. C, secretion of effector SteC-2HA upon pH shift. Strains carrying plasmid psteC-2HA were used for pH shift analysis. Secretion of translocon protein SseB was used as an additional control.
To examine the potential role of SsaQS in secretion of effector proteins, the secretion of epitope-tagged SseJ-2HA was investigated following shift of ambient pH from 5.0 to 7.2, which stimulates effector secretion from the SPI-2 T3SS (4). Bacterial strains were first grown in minimal medium at pH 5.0 for 4 h to activate the T3SS and then incubated in the same medium at pH 5.0 or 7.2 for 1.5 h. Secreted fractions and whole cell lysates were subjected to SDS-PAGE and immunoblotting. In response to the pH shift, the wild-type and ssaQS mutant strains complemented with either ssaQL or ssaQS displayed greatly enhanced and similar levels of SseJ-2HA secretion, respectively (Fig. 2B). In contrast, quantification of the amount of SseJ-2HA secreted by the ssaQS mutant showed that it was ∼44% of the wild-type level (Fig. 2B). Mutation of ssaQS caused a similar reduction in the secreted levels of another effector, SteC-2HA (Fig. 2C). These findings indicate that the translocon-to-effector switch mediated by the SsaL-SpiC-SsaM complex in response to pH shift (4) still occurs in the absence of SsaQS, but the overall efficiency of secretion through the SPI-2 T3SS is reduced in vitro.
SsaQS Is Required for Efficient Translocation of Effectors
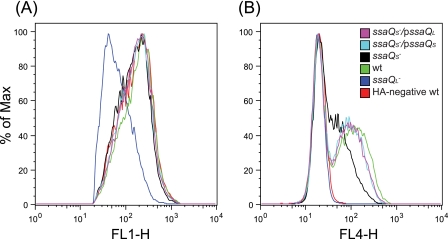
We next examined whether SsaQS contributes to effector translocation in infected cells. To do this, HeLa cells were infected for 13 h with different strains expressing the epitope-tagged effector SseJ-2HA from the bacterial chromosome, and the levels of translocation were quantified by flow cytometry. The ssaQL mutant was unable to translocate SseJ-2HA. The levels of translocated SseJ-2HA from the ssaQS mutant bearing the complementing plasmid pssaQS or pssaQL were similar to those from the wild-type strain, but the ssaQS mutant translocated noticeably less SseJ-2HA (Fig. 3). Quantification of the amount of SseJ-2HA translocated by the ssaQS mutant showed that it was ∼50% of the wild-type level. We concluded that SsaQS is required for efficient translocation of SPI-2 T3SS effectors in infected host cells.
FIGURE 3.
Flow cytometric analysis of translocation of SseJ-2HA. HeLa cells were infected for 13 h with bacterial strains expressing SseJ-2HA from chromosomal DNA except for the HA-negative wild-type strain and labeled with antibodies for flow cytometric assay. A, intracellular bacteria were detected with anti-Salmonella antibody (FL1). B, translocated SseJ-2HA was detected with anti-HA antibody (FL4).
SsaQS Interacts with and Stabilizes SsaQL
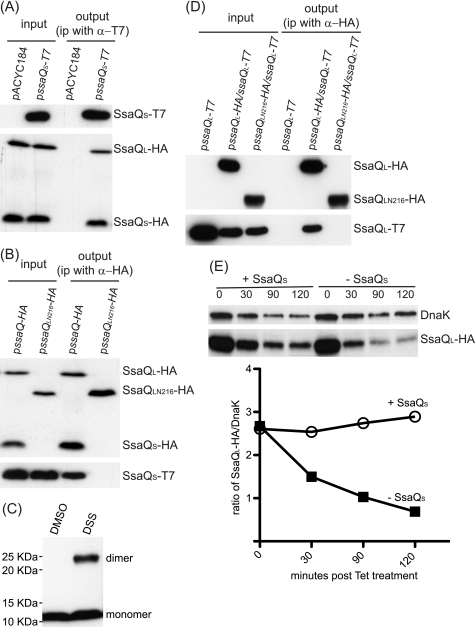
The observation that overexpression of SsaQL compensates for the loss of SsaQS suggested that SsaQS might function as a chaperone, stabilizing SsaQL. To test if SsaQS interacts with SsaQL, the ssaQ mutant strain containing plasmid pssaQ-HA (which produces both SsaQL-HA and SsaQS-HA) and either pssaQS-T7 or the empty vector (pACYC184) was grown in minimal medium at pH 5.0. Lysates were incubated with an antibody against T7 to immunoprecipitate SsaQS-T7. SsaQL-HA and SsaQS-HA were not co-immunoprecipitated in the negative control strain containing plasmids pssaQ-HA and pACYC184. However, both SsaQL-HA and SsaQS-HA were co-immunoprecipitated from the strain containing pssaQ-HA and pssaQS-T7 (Fig. 4A). This result suggested that SsaQS interacts with the C-terminal domain (SsaQS region) of SsaQL. To test this, a plasmid expressing the N-terminal 216 amino acids of SsaQL (pssaQLN216-HA) was transformed into the ssaQ mutant containing plasmid pssaQS-T7, and following growth in minimal medium at pH 5.0, bacterial lysates were subjected to immunoprecipitation with an antibody against the HA tag. As a positive control, this antibody was shown to co-immunoprecipitate SsaQS-T7 in the strain containing plasmids pssaQ-HA and pssaQS-T7 (Fig. 4B). However, the antibody did not immunoprecipitate SsaQS-T7 in the ssaQ mutant containing pssaQLN216-HA and pssaQS-T7 (Fig. 4B). In addition, a cross-linking experiment showed that SsaQS homodimerizes following its production in E. coli (Fig. 4C). These experiments provided evidence that SsaQS interacts with the corresponding region of SsaQL. Next, we tested if SsaQL oligomerizes in the absence of SsaQS. To do this, plasmids expressing SsaQL-T7 and either SsaQL-HA or SsaQLN216-HA were transformed into the ssaQ mutant and subjected to immunoprecipitation with the anti-HA antibody. SsaQL-T7 was co-precipitated in the presence of SsaQL-HA but not by the N-terminal 216 amino acids of SsaQL (Fig. 4D). Therefore, the SsaQS region of SsaQL, but not SsaQS itself, is required for SsaQL to oligomerize.
FIGURE 4.
SsaQS functions as a chaperone for SsaQL. A, SsaQS interacts with SsaQL and itself. Bacterial strain ssaQ pssaQ-HA was cotransformed with pssaQS-T7 or vector pACYC184, grown for 5 h in MgM/MES at pH 5.0, and then lysed for co-immunoprecipitation. Membranes were probed with antibodies against T7 to detect SsaQS-T7 or HA to detect SsaQL-HA and SsaQS-HA. B, the C-terminal region of SsaQL is required for interaction with SsaQS. Bacterial strain ssaQ pssaQS-T7 was cotransformed with pssaQ-HA or pssaQLN216-HA and used for co-immunoprecipitation. C, dimerization of SsaQS. E. coli BL21(DE3) cells containing pET-ssaQS were subjected to treatment with the cross-linker disuccinimidyl suberate (DSS) or DMSO and then analyzed by immunoblotting. D, the C-terminal region of SsaQL is required for self-interaction. The ssaQ mutant (HH225) was transformed with the indicated plasmids and used for immunoprecipitating HA-tagged protein. Samples were analyzed by immunoblotting. E, stabilization of SsaQL requires SsaQS. The ssaQ gene in the wild-type strain was replaced with ssaQM217L-HA to create the ssaQL-HA strain (HH232) and transformed with pssaQS or vector pWSK29 for analysis. Transformants were cultured for 4 h in MgM/MES at pH 5.0, and tetracycline was added to stop protein synthesis. Samples were taken at the indicated time points for analysis. The band intensities were measured with Image J software and normalized with DnaK to construct the stability curve of SsaQL-HA.
To assess the stability of SsaQL in the absence of SsaQS, a bacterial strain was constructed in which the chromosomal ssaQ gene was modified to express SsaQM217L-HA but not SsaQS. This strain was then transformed with pssaQS or the empty vector (pWSK29). The transformants were grown in minimal medium at pH 5.0 to express SsaQL-HA and other SPI-2 proteins. Tetracycline was then added to the growth medium to stop protein synthesis, and samples were taken at different time points to analyze protein stability by SDS-PAGE and immunoblotting. As shown in Fig. 4E, SsaQL was markedly less stable in the absence of SsaQS.
Structural Predictions
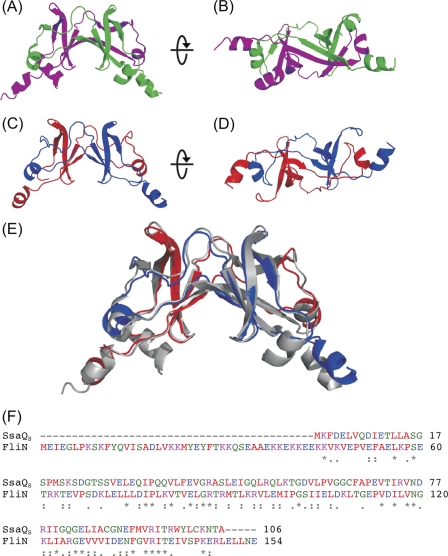
The level of sequence similarity between SsaQS and the flagellar C-ring protein FliN (29) is sufficient for a reliable homology model to be calculated. The template structure of FliN exists as an intimately intertwined dimer that forms a saddle shape (Fig. 5). The homology model of an SsaQS monomer shows the expected extended β-sheet structure and displays significant solvent exposure of hydrophobic residues, suggesting a higher order organization. The ability of SsaQS to oligomerize in solution was confirmed by 1H NMR spectroscopy (data not shown), in which concentration-independent line-widths of SsaQS were consistent with a molecular species between ∼20 and ∼40 kDa. These data indicate that SsaQS probably exists as the intertwined dimer as observed in the crystal structure of FliN.
FIGURE 5.
Alignment and three-dimensional structure of SsaQS and FliN. A and B, two orthogonal views of the ribbon representation of the flagellar rotor protein FliN from T. maritima (Protein Data Bank code 1YAB). Individual monomers are shown in pink and green. C and D, two orthogonal views of the homology model of the SsaQS dimer. Individual monomers are shown in blue and red. E, superposition of the FliN (gray) and SsaQS (blue/red) dimer structures. F, sequence alignment of SsaQS and FliN.
SsaQS Contributes to sv. Typhimurium Virulence
Because SsaQS is required to stabilize SsaQL and because SsaQL is an essential component of the SPI-2 T3SS, we tested if SsaQS contributes to virulence by CI analysis (30) involving mixed infections of the wild-type and ssaQS mutant strains in the sv. Typhimurium/mouse model of systemic disease. Approximately 5 × 102 colony-forming units of each strain were combined and used to inoculate BALB/c mice by the intraperitoneal route. Infection was allowed to proceed for 4 days, at which time mice were killed, and spleens were homogenized and plated onto rich medium to determine the colony-forming units of each strain. The resulting CI was 0.206 ± 0.133, indicating that the ssaQS mutant strain is attenuated in virulence. The virulence defect was fully complemented by introducing pssaQS into the mutant strain and determining its CI in relation to the wild-type strain (CI 0.910 ± 0.212, p = 0.0013) and partially complemented by introducing pssaQL into the mutant strain (CI of 0.448 ± 0.067, p = 0.0171). This showed that SsaQS contributes to sv. Typhimurium virulence in the mouse model of infection. The partial complementation of virulence by overexpression of SsaQL in the ssaQS mutant suggested either that SsaQS has an additional function or that overexpressing SsaQL somehow affects the virulence of sv. Typhimurium. To test this, we measured the virulence of the ssaQL mutant carrying pssaQL compared with the wild-type strain in mice. The resulting CI was 0.317 ± 0.10, significantly lower than 1 (p = 0.0008) but significantly higher than the CI obtained from the ssaQL mutant strain mixed with the wild-type strain (0.013 ± 0.005, p = 0.0009). This indicates that overexpressing SsaQL in sv. Typhimurium affects its virulence in the mouse model of infection.
DISCUSSION
In this work, we discovered that the ssaQ gene of sv. Typhimurium SPI-2 encodes two proteins: the predicted full-length protein SsaQL, composed of 322 amino acids, and the shorter protein SsaQS, comprising the C-terminal 106 residues of SsaQL. SsaQL is essential for SPI-2 T3SS function, whereas SsaQS stabilizes SsaQL and augments the activity of the T3SS.
Site-directed mutagenesis and promoter analysis revealed that SsaQS is a tandem translated product of ssaQ. Tandem translation or “in-frame initiated translation” often occurs in viruses and bacteriophage, where DNA coding capacity is limited, but has rarely been found in bacteria. The few cases reported to date include the widely conserved translational initiation factor IF2 (34), the chemotactic signaling protein CheA (35), the heat shock protein ClpB (36), and the methylation-dependent endonuclease component McrB from E. coli (37). It is also possible that the flagellar secretion system component FliO from Salmonella is tandemly translated, but this has not yet been shown in wild-type cells (38). To our knowledge, SsaQS represents the first example of a bacterial virulence factor produced by tandem translation and of a chaperone produced by this process.
The finding that SsaQL is required for secretion of translocon proteins and effectors is not surprising given its sequence similarity to C-ring components of other bacteria. Homologs of SsaQL, including Spa33 from Shigella and YscQ from Yersinia, are C-ring proteins that interact with components of the secretion machinery, including the ATPase and its regulators (16, 39), and are necessary for formation of the needle structure of the T3SS (16) and assembly of the associated complex between ATPase and the C-ring (17). We have also observed that SsaQL interacts with the putative SPI-2 T3SS ATPase SsaN.3 Therefore, SsaQL is very likely to be an essential C-ring component of the SPI-2 secretion machinery.
In the flagellar C-ring, FliN interacts with the C-terminal region of FliM (FliM248–334), where the two proteins show sequence similarity (40). A similar situation is found in the related HrcQB and HrcQA proteins of the Pseudomonas syringae pv. phaseolicola T3SS (41). Here, the C-terminal region of the smaller HrcQB protein interacts with the C-terminal region of HrcQA. However, the precise role of HrcQB in the function of the P. syringae T3SS is not clear. Structural studies have revealed that FliN forms a tetramer that links molecules of FliM to create a large repeating structure that comprises the lower region of the flagellar C-ring (42). FliN is thus an integral component of the C-ring. The obvious similarities between SsaQS and FliN (Fig. 5) and our finding that SsaQS interacts with an identical region in a larger protein that is predicted to be part of the SPI-2 T3SS C-ring suggested that SsaQS is also a C-ring protein that could act as a bridge between molecules of SsaQL to form a repeating unit at the bottom of the C-ring (42). Although this remains a possibility, two results suggest otherwise. First, it is clear that the SPI-2 T3SS is partially functional in the absence of SsaQS and that the secretion and translocation defects can be completely overcome in vitro and in infected cells by overexpression of SsaQL (Figs. 2 and 3). This contrasts with the situation in the flagellar system, where FliN has an essential role in C-ring formation and flagellum assembly (43, 44). Second, we have shown that SsaQL can oligomerize in the absence of SsaQS. Instead, our data show that SsaQS has a chaperoning function by binding to its corresponding region within SsaQL, stabilizing the larger protein. Consistent with this, SsaQS has a predicted molecular mass of 11.7 kDa and an acidic pI (4.7), features that typify many chaperones (45). It is possible that SsaQL and SsaQS heterodimerize, in which case SsaQS would assist in the proper folding of SsaQL or prevent self-polymerization or degradation of SsaQL before it docks to the T3SS apparatus. Once there, a conformational change induced by interaction with component(s) of the apparatus might lead to the dissociation of the heterodimer. In another scenario, a tetramer could be constructed from SsaQS homodimers interacting with SsaQL dimers. The fact that the two SsaQ variants are cotranslated from the same transcript would favor the formation of a heterodimer, as the two proximal polypeptides would fold and dimerize together as they simultaneously exited from ribosomes. It is conceivable that, during assembly of higher order structures, SsaQL then displaces SsaQS to construct the mature active SPI-2 T3SS.
To our knowledge, SsaQS is the first example of a chaperone for a C-ring protein. It is interesting to note that, in the T3SSs encoded by Salmonella SPI-1, Shigella, and Yersinia, there is no evidence for the presence for a small C-ring protein that would correspond to FliN or SsaQS, suggesting that the C-rings of these T3SSs could assemble through direct oligomerization of their SsaQL homologs (SpaO, Spa33, and YscQ, respectively). Structural studies on SsaQL are now required to reveal the architecture and to help understand the function of the putative C-ring of the SPI-2 T3SS.
Supplementary Material
This work was supported by grants from the Medical Research Council (United Kingdom) and the Wellcome Trust (to D. W. H.).

The on-line version of this article (available at http://www.jbc.org) contains a supplemental table.
X.-J. Yu, P. J. Simpson, M. Liu, S. Matthews, and D. W. Holden, unpublished data.
- SPI-2
- Salmonella pathogenicity island 2
- T3SS
- type III secretion system
- MgM
- magnesium minimal medium
- CI
- competitive index.
REFERENCES
- 1. Chakravortty D., Rohde M., Jäger L., Deiwick J., Hensel M. (2005) EMBO J. 24, 2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deiwick J., Nikolaus T., Erdogan S., Hensel M. (1999) Mol. Microbiol. 31, 1759–1773 [DOI] [PubMed] [Google Scholar]
- 3. Beuzón C. R., Banks G., Deiwick J., Hensel M., Holden D. W. (1999) Mol. Microbiol. 33, 806–816 [DOI] [PubMed] [Google Scholar]
- 4. Yu X. J., McGourty K., Liu M., Unsworth K. E., Holden D. W. (2010) Science 328, 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mazurkiewicz P., Thomas J., Thompson J. A., Liu M., Arbibe L., Sansonetti P., Holden D. W. (2008) Mol. Microbiol. 67, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McLaughlin L. M., Govoni G. R., Gerke C., Gopinath S., Peng K., Laidlaw G., Chien Y. H., Jeong H. W., Li Z., Brown M. D., Sacks D. B., Monack D. (2009) PLoS Pathog. 5, e1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Worley M. J., Nieman G. S., Geddes K., Heffron F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17915–17920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruiz-Albert J., Yu X. J., Beuzón C. R., Blakey A. N., Galyov E. E., Holden D. W. (2002) Mol. Microbiol. 44, 645–661 [DOI] [PubMed] [Google Scholar]
- 9. Hensel M., Shea J. E., Gleeson C., Jones M. D., Dalton E., Holden D. W. (1995) Science 269, 400–403 [DOI] [PubMed] [Google Scholar]
- 10. Ochman H., Soncini F. C., Solomon F., Groisman E. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7800–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones M. A., Wigley P., Page K. L., Hulme S. D., Barrow P. A. (2001) Infect. Immun. 69, 5471–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bispham J., Tripathi B. N., Watson P. R., Wallis T. S. (2001) Infect. Immun. 69, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walthers D., Carroll R. K., Navarre W. W., Libby S. J., Fang F. C., Kenney L. J. (2007) Mol. Microbiol. 65, 477–493 [DOI] [PubMed] [Google Scholar]
- 14. Tomljenovic-Berube A. M., Mulder D. T., Whiteside M. D., Brinkman F. S., Coombes B. K. (2010) PLoS Genet. 6, e1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pallen M. J., Beatson S. A., Bailey C. M. (2005) BMC Microbiol. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morita-Ishihara T., Ogawa M., Sagara H., Yoshida M., Katayama E., Sasakawa C. (2006) J. Biol. Chem. 281, 599–607 [DOI] [PubMed] [Google Scholar]
- 17. Diepold A., Amstutz M., Abel S., Sorg I., Jenal U., Cornelis G. R. (2010) EMBO J. 29, 1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Donnenberg M. S., Kaper J. B. (1991) Infect. Immun. 59, 4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uzzau S., Figueroa-Bossi N., Rubino S., Bossi L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu X. J., Liu M., Holden D. W. (2004) Mol. Microbiol. 54, 604–619 [DOI] [PubMed] [Google Scholar]
- 22. Davis R. H., Botstein D., Roth J. R. (1980) Advanced Bacterial Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23. Wang R. F., Kushner S. R. (1991) Gene 100, 195–199 [PubMed] [Google Scholar]
- 24. Chang A. C., Cohen S. N. (1978) J. Bacteriol. 134, 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valdivia R. H., Falkow S. (1996) Mol. Microbiol. 22, 367–378 [DOI] [PubMed] [Google Scholar]
- 26. Poh J., Odendall C., Spanos A., Boyle C., Liu M., Freemont P., Holden D. W. (2008) Cell. Microbiol. 10, 20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beuzón C. R., Méresse S., Unsworth K. E., Ruíz-Albert J., Garvis S., Waterman S. R., Ryder T. A., Boucrot E., Holden D. W. (2000) EMBO J. 19, 3235–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 29. Brown P. N., Mathews M. A., Joss L. A., Hill C. P., Blair D. F. (2005) J. Bacteriol. 187, 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beuzón C. R., Unsworth K. E., Holden D. W. (2001) Infect Immun. 69, 7254–7261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schuch R., Maurelli A. T. (2001) Infect. Immun. 69, 2180–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garmendia J., Beuzón C. R., Ruiz-Albert J., Holden D. W. (2003) Microbiology 149, 2385–2396 [DOI] [PubMed] [Google Scholar]
- 33. McCarthy J. E., Brimacombe R. (1994) Trends Genet. 10, 402–407 [DOI] [PubMed] [Google Scholar]
- 34. Plumbridge J. A., Deville F., Sacerdot C., Petersen H. U., Cenatiempo Y., Cozzone A., Grunberg-Manago M., Hershey J. W. (1985) EMBO J. 4, 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith R. A., Parkinson J. S. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 5370–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park S. K., Kim K. I., Woo K. M., Seol J. H., Tanaka K., Ichihara A., Ha D. B., Chung C. H. (1993) J. Biol. Chem. 268, 20170–20174 [PubMed] [Google Scholar]
- 37. Ross T. K., Achberger E. C., Braymer H. D. (1989) J. Bacteriol. 171, 1974–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schoenhals G. J., Kihara M., Macnab R. M. (1998) J. Bacteriol. 180, 2936–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jackson M. W., Plano G. V. (2000) FEMS Microbiol. Lett. 186, 85–90 [DOI] [PubMed] [Google Scholar]
- 40. Mathews M. A., Tang H. L., Blair D. F. (1998) J. Bacteriol. 180, 5580–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fadouloglou V. E., Tampakaki A. P., Glykos N. M., Bastaki M. N., Hadden J. M., Phillips S. E., Panopoulos N. J., Kokkinidis M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sarkar M. K., Paul K., Blair D. F. (2010) J. Biol. Chem. 285, 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao R., Pathak N., Jaffe H., Reese T. S., Khan S. (1996) J. Mol. Biol. 261, 195–208 [DOI] [PubMed] [Google Scholar]
- 44. Minamino T., Macnab R. M. (1999) J. Bacteriol. 181, 1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bennett J. C., Hughes C. (2000) Trends Microbiol. 8, 202–204 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.