Abstract
The mammalian target of rapamycin (mTOR) is essential for skeletal myogenesis through controlling distinct cellular pathways. The importance of the canonical mTOR complex 1 signaling components, including raptor, S6K1, and Rheb, had been suggested in muscle maintenance, growth, and metabolism. However, the role of those components in myogenic differentiation is not entirely clear. In this study we have investigated the functions of raptor, S6K1, and Rheb in the differentiation of C2C12 mouse myoblasts. We find that although mTOR knockdown severely impairs myogenic differentiation as expected, the knockdown of raptor, as well as Rheb, enhances differentiation. Consistent with a negative role for these proteins in myogenesis, overexpression of raptor or Rheb inhibits C2C12 differentiation. On the other hand, neither knockdown nor overexpression of S6K1 has any effect. Moreover, the enhanced differentiation elicited by raptor or Rheb knockdown is accompanied by increased Akt activation, elevated IRS1 protein levels, and decreased Ser-307 (human Ser-312) phosphorylation on IRS1. Finally, IRS1 knockdown eliminated the enhancement in differentiation elicited by raptor or Rheb knockdown, suggesting that IRS1 is a critical mediator of the myogenic functions of raptor and Rheb. In conclusion, the Rheb-mTOR/raptor pathway negatively regulates myogenic differentiation by suppressing IRS1-PI3K-Akt signaling. These findings underscore the versatility of mTOR signaling in biological regulations and implicate the existence of novel mTOR complexes and/or signaling mechanism in skeletal myogenesis.
Keywords: Akt PKB, Cell Differentiation, Insulin-like Growth Factor (IGF), mTOR, Signal Transduction, Skeletal Muscle, Insulin Receptor Substrate (IRS)
Introduction
During embryonic skeletal myogenesis, cells in somites commit to myogenic lineage and become myoblasts, which differentiate and fuse to form multinucleated myofibers (1). This is a highly coordinated process where various environmental cues and signaling pathways integrate to regulate the formation of skeletal muscle (2, 3). This process is largely recapitulated by the in vitro differentiation of myoblasts, such as the C2C12 mouse satellite cell line. Upon growth factor withdrawal, these cells produce insulin-like growth factor II (IGF-II),2 which in an autocrine fashion stimulates myogenic differentiation (4). One of the critical pathways downstream of myogenic IGF signaling is the PI3K-Akt pathway (5, 6), and insulin receptor substrate 1 (IRS1) is a well-established mediator of IGF receptor activation of downstream signaling (7).
mTOR, the mammalian target of rapamycin, has long been recognized as a nutrient- and energy-sensing signaling hub regulating a wide spectrum of cellular processes including proliferation, growth, survival, differentiation, and metabolism (8). It nucleates two distinct biochemical complexes: the raptor-associated mTORC1 is acutely sensitive to rapamycin, and it targets S6K1 and 4E-BP1 to regulate translation initiation, among other functions; the rictor-associated mTORC2 is a kinase for the multifunctional kinase Akt and it also regulates cytoskeleton reorganization (9). Although mTORC2 was initially characterized as the rapamycin-insensitive complex, prolonged rapamycin treatment inhibits mTORC2 in some cellular contexts (10).
Accumulative evidence has revealed the mTOR role as a master regulator of skeletal myogenesis. mTOR regulates the initiation of myoblast differentiation in vitro and early stage of muscle regeneration in vivo by controlling the production of IGF-II at two levels, transcriptional regulation through a muscle-specific enhancer (11, 12) and post-transcriptional regulation through the microRNA miR-125 (13). Myocyte fusion is also regulated by mTOR via a MyoD/microRNA-1/HDAC4/Follistatin pathway (14). Interestingly, mTOR utilizes distinct mechanisms in regulating different stages of myogenesis: the kinase activity of mTOR is entirely dispensable for the early stage of myogenesis and IGF-II production (11–13), whereas myotube and myofiber maturation, involving late-stage fusion, requires mTOR kinase activity (14, 15).
Whereas all the myogenic functions of mTOR mentioned above are sensitive to rapamycin, it is not clear whether mTORC1 mediates those functions. Efforts to directly examine the mTOR role in vivo with skeletal muscle-specific gene knock-out have been invaluable in determining the physiological functions of mTOR and its binding proteins. Consistent with their essential roles in muscle development, muscles depleted of either mTOR (16) or raptor (17) display severe muscular dystrophy during postnatal development. Rictor, on the other hand, appears to be dispensable for muscle development as no defect is detectable in rictor-depleted muscles (17). However, it is important to note that all the aforementioned gene deletions are induced by Cre recombinase expression driven by the human skeletal actin (HSA) promoter, which is only active in differentiated myofibers and not in satellite cells (18). Thus, any role of mTOR, raptor, or rictor in early stages of myogenesis, in particular initiation of differentiation, could conceivably have been missed in those investigations. Systemic s6k1 deletion leads to skeletal muscle atrophy, but the s6k1−/− myofibers, albeit smaller than wt myofibers in diameter, contain normal myonuclei numbers (19), suggesting a lack of myogenic defect.
The depletion of raptor by RNAi in human skeletal myoblasts has been reported to have little effect on differentiation although it augments the inhibitory effect of myostatin (20). In C2C12 myoblasts, rictor and mTORC2, rather mTORC1, have been suggested to be the mediator of rapamycin inhibition of differentiation (21). To further probe into the role of mTORC1 in myogenic differentiation, we investigated the effects of manipulating protein levels of the canonical mTORC1 signaling components in C2C12 cells. We have found that the rapamycin-sensitive mTOR function in myogenic differentiation is independent of the canonical mTORC1. Instead, raptor, along with the activator of mTORC1, Rheb, negatively regulates myoblast differentiation through suppression of IRS1 and inhibition of Akt activation.
EXPERIMENTAL PROCEDURES
Antibodies and Other Reagents
Anti-MHC (MF20) and anti-myogenin (F5D) were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health and maintained by The University of Iowa, Department of Biological Sciences. Anti-tubulin was from Abcam. All other primary antibodies were from Cell Signaling Technology. All secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc. Rapamycin was from LC labs. Gelatin was from Sigma-Aldrich.
Plasmids
pRK5-HA-raptor was obtained from Addgene (22). pRK7-Flag-Rheb was a generous gift from the Blenis laboratory (23).
Cell Culture and Transfection
C2C12 myoblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 1 g/liter glucose with 10% fetal bovine serum at 37 °C with 7.5% CO2. Transfection of myoblasts was performed using TransIT-LT1 (Mirus) following the manufacturer's recommendations. To induce differentiation, cells were plated on tissue culture plates coated with 0.2% gelatin and grown to 100% confluence before switching to differentiation medium (DMEM containing 2% horse serum). The cells were replenished with fresh differentiation medium daily for 3 days.
Western Blotting
Cells were lysed in 50 mm Tris-HCl, pH 7.2, 150 mm NaCl, 1% Nonidet P-40, and 1% protease inhibitor mixture (Sigma). The lysates were cleared by micro-centrifugation at 14,000 rpm, and then mixed with 2× SDS sample buffer. Proteins were resolved by SDS-PAGE and transferred onto PVDF membranes (Millipore), which were then incubated with various antibodies following the manufacturer's recommendations. Detection of horseradish peroxidase-conjugated secondary antibodies was performed with Western LightningTM Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences, Inc.), and images were developed on x-ray films. Quantification of Western band intensities was performed by densitometry using the software Image J.
Immunofluorescence Microscopy and Quantitative Analysis of Myocytes
C2C12 cells differentiated in 12-well plates were fixed and stained for MHC and DAPI as previously described (15). The stained cells were examined with a Leica DMI 4000B fluorescence microscope, and the fluorescent images were captured using a RETIGA EXi camera, and analyzed with Q-capture Pro51 software (Q-ImagingTM). The differentiation and fusion indexes were calculated as the percentage of nuclei in MHC-positive myocytes and in myotubes with ≥2 nuclei, respectively. Each data point was generated from at least 200 randomly chosen MHC-positive cells or myotubes.
Lentivirus-mediated RNAi
shRNAs in the pLKO.1-puro vector for knocking down were purchased from Sigma-Aldrich (MISSION® TRC). Lentivirus packaging and testing were performed as previously described (24). The Sigma clone ID for the shRNA constructs used in this study are: mTOR #1, NM_020009.1–7569s1c1; mTOR #2, NM_020009.1–5493s1c1; rictor #1, NM_030168.2–6240s1c1, rictor #2, NM_030168.2–5030s1c1; Rheb #1, NM_053075 .2–740s1c1, Rheb #2, NM_053075.2–339s1c1; IRS1 #1, NM_010570.4–2308s21c1, IRS1 #2, NM_010570.2–3585s1c1; S6K1 #1, NM_028259.1–264s1c1; S6K1 #2, NM_028259.1–616s1c1; raptor #1, NM_028898.1–3729s1c1. Raptor #2 shRNA was cloned by inserting the following sequence into pLKO.1puro: 5′CCGGG GCTAGTCTGTTTCGAAATTTCTTCCTGTCAAAATTTCGAAACAGACTAGCCTTTTTG3′. C2C12 cells were transduced with lentiviruses in growth medium containing 8 μg/ml polybrene, selected in 3 μg/ml puromycin for 2 days, followed by plating into 12-well plates for differentiation.
Statistical Analysis
All data are presented as mean ± S.D. of at least three sets of independent experiments. Whenever necessary, statistical significance of the data were analyzed by performing one-sample or paired t-tests. The specific types of tests and the p values, when applicable, are indicated in figure legends.
RESULTS
mTOR and Raptor Have Opposite Roles in Regulating Myoblast Differentiation
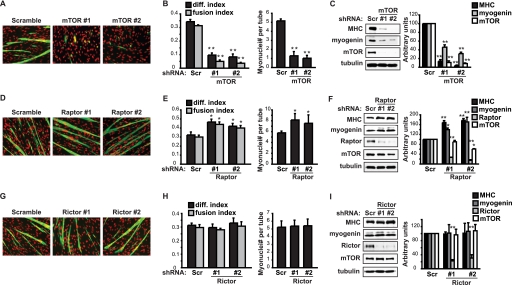
To directly examine the role of mTOR, raptor and rictor in myoblast differentiation, lentivirus-delivered shRNA-mediated knockdown of each of those proteins was carried out in C2C12 myoblasts, which were then induced to differentiate by serum withdrawal. Consistent with the inhibitory effect of rapamycin in myogenesis, mTOR knockdown drastically inhibited C2C12 cell differentiation, as evidenced by both severely impaired myotube formation (Fig. 1, A and B) and blocked expression of the differentiation markers myogenin and myosin heavy chain (MHC) (Fig. 1C) by Day 3 of differentiation induction. Because mTOR is known to regulate cell growth and proliferation, and the capacity of myoblast differentiation is correlated to cell density, we took care to equalize the cell numbers between mTOR knockdown samples and control samples expressing a scrambled hairpin sequence at the initiation of differentiation (Day 0). The cell numbers across samples remained similar on Day 3 of differentiation (see Fig. 1A, DAPI stain). Hence, the differentiation defect in mTOR knockdown cells is unlikely to be a consequence of growth defect.
FIGURE 1.
mTOR and raptor have opposite roles in myoblast differentiation. C2C12 myoblasts were transduced overnight with lentiviruses expressing shRNAs for mTOR (A–C), raptor (D–F), or rictor (G–I) (Scramble or Scr as a non-targeting control), and subjected to puromycin selection for 2 days followed by differentiation for 3 days. A, D, G, differentiated myocytes were stained for MHC (green) and DAPI (red). B, E, H, myocytes were quantified for differentiation index, fusion index, and average myotube size (myonuclei number per myotube). C, F, I, cells were lysed for Western analyses, and band intensities were quantified by densitometry and normalized to tubulin control. All data shown are mean ± S.D. (n = 3 for each condition). For B, E, H, paired t test was performed to compare each data to Scramble control. For C, F, I, one-sample t test was performed to compare each data to Scramble control. *, p < 0.05; **, p < 0.01.
Surprisingly, raptor knockdown led to enhanced differentiation (Fig. 1D), with elevated differentiation and fusion indexes, as well as a larger average size of myotubes (Fig. 1E). Myogenin and MHC expression were also increased by raptor knockdown (Fig. 1F). A modest decrease of mTOR levels was observed in raptor knockdown cells (Fig. 1F), consistent with the proposed inter-dependence of protein stability between mTOR and raptor (22). The positive effect of raptor on differentiation was evident despite the partial loss of mTOR. These observations suggest that the rapamycin-sensitive function of mTOR in myogenic differentiation is independent of the raptor-defined mTORC1. Instead, raptor appears to have a negative role in myogenesis.
Rictor knockdown, on the other hand, did not affect myoblast differentiation (Fig. 1, G–I). The efficiency of rictor knockdown was high and comparable to that of raptor knockdown (Fig. 1I), but we cannot formally rule out the possibility that the residual rictor protein is functionally sufficient. In all the knockdown experiments two independent shRNAs were used for each gene, yielding consistent results, thus ruling out off-target effects.
Raptor and Rheb Negatively Regulate Myoblast Differentiation
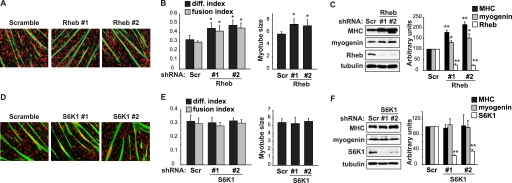
To probe into the potentially negative role of raptor, we considered the other components in the canonical mTORC1 pathway. To that end, we knocked down the activator of mTORC1, Rheb (9), and examined its effect on myoblast differentiation. Rheb-depleted cells differentiated more robustly, with higher differentiation and fusion indexes, as well as increased average myotube size, compared with control cells (Fig. 2, A and B). The expression of myogenin and MHC was also elevated (Fig. 2C). Rheb and raptor, therefore, might reside in the same pathway as negative regulators of myogenesis, in contrast to the mTOR positive role.
FIGURE 2.
Rheb, but not S6K1, negatively regulates myoblast differentiation. C2C12 myoblasts were transduced overnight with lentiviruses expressing shRNAs for Rheb (A–C) or S6K1 (D–F) (Scramble or Scr as a non-targeting control), and subjected to puromycin selection for 2 days followed by differentiation for 3 days. A and D, differentiated myocytes were stained for MHC (green) and DAPI (red). B and E, myocytes were quantified for differentiation index, fusion index, and average myotube size (myonuclei number per myotube). C and F, cells were lysed for Western analyses, and band intensities were quantified by densitometry and normalized to tubulin control. All data shown are mean ± S.D. (n = 3 for each condition). For B & E, paired t test was performed to compare each data to Scramble control. For C and F, one-sample t test was performed to compare each data to Scramble control. *, p < 0.05; **, p < 0.01.
On the other hand, knockdown of S6K1, the immediate downstream effector of mTORC1, did not affect differentiation (Fig. 2, D–F). This is consistent with our previous conclusion that S6K1 is dispensable for myoblast differentiation (25), and also with the observation that s6k1−/− mice form normal numbers of myofibers with wild-type myonuclei number (19). However, we did not observe smaller diameter of S6K1-depleted myotubes (Fig. 2D), in contrast to the observations that s6k1−/− myofibers are smaller during both development (19) and regeneration (12). While we cannot rule out the possibility that residual S6K1 in the knockdown cells is fully functional, given the high efficiency of knockdown (Fig. 2F), it is more likely that the in vitro culture system does not have the resolution to reveal a modest myotube growth defect.
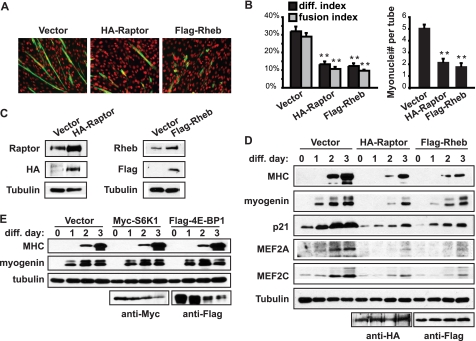
To further confirm the function of raptor and Rheb, recombinant HA-raptor or Flag-Rheb was overexpressed in C2C12 myoblasts. Consistent with their negative roles in myogenesis, both raptor and Rheb overexpression impaired myotube formation (Fig. 3A), as quantified by differentiation index, fusion index, and average size of myotubes (Fig. 3B). Stable expression of the recombinant protein resulted in a total protein level that was at least 2-fold of that of endogenous protein, for both raptor and Rheb (Fig. 3C). Furthermore, the expression of an array of myogenic markers was found to be decreased by raptor and Rheb expression (Fig. 3D). At the same time, overexpression of S6K1, or 4E-BP1 (another target of mTORC1 in cell growth regulation), did not have any effect on myogenic differentiation (Fig. 3E). These data are not only consistent with those two mTORC1 effectors being dispensable, but also help exclude nonspecific effects of protein overexpression on differentiation. Taken together, our results have revealed a negative regulation of myogenic differentiation by the canonical mTORC1 signaling components raptor and Rheb.
FIGURE 3.
Raptor and Rheb negatively regulate myoblast differentiation. A, C2C12 myoblasts were transfected with HA-raptor or Flag-Rheb together with pCDNA3 (vector), selected with G418 for 2 days, and then induced to differentiate for 3 days. Cells were fixed and stained for MHC (green) and DAPI (red). B, myocytes in A were quantified for differentiation index, fusion index, and average myotube size (myonuclei number per myotube). C, cells were transfected as in A, selected with G418 for 10 days to establish stably transfected pools. Expression of recombinant raptor and Rheb was assessed by Western blotting. D, stably transfected cells were induced to differentiate for 3 days, and lysed for Western blotting at the indicated time points. E, myoblasts were transfected with Myc-S6K1 or Flag-4E-BP1, selected with G418 for 2 days, and then induced to differentiate for 3 days. Differentiated myocytes were lysed for Western blotting. Tubulin served as a loading control. Each experiment was repeated at least three times with representative blots shown or mean ± S.D. For data in B, paired t test was performed to compare each data to vector control. **, p < 0.01.
mTOR/Raptor Negatively Regulates Akt Activation
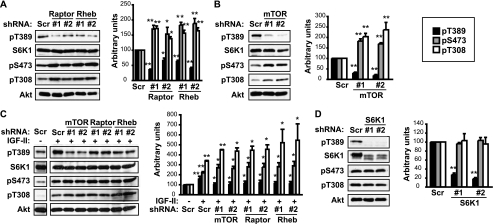
To decipher the mechanism underlying the negative roles of raptor and Rheb in myogenic mTOR signaling, we considered the well-established feedback inhibition of PI3K/Akt signaling by mTORC1 activation in a variety of cellular contexts (26). Indeed, raptor and Rheb knockdown each increased Akt phosphorylation on both Ser-473 and Thr-308, to statistically significant degrees (Fig. 4A). Enhanced Akt phosphorylation was also seen in raptor-deleted mouse skeletal muscles in vivo (17), which accompanied muscular dystrophy, contrary to the enhanced differentiation we observed here. As expected, S6K1 phosphorylation on Thr389 was dampened by the knockdown of either protein (Fig. 4A). Interestingly, mTOR knockdown also led to increased Akt phosphorylation (Fig. 4B), as was reported for in mTOR-depleted muscle in vivo (16). This is consistent with a negative feedback regulation of Akt by mTOR/raptor and at the same time suggests that mTOR might not be the main kinase for Akt in muscle cells. Indeed, mTORC2-independent phosphorylation of Akt has been reported in mouse skeletal muscles (17).
FIGURE 4.
mTOR/raptor negatively regulates Akt activation. C2C12 myoblasts were transduced overnight with lentiviruses expressing shRNAs for raptor or Rheb (A), mTOR (B), or S6K1 (D) (Scr as a non-targeting control), and subjected to puromycin selection for 2 days followed by differentiation for 3 days and lysis for Western blotting. C, cells were transduced with lentiviruses expressing shRNAs for raptor, Rheb or mTOR, and drug-selected as described above, followed by incubation in differentiation medium for 12 h in the presence of 300 ng/ml IGF-II, and then lysis for Western blotting. The scr/no IGF-II lane was from the same Western blot as the +IGF-II lanes for each protein blotted (the separation of lanes in this figure is due to removal of other lanes irrelevant to the experiment), therefore, the band intensities could be directly compared. Band intensities were quantified and normalized to tubulin control. Black bars: pT389-S6K1; gray bars: pS473-Akt; white bars: pT308-Akt. All data shown are mean ± S.D. (n = 3 for each condition). For A, B, and D, one-sample t test was performed to compare each data to Scr control. For C, the “+IGF-II” scramble samples were compared with –IGF-II scramble control by one-sample t test; the other +IGF-II samples were compared with their corresponding +IGF-II scramble samples by paired t test. *, p < 0.05; **, p < 0.01.
IGF-II is a well-established autocrine factor that promotes differentiation of C2C12 cells (4), and mTOR regulates its expression during initiation of differentiation (11, 13). Therefore, we wondered what the effect of raptor and Rheb knockdown might be in the presence of exogenous IGF-II. To minimize the complication from endogenous IGF-II, we chose a time point before IGF-II expression became significant (11) and supplemented the cell media with recombinant IGF-II during the first 12 h of differentiation. In the presence of exogenous IGF-II, the effects of knocking down mTOR, raptor and Rheb on pThr389-S6K1, pSer473-Akt and pThr308-Akt (Fig. 4C) were very similar to those without exogenous IGF-II (Fig. 4, A and B), although the degree of change varied somewhat. This observation is consistent with IGF-II (via IGF-I receptor) acting upstream of Akt and IRS1, the point of raptor/Rheb action (see below).
We also examined whether S6K1 mediated the regulation of Akt phosphorylation. As shown in Fig. 4D, knockdown of S6K1 did not affect Akt phosphorylation on either Ser-473 or Thr-308. Therefore, enhanced Akt activation in raptor or Rheb-depleted myocytes is likely a direct consequence of mTOR/raptor inactivation, rather than mediated by S6K1.
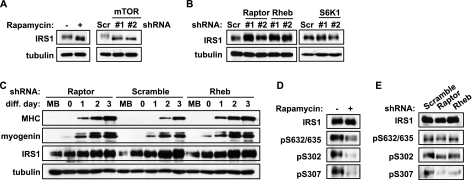
mTOR/raptor Negatively Regulates IRS1 Protein Levels
Because activation of mTORC1 is known to induce feedback inhibition of the insulin/IGF-PI3K-Akt pathway by modulating IRS1 levels (27), we went on to examine IRS1 in C2C12 cells. Acute (30 min) rapamycin treatment in myoblasts led to increased IRS1 protein levels, and so did mTOR knockdown (Fig. 5A), suggesting that mTORC1 activity may indeed negatively regulate the level of IRS1 protein. Raptor and Rheb knockdown also each increased IRS1 levels, whereas S6K1 knockdown had no effect (Fig. 5B). Therefore, a Rheb-mTOR/raptor pathway negatively regulates IRS1 levels in myoblasts independently of S6K1. It should be pointed out that, since mTOR is a regulator of multiple aspects of myogenesis, its inactivation by knockdown or rapamycin inhibits differentiation despite increased IRS1 levels.
FIGURE 5.
mTOR/raptor negatively regulates IRS1 protein levels. A, C2C12 myoblasts were treated with 50 nm rapamycin for 30 min (left panels), or infected overnight with mTOR lentivirus followed by 2-day puromycin selection (right panels), before cell lysis and Western analyses. B, myoblasts were transduced overnight with lentiviruses expressing shRNA for raptor or Rheb (left panels), or S6K1 (right panels), followed by 2-day puromycin selection before cell lysis and Western analyses. C, myoblasts were infected overnight with lentiviruses expressing shRNA for raptor or Rheb followed by 2-day puromycin selection; myoblasts (MB) or time-course differentiating myocytes (diff. day 0–3) were lysed for Western analyses. D, 3-day differentiated myocyte were treated with 50 nm rapamycin for 30 min and lyzed for Western analyses. E, 3-day differentiated myocyte depleted with raptor or Rheb as described in C were lysed for Western blot. Scramble (Scr) was a non-targeting shRNA control. Tubulin served as a loading control. Results were repeated at least three times with representative blots shown.
Interestingly, the IRS1 protein was naturally increased upon myogenic differentiation (Fig. 5C, Scramble), consistent with a positive role this protein plays in myogenesis. Importantly, the elevation of IRS1 in myoblasts upon raptor/Rheb knockdown correlated well with enhanced myogenic marker expression in these cells during the early phase of differentiation (Fig. 5C), suggesting that raptor/Rheb may regulate differentiation through controlling IRS1 levels. Notably, while raptor and Rheb knockdown increased IRS1 levels in myoblasts, in differentiated myotubes the knockdown no longer had any effect on the naturally elevated IRS1 protein level (Fig. 5C). It is thus possible that endogenous raptor and/or Rheb activity is suppressed at the initiation of differentiation to allow IRS1 accumulation. The protein levels of raptor and Rheb were not changed during differentiation, nor was the interaction between raptor and mTOR (data not shown).
We noticed that in addition to IRS1 protein level change, rapamycin and mTOR knockdown down-shifted IRS1 mobility on SDS-PAGE (Fig. 5A), as would be expected when the serine residues on IRS1 were drastically de-phosphorylated. However, raptor or Rheb knockdown increased IRS1 levels without detectable alteration of its mobility (Fig. 5B). This could potentially be explained by the existence of mTOR-dependent (rapamycin-sensitive) inputs other than Rheb/raptor governing IRS1 phosphorylation, but the Rheb/raptor input was apparently sufficient to modulate IRS1 protein levels. We then asked whether Rheb/raptor regulated IRS1 through the reported rapamycin-sensitive serine phosphorylation sites (28–31). Because of the higher IRS1 levels upon differentiation, assessment of IRS1 serine phosphorylation was done in myotubes, which was more reliable than in myoblasts, even though the relative degrees of IRS1 phosphorylation were lower in myotubes. As expected, all three reported mTORC1 sites were sensitive to acute (30 min) rapamycin treatment (Fig. 5D). However, only phosphorylation on Ser-307 (equivalent to Ser-312 of human IRS1) was clearly inhibited by raptor or Rheb knockdown (Fig. 5E). It has been reported that phosphorylation of Ser-312 on human IRS1 is responsible for IRS1 degradation (32). Hence, the raptor/mTOR activity, downstream of Rheb, may modulate IRS1 levels by targeting Ser-307 (human Ser-312) during myogenesis. Taken together, our results strongly suggest that the Rheb-mTOR/raptor pathway negatively regulates IRS1 and Akt signaling in myogenic differentiation.
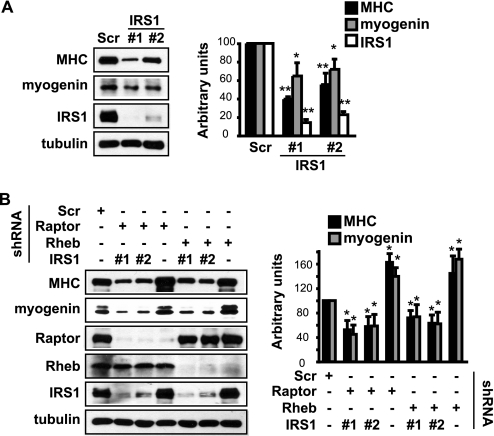
The Function of Raptor and Rheb in Myogenesis Is Mediated by IRS1
Next, we set out to validate the functional significance of Rheb/raptor regulation of IRS1 in myogenesis. Knockdown of IRS1 in C2C12 cells impaired differentiation as evidenced by suppressed myogenin and MHC expression (Fig. 6A), consistent with an indispensable role of IRS1 in muscle differentiation. If the increased IRS1 levels were responsible for the enhanced myogenic differentiation upon Rheb or raptor knockdown, one would expect that simultaneous knockdown of IRS1 and raptor or Rheb would eliminate the positive effect of raptor/Rheb depletion. That was indeed what we observed: IRS1 depletion suppressed the differentiation enhancement elicited by raptor or Rheb depletion, as shown by decreased MHC and myogenin expression in double knockdown cells compared with raptor or Rheb knockdown cells (Fig. 6B). These observations are in full agreement with IRS1 being a critical target in the negative regulation of myogenesis by the Rheb-raptor/mTOR pathway.
FIGURE 6.
The function of raptor and Rheb in myogenesis is mediated by IRS1. A, C2C12 myoblasts were transduced overnight with lentiviruses expressing shRNA for IRS1, and subjected to 2-day puromycin selection followed by 3-day differentiation. Differentiated myocytes were lysed for Western analyses. B, myoblasts were co-infected overnight with lentiviruses expressing shRNAs for IRS1 and raptor or Rheb, and treated as in A. Scramble (Scr) was a non-targeting shRNA control. Tubulin served as a loading control. Western bands were quantified and normalized to tubulin control. All data shown are mean ± S.D. (n = 3 for each condition). One-sample t test was performed to compare each data to Scramble control. *, p < 0.05; **, p < 0.01.
DISCUSSION
Although rapamycin had long been known to inhibit myoblast differentiation (5, 25, 33, 34) and mTOR was established as a master regulator controlling different stages of skeletal myogenesis, the role of the rapamycin-sensitive mTORC1 (with raptor as a defining component) in myogenesis remained to be fully defined. In this study we find that raptor, together with mTOR, as well as the mTORC1 activator Rheb, negatively regulates myoblast differentiation. This is in stark contrast to the reported positive role of raptor in skeletal muscle maintenance and metabolic functions (17). Most likely, the use of the HSA promoter to drive Cre-dependent raptor deletion in that study led to normal myogenic differentiation prior to raptor depletion, thus, an early myogenic function of raptor would have been missed. The only function of Rheb in skeletal muscle reported so far is its role in stimulating muscle hypertrophy when overexpressed (35), analogous to the canonical mTORC1 function in cell growth regulation. The negative regulation of myogenic differentiation by the Rheb-mTOR/raptor pathway suggests a homeostatic role of mTOR, and attests to the versatility of mTOR signaling in biological regulation.
Curiously, two other studies did not find any significant effect of knocking down raptor on myoblast differentiation, one in human myoblasts (20) and one in C2C12 cells (21). This apparent discrepancy with our observation could potentially be explained by differences in the degrees of differentiation in different experimental systems. When myoblasts are maximally differentiated in culture, it may be difficult to observe further increase of differentiation upon knockdown of a negative regulator. During the preparation of this manuscript, Jaafar et al. also reported a negative role of mTORC1 in vasopressin-induced differentiation of rat L6 myoblasts (36), in agreement with our findings in C2C12 cells.
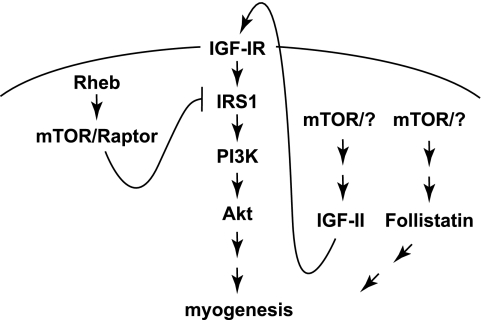
We have shown that the Rheb-mTOR/raptor pathway inhibits IRS1-PI3K-Akt signaling by suppressing IRS1 protein levels, and that the down-regulation of IRS1 is likely responsible for the inhibitory function of Rheb and raptor in myogenesis (Fig. 7). The negative feedback regulation of insulin/IGF signaling by mTORC1 through serine phosphorylation and/or stability of the adaptor protein IRS1 has been widely reported (27, 37, 38), but for the first time this feedback pathway is now found to impact myogenic differentiation. In our current experimental system, IRS1 is believed to mediate autocrine IGF-II signaling to PI3K-Akt (4–7). The functional importance of IRS1 in myogenesis has been confirmed by the smaller myofiber size in IRS1-depleted skeletal muscle (39), and by the effect of IRS1 RNAi on inhibiting myoblast differentiation ((40) and our current study). mTOR/raptor suppresses the protein level of IRS1 in myoblasts, possibly through phosphorylation of IRS1 at Ser-307 (Ser-312 in human IRS1). This suppression is removed upon myogenic differentiation to allow accumulation of IRS1 and facilitation of PI3K-Akt signaling. The mechanism by which the de-repression of IRS1 occurs is not clear, but it is conceivable that down-regulation of mTORC1 activity toward IRS1 may be responsible. The phosphorylation of Ser-307 (human Ser-312) has been shown to be at least partially dependent on S6K1 in some cell types as well as fat tissues (28, 30), but a direct role of S6K1 on this site has not been reported for muscles or myoblasts. Our data suggest that mTORC1 itself, rather than S6K1, may be the kinase for Ser-307 in muscle cells, perhaps similar to Ser-632/635 (human Ser-636/639) (29). The exclusion of S6K1 as the mediator of mTORC1 negative regulation of IRS1 by our data corroborates well with the fact that S6K1 activity increases upon myogenic differentiation (5, 25, 33, 34).
FIGURE 7.
mTOR regulates skeletal myogenesis through multiple pathways. See text for details.
It is intriguing that the negative function of raptor in myogenesis involves both mTOR and Rheb - the canonical mTORC1 well accepted as a positive regulator of cell growth and other cellular functions. The lack of positive contribution from raptor and Rheb to myogenic differentiation also implicates the existence of novel mTOR complexes and/or mechanisms responsible for rapamycin-sensitive functions of mTOR in myogenesis. One possible mechanism was proposed by Shu and Houghton to be through mTORC2 and Akt (21). There are likely other yet-to-be-identified mechanisms. At least two distinct mechanisms underlie known mTOR myogenic signaling, one is dependent on the kinase activity of mTOR and regulates myocyte fusion through a MyoD/microRNA-1/HDAC4/follistatin pathway (14, 15), and the other controls IGF-II production in a kinase-independent manner (11, 13) (Fig. 7). Identification of the putative myogenic mTOR complexes and dissection of signaling mechanisms at or upstream of mTOR are of great interest in our future investigations.
This work was supported, in whole or in part, by Grant AR048914 (to J. C.) from the National Institutes of Health.
- IGF-II
- insulin-like growth factor II
- IRS1
- insulin receptor substrate 1
- mTOR
- mammalian target of rapamycin
- MHC
- myosin heavy chain.
REFERENCES
- 1. Buckingham M. (2001) Curr. Opin. Genet. Dev. 11, 440–448 [DOI] [PubMed] [Google Scholar]
- 2. Naya F. J., Olson E. (1999) Curr Opin Cell Biol. 11, 683–688 [DOI] [PubMed] [Google Scholar]
- 3. Lassar A., Münsterberg A. (1994) Curr Opin Cell Biol. 6, 432–442 [DOI] [PubMed] [Google Scholar]
- 4. Florini J. R., Magri K. A., Ewton D. Z., James P. L., Grindstaff K., Rotwein P. S. (1991) J. Biol. Chem. 266, 15917–15923 [PubMed] [Google Scholar]
- 5. Coolican S. A., Samuel D. S., Ewton D. Z., McWade F. J., Florini J. R. (1997) J. Biol. Chem. 272, 6653–6662 [DOI] [PubMed] [Google Scholar]
- 6. Kaliman P., Canicio J., Shepherd P. R., Beeton C. A., Testar X., Palacín M., Zorzano A. (1998) Mol. Endocrinol. 12, 66–77 [DOI] [PubMed] [Google Scholar]
- 7. Taguchi A., White M. F. (2008) Annu. Rev. Physiol. 70, 191–212 [DOI] [PubMed] [Google Scholar]
- 8. Zoncu R., Efeyan A., Sabatini D. M. (2011) Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laplante M., Sabatini D. M. (2009) J. Cell Sci. 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 11. Erbay E., Park I. H., Nuzzi P. D., Schoenherr C. J., Chen J. (2003) J. Cell Biol. 163, 931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ge Y., Wu A. L., Warnes C., Liu J., Zhang C., Kawasome H., Terada N., Boppart M. D., Schoenherr C. J., Chen J. (2009) Am. J. Physiol. Cell Physiol. 297, C1434–C1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ge Y., Sun Y., Chen J. (2011) J. Cell Biol. 192, 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun Y., Ge Y., Drnevich J., Zhao Y., Band M., Chen J. (2010) J. Cell Biol. 189, 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park I. H., Chen J. (2005) J. Biol. Chem. 280, 32009–32017 [DOI] [PubMed] [Google Scholar]
- 16. Risson V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A., Freyssenet D., Tanti J. F., Le-Marchand-Brustel Y., Ferrier B., Conjard-Duplany A., Romanino K., Bauché S., Hantaï D., Mueller M., Kozma S. C., Thomas G., Rüegg M. A., Ferry A., Pende M., Bigard X., Koulmann N., Schaeffer L., Gangloff Y. G. (2009) J. Cell Biol. 187, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M., Zorzato F., Hall M. N., Rüegg M. A. (2008) Cell Metab. 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 18. Nicole S., Desforges B., Millet G., Lesbordes J., Cifuentes-Diaz C., Vertes D., Cao M. L., De Backer F., Languille L., Roblot N., Joshi V., Gillis J. M., Melki J. (2003) J. Cell Biol. 161, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohanna M., Sobering A. K., Lapointe T., Lorenzo L., Praud C., Petroulakis E., Sonenberg N., Kelly P. A., Sotiropoulos A., Pende M. (2005) Nat. Cell Biol. 7, 286–294 Epub 2005 Feb 2020 [DOI] [PubMed] [Google Scholar]
- 20. Trendelenburg A. U., Meyer A., Rohner D., Boyle J., Hatakeyama S., Glass D. J. (2009) Am. J. Physiol. Cell Physiol. 296, C1258–C1270 [DOI] [PubMed] [Google Scholar]
- 21. Shu L., Houghton P. J. (2009) Mol. Cell. Biol. 29, 4691–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 23. Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 24. Yoon M. S., Chen J. (2008) J. Cell Sci. 121, 282–289 [DOI] [PubMed] [Google Scholar]
- 25. Erbay E., Chen J. (2001) J. Biol. Chem. 276, 36079–36082 [DOI] [PubMed] [Google Scholar]
- 26. Um S. H., D'Alessio D., Thomas G. (2006) Cell Metab. 3, 393–402 [DOI] [PubMed] [Google Scholar]
- 27. Manning B. D. (2004) J. Cell Biol. 167, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shah O. J., Hunter T. (2006) Mol. Cell. Biol. 26, 6425–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tzatsos A., Kandror K. V. (2006) Mol. Cell. Biol. 26, 63–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 31. Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. (2004) J. Cell Biol. 166, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greene M. W., Sakaue H., Wang L., Alessi D. R., Roth R. A. (2003) J. Biol. Chem. 278, 8199–8211 [DOI] [PubMed] [Google Scholar]
- 33. Conejo R., Valverde A. M., Benito M., Lorenzo M. (2001) J. Cell. Physiol. 186, 82–94 [DOI] [PubMed] [Google Scholar]
- 34. Cuenda A., Cohen P. (1999) J. Biol. Chem. 274, 4341–4346 [DOI] [PubMed] [Google Scholar]
- 35. Goodman C. A., Miu M. H., Frey J. W., Mabrey D. M., Lincoln H. C., Ge Y., Chen J., Hornberger T. A. (2010) Mol. Biol. Cell 21, 3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jaafar R., Zeiller C., Pirola L., Di Grazia A., Naro F., Vidal H., Lefai E., Némoz G. (2011) J. Biol. Chem. 286, 22609–22621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shah O. J., Hunter T. (2005) Cell Cycle. 4, 46–51 [DOI] [PubMed] [Google Scholar]
- 38. Harrington L. S., Findlay G. M., Lamb R. F. (2005) Trends Biochem. Sci. 30, 35–42 [DOI] [PubMed] [Google Scholar]
- 39. Long Y. C., Cheng Z., Copps K. D., White M. F. (2011) Mol. Cell. Biol. 31, 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee C. S., Yi J. S., Jung S. Y., Kim B. W., Lee N. R., Choo H. J., Jang S. Y., Han J., Chi S. G., Park M., Lee J. H., Ko Y. G. (2010) Cell Death Differ. 17, 1254–1265 [DOI] [PubMed] [Google Scholar]