Abstract
Pigment epithelium-derived factor (PEDF) is an intrinsic anti-angiogenic factor and a potential anti-tumor agent. The tumoricidal mechanism of PEDF, however, has not been fully elucidated. Here we report that PEDF induces the apoptosis of TC-1 and SK-Hep-1 tumor cells when they are cocultured with bone marrow-derived macrophages (BMDMs). This macrophage-mediated tumor killing is prevented by blockage of TNF-related apoptosis-inducing ligand (TRAIL) following treatment with the soluble TRAIL receptor. PEDF also increases the amount of membrane-bound TRAIL on cultured mouse BMDMs and on macrophages surrounding subcutaneous tumors. PEDF-induced tumor killing and TRAIL induction are abrogated by peroxisome proliferator-activated receptor γ (PPARγ) antagonists or small interfering RNAs targeting PPARγ. PEDF also induces PPARγ in BMDMs. Furthermore, the activity of the TRAIL promoter in human macrophages is increased by PEDF stimulation. Chromatin immunoprecipitation and DNA pull-down assays confirmed that endogenous PPARγ binds to a functional PPAR-response element (PPRE) in the TRAIL promoter, and mutation of this PPRE abolishes the binding of the PPARγ-RXRα heterodimer. Also, PPARγ-dependent transactivation and PPARγ-RXRα binding to this PPRE are prevented by PPARγ antagonists. Our results provide a novel mechanism for the tumoricidal activity of PEDF, which involves tumor cell killing via PPARγ-mediated TRAIL induction in macrophages.
Keywords: Apoptosis, Cancer Therapy, Immunology, Macrophages, Transcription Enhancers
Introduction
Pigment epithelium-derived factor (PEDF)2 is a 50-kDa secreted glycoprotein with multiple biologic activities, including induction of neural differentiation, anti-angiogenesis, and anti-inflammation (1–3). PEDF is widely expressed in human fetal and adult tissues (4), whereas PEDF expression is down-regulated during malignant progression of human glioma, hepatoma, breast cancer, ovarian cancer, lung cancer, and melanoma (5–10). Animal studies have demonstrated that gene delivery of PEDF to tumors leads to suppressed tumor growth and metastasis (11, 12). Meanwhile, the PEDF-mediated antitumor effect is correlated to reduced microvessel density both in the tumor and in the surrounding healthy tissue (11–13). In culture, PEDF displays direct antitumor action in ovarian cancer, melanoma, and osteosarcoma, as evident by the block of cell proliferation, repressed cell migration, and induction of cell apoptosis (8, 10, 14). Therefore, PEDF is considered as a potential agent for treatment of solid tumors.
Inside a solid tumor, in addition to dysregulated blood vessels, stromal cells (including fibroblasts and infiltrating immune cells) also contribute to promote tumor growth, tumor-associated angiogenesis, and metastasis (15, 16). In addition, the most abundant tumor-infiltrating innate immune cells are macrophages, also termed tumor-associated macrophages (TAMs) (15, 16). TAMs are considered an attractive target for therapeutic intervention. These proposed strategies include activation of TAMs to a tumoricidal state (17–19), repression of the tumor-supportive activities of TAMs (20), and depletion of TAMs (21–23). A recent animal study showed that overexpression of PEDF in a rat prostate tumor by transient transfection of a PEDF-expressing plasmid caused increased macrophage recruitment and expression of inducible nitric-oxide synthase in macrophages (12). These findings indicate that PEDF may able to modify the gene expression profile of TAMs.
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is a transmembrane protein that acts as an apoptosis-inducing ligand by binding to death receptor 5 (DR5) expressed on murine cells or DR4 and DR5 expressed on human cells (24, 25). TRAIL is best known as a tumor suppressor because TRAIL-null mice demonstrate an increase in susceptibility to tumor initiation and metastasis (26). TRAIL expression can be up-regulated by several cytokines. For example, interferon (IFN)-γ or -α stimulate tumoricidal activity in human monocytes via induction of TRAIL (17, 27). In theory, these tumoricidal effects can be promoted not only by inducing the TRAIL receptor on tumor cells but also by inducting expression of TRAIL on immune cells.
Peroxisome proliferator-activated receptor γ (PPARγ) is a ligand-activated transcription factor, and its major sources of endogenous ligands are fatty acid derivatives and oxysterols derived from the cholesterol biosynthetic pathway (28, 29). PPARγ heterodimerizes with the 9-cis-retinoic acid receptor (RXR) to form a transcription factor that binds to peroxisome proliferator response elements (PPREs) in the promoter region of target genes (30). PPARγ modulates many functions of macrophages, such as promoting cell differentiation (31), regulating cholesterol homeostasis (32), and antagonizing expression of proinflammatory genes (32, 33). Recent findings indicate that PPARγ activation functions as a signaling mechanism for the anti-angiogenesis and anti-inflammatory function of PEDF (34, 35). The participation of PPARγ in the anti-tumor function of PEDF is not clear. On the other hand, an increase of PPARγ activity in TAMs has been reported as a strategy to overcome TAM-mediated cytotoxic T-lymphocyte suppression in the tumor microenvironment (36). These observations suggest that PPARγ has the potential to modulate the function of TAMs and is thus a possible mediator of the antitumor effect of PEDF.
In the present in vitro and animal study, we demonstrated that a TRAIL-mediated tumoricidal activity of macrophages can be stimulated by PEDF. Our observations also indicated that PPARγ mediates this effect. We identified a PPRE located within the promoter of the human TRAIL gene and determined that the binding of the PPARγ-RXRα heterodimer can be induced by PEDF treatment in macrophages.
EXPERIMENTAL PROCEDURES
Materials
PKH-26 was purchased from Sigma. G3335, GW9662, and benzyloxycarbonyl-VAD(OMe)-fluoromethylketone were purchased from Calbiochem. DNA restriction and modification enzymes were obtained from New England Biolabs (Beverly, MA) and Promega (Madison, WI). TRAIL-R2-Fc (ALX-522-067), Fas-Fc (ALX-522-002), and TNF-R2-Fc (ALX-522-014) were purchased from Alexis (San Diego, CA). Antibodies to PPARα (sc-1985), PPARβ (sc-1986), PPARγ (sc-7273), F4/80 (sc-25830), RXRα (sc-46659), paxillin (sc-365174), and TRAIL (sc-6079) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). RXRβ (GTX89670) and RXRγ (GTX15518) were purchased from GenTex (San Antonio, TX). The DR5 (ab8416) and activated caspase-3 antibodies (ab2302) were obtained from Abcam (Cambridge, UK). The β-actin (MAB1501) antibody was obtained from Millipore (Bedford, MA). Recombinant human TRAIL was from R&D Systems (Minneapolis, MN). PEDF was purified from human plasma via collagen I-Sepharose resin as described previously (37) and was analyzed by SDS-PAGE and Western blot analysis using an anti-PEDF antibody.
Bone Marrow-derived Macrophage (BMDM) Isolation, Cell Culture, and Treatments
Five-week-old C57BL/6 mice were housed under a constant 12-h light/12-h dark cycle and were allowed free access to standard food and water. The experiments were approved by the Mackay Memorial Hospital Review Board for Animal Investigation. Mice were anesthetized via intraperitoneal injection of a mixture of zoletil (6 mg/kg) and xylazine (3 mg/kg) and subsequent cervical dislocation. To isolate BMDMs, femora were aseptically removed and dissected free of adhering tissues, and then the marrow cavities were flushed by injection of Dulbecco's modified Eagle's medium (DMEM)/F-12 medium (Invitrogen). Collected bone marrow cells were incubated in a 100 × 15-mm Petri dish in DMEM/F-12 medium supplemented with 20% heat-inactivated fetal bovine serum (FBS), 1% l-glutamine, 1% HEPES, and 10 ng/ml recombinant mouse macrophage colony- stimulating factor (M-CSF; R&D Systems) for 7 days in 5% CO2 at 37 °C. Treatments with PEDF (200 ng/ml unless otherwise specified) or inhibitors were performed on BMDMs (4 × 105 cells/well of a 6-well plate) seeded in serum-free DMEM/F-12 medium.
THP-1 cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FBS. Differentiation of THP-1 cells was induced by resuspending the cells in fresh medium containing 50 nm phorbol 12-myristate 13-acetate for 48 h, as in our previous report (38). Treatments with PEDF and inhibitors were performed on cells (5 × 105 cells/well of a 6-well plate) seeded in serum-free RPMI 1640 medium with 50 nm phorbol 12-myristate 13-acetate.
For primary culture of human monocytes/macrophages, peripheral blood mononuclear cells were obtained from heparinized whole blood by density gradient centrifugation on Ficoll-Hypaque (Histopaque, Sigma-Aldrich). The monocytes were differentiated into macrophages on culture plates (Costar, Cambridge, MA) for 7 days at 37 °C with 7.5% CO2, as in our previous study (38). Treatments with PEDF or inhibitors were performed on cells seeded in serum-free α-minimal essential medium (Invitrogen).
TC-1 cells, which were established by transforming primary lung epithelial cells of C57/BL6 mice with HPV-16 E6 and E7 as well as c-Ha-ras oncogenes were cultured on RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. Human hepatoma SK-Hep-1 and HuH-7 cells were maintained in DMEM with 10% FBS and 1% penicillin/streptomycin. The cells were grown at 37 °C in 5% CO2 in a humidified environment.
Animal Studies
C57BL/6 mice were inoculated subcutaneously with 1 × 106 TC-1 cells into their left flanks. At day 21, when the tumor size reached ∼100 mm3, the animals were randomized into five groups (n = 6) and given PEDF (0.6 μg resolved in 100 μl of PEDF solvent), PEDF solvent (20 mm Tris/HCl, pH 7.4, 100 mm NaCl), PEDF plus 500 ng of TRAIL-R2-Fc chimera, or control IgG by peritumoral injection twice per day for 1 day. One group of mice was intraperitoneally injected with GW9662 (0.5 mg/kg body weight dissolved in phosphate-buffered saline) at 6 h before peritumoral injection of PEDF. For macrophage isolation, mice were euthanized at 24 h after PEDF injection, and tumors (including their capsule) were harvested, dissected into small pieces, and digested with 400 units/ml collagenase type IV, 0.05 mg/ml collagenase type I, and 0.01 mg/ml DNase I (Roche Applied Science) dissolved in DMEM/F-12 at 37 °C for 1 h. A cell suspension pooled from six tumors was sorted for macrophages using CD11b magnetic beads (5 μl of beads/107 cells; Miltenyi Biotec, Auburn, CA) at 4 °C for 20 min. CD11b-positive cells were collected for the evaluation of TRAIL expression by flow cytometry.
For histology analysis, mouse tumor tissues were surgically excised at 48 h after PEDF injection, fixed with 4% paraformaldehyde, embedded in paraffin, and serially sectioned. Apoptotic cells were detected by a TUNEL assay (In Situ Cell Death Detection Kit, Roche Applied Science) according to the manufacturer's instructions. After TUNEL staining, slides were incubated in blocking buffer (5% normal goat serum in PBS, 0.05% Tween 20) for 30 min. The slides were then incubated with the F4/80 primary antibody (1:150) at 37 °C for 1 h. After washing three times with PBS, 0.05% Tween 20, the slides were incubated with rhodamine-conjugated goat anti-rabbit IgG (1:500) for 60 min at room temperature. After washing three times, sections were observed under a Zeiss epifluorescence microscope (×200, 10 fields/tumor section; Carl Zeiss MicroImaging, Thornwood, NY). Images were recorded using Zeiss software.
Fluorescent Labeling of Cells and Evaluation of Apoptosis in Vitro
Tumor cells were labeled with the fluorescent membrane stain PKH-26 (final concentration 1 μm) according to the manufacturer's instructions and used as target cells in the cytotoxicity assay. The PKH-26-labeled target cells were incubated with BMDMs (4 × 105 cells, 2 ml of 10% FBS, DMEM/F-12 medium) for 16 h. For treatment, cells were incubated in serum-free DMEM/F-12 medium and treated with PEDF for 24 h. The apoptosis of the target cells was determined by in situ staining using a TACS annexin V-FITC kit (R&D Systems) according to the manufacturer's protocol.
Flow Cytometry Analysis
Macrophages were stained with PE-conjugated anti-mouse CD253 (TRAIL) antibody (dilution 1:50; catalog no. 109305; BioLegend, San Diego, CA) and PerCP-conjugated anti-mouse F4/80 antibody (dilution 1:50; catalog no. 123126; BioLegend) for quantification of macrophages. PE-conjugated Rat IgG2a, κ Isotype Ctrl antibody (catalog no. 400508; BioLegend), and PerCP Rat IgG2a, κ Isotype Ctrl antibody (catalog no. 400530; BioLegend) were used as isotype controls. Stained cells were analyzed by flow cytometry (FACScaliber; Beckman Coulter, Brea, CA) using CellQuest software.
Measurement of the Soluble TRAIL
BMDMs were treated with PEDF or its solvent for 24 h, 50 μl of the conditioned medium was analyzed for mouse TRAIL by an enzyme-linked immunosorbant assay kit (E90139Mu; Life Science Inc., Cleveland, OH), according to the manufacturer's method.
TRAIL Promoter Cloning
Fragments of the human TRAIL promoter were cloned from human genomic DNA (Promega) by PCR using the primers 5′-gtagactcatttacagatagaaggcaag-3′ (forward) and 5′-gtaagtcagccaggcagccggtcactg-3′ (reverse), yielding a fragment spanning from −1594 to +137 of the promoter sequence (GenBankTM accession number AF178756). Nucleotide numbers indicated for the human TRAIL promoter relates to the transcriptional initiation site (39). After the fragments were subcloned into the pSC-A vector, sequences were checked and then cloned into the SmaI site of the pGL3-Basic vector (Promega) to produce the pGL3-TRAILp plasmid. The PvuII restriction site at nucleotide −914 and the NdeI restriction site at nucleotide −482 were used to generate the hTRAILp-Luc deletion constructs. The resulting products were blunt cloned into the SmaI site of the pGL3-Basic vector, creating the reporter vectors TRAILp-1-Luc and TRAILp-2-Luc. pGL3-TRAIL-m was generated from pGL3-TRAILp by site-directed mutagenesis (QuikChangeTM mutagenesis kit; Stratagene, La Jolla, CA) using primer 5′-GAGAAAAACCACATATGGAACATTCAGGTC-3′ (GT to CA mutation underlined). The presence of the mutations was confirmed by sequencing.
Western Blot Analysis
Cell lysis, fractionation, and SDS-PAGE were performed as described previously (40). Antibodies used in this study were for PPARα (-fold dilution, 1:1000), PPARβ (1:1000), PPARγ (1:500), RXRα (1:1000), F4/80 (1:500), TRAIL (1:2000), activated caspase-3 antibody (1:500), and β-actin (1:10,000). Proteins of interest were detected using the appropriate IgG-HRP secondary antibody (Santa Cruz Biotechnology) and ECL reagent (Amersham Biosciences). X-ray films were scanned on a model GS-700 imaging densitometer (Bio-Rad) and analyzed using Labworks 4.0 software. For quantification, blots of at least three independent experiments were used.
Semiquantitative Reverse Transcription (RT)-PCR and Quantitative Real-time PCR
Total RNA was extracted from cells using TRIzol (Invitrogen). Synthesis of cDNA was performed with 1 μg of total RNA at 50 °C for 50 min using oligo(dT) primers and reverse transcriptase (Superscript III; Invitrogen). The amplification mixture (final volume, 20 μl) contained 1× Taq polymerase buffer, 0.2 mm dNTPs, 1.5 mm MgCl2, 1 μm primer pair, and 0.5 unit of TaqDNA polymerase. Primer sequences are shown in supplemental Table 1. cDNA was synthesized in an 18–22-cycle amplification reaction (denaturation, 20 s at 94 °C; annealing, 30 s at 57 °C; and polymerization, 40 s at 72 °C). The number of cycles for the primer set was chosen to be in the linear range of amplification. The PCR products were electrophoresed in a 2% agarose gel containing ethidium bromide and visualized by UV illumination.
For real-time PCR detection of RNA transcripts, the cDNA was analyzed in an ABI PRISM 7500 Fast sequence detection system (Applied Biosystems, Foster City, CA). Amplification was carried out in a total volume of 40 μl containing 3 pmol of primers, serially diluted RT product, and SYBR Green I PCR Master Mix reagents (Applied Biosystems) with the following cycling conditions: initial denaturation at 95 °C for 5 min, followed by 40 cycles at 94 °C for 30 s, 57 °C for 20 s, and 72 °C for 30 s and a 5-min terminal incubation at 72 °C. The data were calculated with ΔΔCt. Primer sequences are shown in supplemental Table 1. All determinations were measured in triplicate. The cycle threshold (Ct) values corresponded to the PCR cycle number at which fluorescence emission in real time reached a threshold above the base-line emission and were analyzed using GeneAmp 7700 SDS software (Applied Biosystems). The Ct value of the PCR product of interest (human TRAIL in these experiments) and a control mRNA (GAPDH) was then used to calculate relative quantities of mRNA between samples.
Transfection and Luciferase Assay
HuH-7 cells (1.5 × 105 cells/well of a 12-well plate) were plated for 16 h and transfected with plasmid DNA by the calcium phosphate coprecipitation method. Transient transfection was conducted in triplicate in 12-well plates, and 0.5 μg of plasmids were used in each well. To antagonize PPARγ activity, cells were treated with 20 μm GW9662 during transfection. The luciferase assay, including transient transfection studies for THP-1 macrophages, was performed as in our previous report (38). All DNA solutions also contained 0.1 ng/well of the SV40 Renilla luciferase reporter plasmid, which was used as an internal control for transfection efficiency. The luciferase assay was conducted using the dual luciferase substrate system (Promega) with a luminometer. The luciferase activity was normalized to Renilla luciferase, and a mean value together with an S.E. value of the triplicate samples was used to determine the reporter activity. Each experiment was repeated at least three times.
Transfection of Short Interfering RNA (siRNA)
Subconfluent BMDMs were transfected with a mixture of mouse PPARγ siRNAs (sc-29456; Santa Cruz Biotechnology, Inc.) using Lipofectamine RNAiMAX reagent (Invitrogen) in serum-free medium according to the manufacturer's instructions. A species-specific siCONTROL nontargeting siRNA (Dharmacon Research, Lafayette, CO) was utilized as a negative control. The final concentration of siRNA was 1 nm. At 24 h after siRNA transfection, cells were resuspended in new medium for a 24-h recovery period. Cell viability was not altered by transfection reagent alone or transfection with siRNA, as examined by the trypan blue exclusion assay.
Chromatin Immunoprecipitation (ChIP)
ChIP analysis was performed using the ChIP assay kit (Upstate Biotechnology, Inc., Charlottesville, VA) according to the manufacturer's recommendations. In brief, 1 × 107 THP-1 macrophages were treated with PEDF for 16 h and then collected after 1% formaldehyde treatment for 10 min to cross-link DNA-binding proteins to the DNA. The chromatin DNA was extracted by SDS-containing buffer and broken into fragments of 400–1200 bp in length by sonication. Before immunoprecipitation, chromatin samples were precleared using a protein A-agarose slurry containing salmon sperm DNA and BSA (Upstate Biotechnology) and then immunoprecipitated overnight at 4 °C with antibodies specific for PPARγ (10 μg/1 × 106 cells) or a normal rabbit IgG control antibody (Upstate Biotechnology, Inc.) as a control, followed by immunoprecipitation with protein A-agarose. The recovered protein-nucleic acid complexes were incubated for 4 h with 0.4 m sodium chloride at 65 °C to reverse the cross-links. DNA in the immunoprecipitation product was amplified via PCR with the ChIP assay primers (shown in supplemental Table 1) that covered the PPRE site in the human TRAIL gene promoter. The PCR products were calculated with ΔΔCt. Immunoprecipitation with preimmune mouse IgG served as a negative control, and PCR using the same the primers and the input DNA (preimmunoprecipitation samples) was used as an internal control.
DNA Pull-down Assay
The pull-down assay, which was similar to electrophoretic mobility shift assays, was performed as previously described with modifications (41). Briefly, nuclear extracts from THP-1 macrophages were prepared using the NE-PER nuclear and cytoplasmic extraction kit (Pierce). Double-stranded biotinylated oligonucleotides (500 nm) containing the PPRE sequence (shown in supplemental Table 2) were mixed with 200 μg of nuclear extracts at 4 °C for 1 h with rocking. After incubation, the mixture was then bound to 20 μl of 50% streptavidin-agarose beads (Invitrogen). The complexes were washed five times with buffer (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 300 mm sucrose, 0.5 mm dithiothreitol, 2.5% glycerol, 0.5% Nonidet P-40, and protease inhibitor mixture) at 4 °C. Subsequently, the complexes were resuspended in 100 μl of Laemmli sample buffer. After being heated at 95 °C for 10 min, the supernatant was collected for Western blot analysis.
Statistical Analysis
Results are presented as the means ± S.E. Analysis of variance was used for statistical comparisons. p < 0.05 was considered significant.
RESULTS
PEDF-stimulated BMDMs Exert TRAIL-mediated Tumoricidal Activity
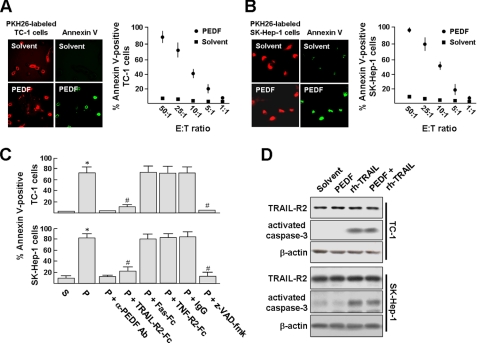
We designed an in vitro assay to test whether PEDF could enhance the antitumor activity of macrophages. In this assay, BMDMs were cocultured with TC-1 cells or SK-Hep-1 cells (at an effector/target ratio of 25:1) for 16 h, and then PEDF or its solvent was added to the culture for 24 h. TC-1 cells or SK-Hep-1 cells were labeled by preincubation with the fluorescent dye PKH-26 (red). Cell apoptosis was visualized via in situ staining using annexin V-FITC (green). As showed in Fig. 1, A and B, PEDF treatment rendered a great majority of TC-1 and SK-Hep-1 cells to stain positive for annexin V, indicating the induction of apoptosis in these cells. In contrast, solvent treatment induced no apoptosis. The percentages of TC-1 and SK-Hep-1 cells undergoing apoptosis dropped in accordance with the effector/target ratio, indicating that TC-1 and SK-Hep-1 cell apoptosis is induced by BMDMs.
FIGURE 1.
PEDF stimulates macrophage-mediated cytotoxicity in a TRAIL-dependent manner. A and B, left panels, BMDMs were cocultured with PKH-26-labeled TC-1 cells or SK-Hep-1 cells (red) at an effector/target ratio of 25:1 for 16 h and then stimulated with PEDF or its solvent for 24 h. Cell apoptosis was visualized via in situ staining using annexin V-FITC (green). Original magnification was ×400. Right panels, PKH-26-labeled TC-1 or SK-Hep-1 target cells were cultured with BMDMs at various effector/target ratios and then stimulated with PEDF. Percentages of annexin V-FITC-positive TC-1 and SK-Hep-1 cells were then determined and presented as the mean ± S.D. (error bars) of triplicate cultures. Experiments were repeated at least three times. C, BMDMs cocultured with TC-1 or SK-Hep-1 cells at an effector/target ratio of 25:1 were stimulated with PEDF (P) or PEDF solvent (S) for 6 h and then treated with soluble TRAIL-R2-Fc (20 ng/ml), Fas-Fc (20 ng/ml), TNF-R2-Fc (20 ng/ml), isotype control IgG1, or 20 μm benzyloxycarbonyl-VAD(OMe)-fluoromethylketone. After 18 h, apoptotic rates were determined by in situ annexin V-FITC staining and presented as the mean ± S.D. of triplicate cultures. To validate the PEDF specificity, PEDF was depleted from its solvent using an anti-PEDF antibody·protein A-Sephadex complex. *, p < 0.002 versus solvent; #, p < 0.005 versus PEDF. D, TC-1 and SK-Hep-1 cells express death receptor TRAIL-R2 (DR5) and are susceptible to TRAIL-mediated apoptosis. Expression of TRAIL-R2 in tumor cells was determined by Western blotting. TRAIL-mediated apoptosis was assessed by culturing cells with 200 ng/ml recombinant human soluble FLAG-tagged TRAIL (rh-TRAIL) for 24 h. The levels of activated caspase-3 (∼17 kDa) were determined by Western blotting. Representative results from three separate experiments are shown.
To identify the apoptosis-inducing ligands involved, we performed blocking experiments using soluble Fc fusion proteins for TNFα, TRAIL, and FasL. Fig. 1C shows that TRAIL-R2-Fc almost completely blocked the killing of TC-1 cells by PEDF-stimulated BMDMs, yet neither Fas-Fc nor TNF-R2-Fc affected the BMDM-mediated cytotoxicity. Likewise, TRAIL-R2-Fc blocked the killing of SK-Hep-1 cells by PEDF-stimulated BMDMs. Again, Fas-Fc and TNF-R2-Fc showed no such blocking effect. In addition, the reduction of viable TC-1 and SK-Hep-1 cells was completely abrogated by the broad spectrum caspase inhibitor benzyloxycarbonyl-VAD(OMe)-fluoromethylketone (20 μm), indicating the involvement of caspase activity in the tumor cell-killing effect of PEDF. In a parallel control experiment, when the PEDF was depleted from its solvent using an anti-PEDF antibody·protein A-Sephadex complex, BMDMs did not kill tumor cells. Western blot analysis revealed that cleavage of procaspase-3 in TC-1 cells and SK-Hep-1 cells was induced by 200 ng/ml recombinant human TRAIL but not by PEDF (Fig. 1D). In addition, the presence of TRAIL-R2 (DR5) in TC-1 cells and SK-Hep-1 cells were confirmed by Western blot analysis, and the levels of TRAIL-R2 were not affected by PEDF. Thus, we concluded that TRAIL was responsible for the cytotoxicity of PEDF-stimulated BMDMs.
PEDF Induces Surface Expression of TRAIL on BMDMs
We examined the induction of TRAIL gene expression by PEDF in BMDMs (Fig. 2A). By 24 h after stimulation with 100 and 200 ng/ml PEDF, TRAIL mRNA levels were increased in a dose-dependent manner, but those of TNF-α and FasL were unaffected. The mRNA level of TRAIL was not further increased in BMDMs incubated with 400 ng/ml PEDF (data not shown). PEDF also dose- and time-dependently increased TRAIL protein levels as assessed by Western blot analysis (Fig. 2, B and C). The TRAIL protein was induced after 8 h and maintained for 12–24 h after PEDF stimulation.
FIGURE 2.
PEDF induces expressions of TRAIL and PPARγ in BMDMs. A, BMDMs were treated with PEDF at the indicated concentrations for 24 h, and the cells were then processed for RT-PCR analysis. GAPDH expression was examined for normalization purposes. B, cells were treated as described above, and proteins were detected by Western blot analysis with antibodies as indicated. Representative blots (left panels) and densitometric analysis with S.D. (error bars) (right panels) of three independent experiments are shown. *, p < 0.05 versus untreated cells. C, BMDMs were exposed to PEDF for the time indicated and then harvested for Western blotting with antibodies against TRAIL (∼33.4 kDa) and PPARγ (∼57.6 kDa). Equal protein loading was confirmed by reprobing the membranes with a β-actin antibody. Representative blots and densitometric analyses with S.D. from four separate experiments are shown. D, PEDF-treated BMDMs were assessed for cell surface TRAIL expression. BMDMs were treated with PEDF or its solvent for 24 h. Cell membrane and cytosolic fractions were isolated as described under “Experimental Procedures” and then subjected to Western blot analysis. A representative result from two independent experiments is shown. E, BMDMs were treated with PEDF or solvent control for 24 h. The cells were then incubated with PE-conjugated isotypic control or PE-conjugated TRAIL antibody and analyzed by flow cytometry.
Because TRAIL is a cell surface protein, we examined the levels of TRAIL on the cell surface of BMDMs after PEDF treatment for 24 h. Cell lysates were fractionated, and Western blot analysis showed that stimulation with PEDF caused a marked increase in TRAIL levels within the cell membrane fraction (Fig. 2D). Flow cytometry analysis also revealed that PEDF increased the TRAIL staining in BMDMs as compared with controls, including solvent and solvent derived from PEDF depletion via treatment with the anti-PEDF antibody·protein A-Sephadex complex (Fig. 1E; 55.2% versus 8.8 and 15.0%, respectively). This confirmed that PEDF increased TRAIL levels on the cell surface of BMDMs. At the same time, we examined the apoptosis-inducing activity of conditioned medium from PEDF-stimulated BMDMs and found that TC-1 and SK-Hep-1 cells incubated with this medium for 24 h did not induce apoptosis. In addition, the concentrations of soluble TRAIL in conditioned medium were below the enzyme-linked immunosorbant assay detection limit, 0.16 ng/ml. This suggests that membrane-bound TRAIL on BMDMs plays a predominant role in the induction of apoptosis in TC-1 and SK-Hep-1 cells.
PPARγ Mediates PEDF-induced TRAIL Expression and Tumor Cell Killing by BMDMs
To explore the molecular mechanism that mediated TRAIL expression, we determined whether PEDF could induce PPARγ expression in BMDMs. RT-PCR and Western blot analyses revealed that PEDF induced PPARγ mRNA and protein expression in a dose-dependent manner (Fig. 2, A and B). PEDF treatment did not affect the protein expression of PPARα or PPARβ. Time course analysis further revealed that PPARγ protein levels increased with the same kinetics as that of TRAIL (Fig. 2C).
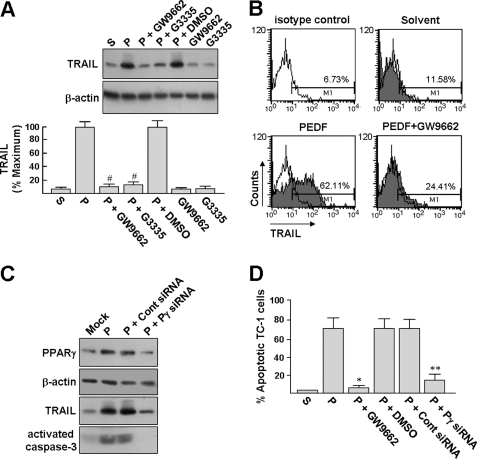
We next analyzed whether the PPARγ antagonists GW9662 and G3335 could prevent TRAIL induction promoted by PEDF. Western blot analysis revealed that BMDMs pretreated with PPARγ antagonists (10 μm, 1 h) but not DMSO (vehicle control) abrogated PEDF-induced TRAIL protein accumulation in BMDMs (Fig. 3A). Flow cytometric analysis confirmed that the surface TRAIL expression in BMDMs induced by PEDF was significantly reduced by GW9662 pretreatment (Fig. 3B; 62.11% versus 24.41%). Transfection of a PPARγ siRNA into BMDMs substantially reduced the ability of PEDF to induce PPARγ and TRAIL in BMDMs, as compared with transfection with a control siRNA (Fig. 3C). These findings indicated that PEDF, by way of PPARγ signaling, induced the expression of TRAIL in BMDMs.
FIGURE 3.
PEDF mediates the induction of TRAIL expression by PPARγ. A, PPARγ antagonists suppress PEDF-induced TRAIL expression. BMDMs were pretreated with 10 μm GW9662 or G3335 for 1 h and then treated with or without 200 ng/ml PEDF (P) for an additional 24 h. Cells were harvested for Western blot analysis. Loading equality was confirmed with antibodies against β-actin. Representative blots and densitometric analyses with S.D. (error bars) from three separate experiments are shown. #, p < 0.005 versus PEDF + DMSO. B, surface expression of TRAIL was quantified by flow cytometry. BMDMs were exposed to PEDF or PEDF solvent for 24 h or pretreated with 10 μm GW9662 for 1 h before exposure to PEDF for an additional 24 h. The cells were then stained with PE-conjugated isotypic control or anti-TRAIL antibody for analysis by flow cytometry. Data shown are from one representative experiment of four. C, PPARγ siRNA abrogates PEDF-induced TRAIL expression. BMDMs were transfected with a PPARγ siRNA or control siRNA for 16 h and allowed to recover for a further 24 h. Mock, cells were treated with transfection reagents alone. After the respective treatment, both BMDMs and siRNA-transfected BMDMs were exposed to PEDF for 24 h, and the cells were then harvested for Western blot analysis (blots 1–3). The siRNA-transfected BMDMs were also used for coculture with TC-1 cells at an effector/target ratio of 25:1 for 16 h and then exposed to PEDF for a further 24 h, followed by detection of activated caspase-3 by Western blot analysis. Representative results from three separate experiments are shown. D, PPARγ antagonist and siRNA block the BMDM-mediated cytotoxicity induced by PEDF, GW9662, and siRNAs pretreatments were performed as described above, followed by PEDF treatment for an additional 24 h. BMDM-mediated cytotoxicity was performed at an effector/target ratio of 25:1, and cell apoptosis was detected by annexin V-FITC staining as described in the legend to Fig. 1A. *, p < 0.001 versus PEDF + DMSO. **, p < 0.02 versus control siRNA + PEDF.
The signaling role of PPARγ for PEDF-induced tumor cell killing was also assayed using the coculture system described above. Pretreatment of the cocultured cells with GW9662 before PEDF treatment prevented almost all of the TC-1 cell apoptosis (Fig. 3D). Also, BMDMs pretreated with PPARγ-specific siRNA significantly reduced PEDF-induced TC-1 cell apoptosis, as compared with transfection with a control siRNA. As a control, immunoblot results revealed that TRAIL protein cannot be detected in cell extracts from PEDF-treated TC-1 cells (supplemental Fig. S1). In addition, Western blot analysis showed that the PPARγ siRNA pretreatment significantly reduced PEDF-induced procaspase-3 cleavage when compared with pretreatment with the control siRNA (Fig. 2C, blot 4), supporting the essential role of PPARγ in PEDF-induced apoptotic signaling. On average, pretreatment of coculture cells with GW9662 or pretreatment of BMDMs with PPARγ-specific siRNA significantly reduced PEDF-induced TC-1 cell apoptosis from 64 ± 10% to 5 ± 2 and 11 ± 6%, respectively (Fig. 3D). These results clearly showed the signaling role of PPARγ in PEDF-induced TRAIL expression and tumor cell killing by BMDMs.
PEDF Induces TRAIL Expression in Macrophages in the Tumor Stroma
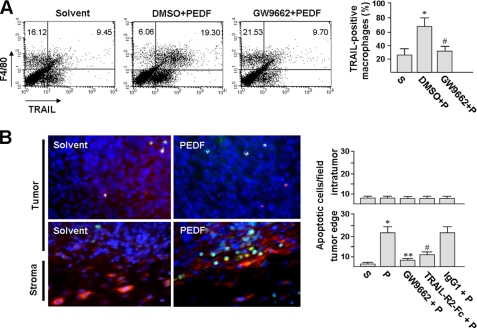
After observing TRAIL induction in cultured macrophages, we were interested in the potential of PEDF to induce TRAIL in tumor macrophages in vivo. C57BL/6 mice were inoculated subcutaneously with 1 × 106 TC-1 cells. After 21 days, when the tumor volume reached 100 mm3, the mice were injected subcutaneously at four sites around the tumor with a total of 0.6 μg of PEDF, twice with 12 h in between. At 24 h after the second PEDF injection, macrophages were isolated from the tumor, and the percentages of TRAIL-positive macrophages were assayed by flow cytometry. Results revealed a significant increase in TRAIL-positive macrophages from the PEDF treatment group as compared with the solvent control group (Fig. 4A; 68.3 ± 11% versus 25.7 ± 8%). Pretreatment 6 h prior to PEDF injection with intraperitoneal injection of GW9662 prevented PEDF-induced TRAIL expression in macrophages (32.1 ± 6%). Therefore, we proposed that PEDF could induce TRAIL in tumor macrophages, and this effect was mediated via cellular PPARγ.
FIGURE 4.
PEDF causes a TRAIL-mediated antitumor effect in a murine model. A, PEDF induces TRAIL expression in stromal macrophages. C57BL/6 mice with established TC-1 tumors were intraperitoneally injected with GW9662 or DMSO vehicle for 6 h, followed by peritumoral injections with PEDF (P) or PEDF solvent (S) control for a further 24 h as described under “Experimental Procedures.” After treatment, the mice were euthanized, and the tumors (n = 6) were processed for macrophage isolation. The freshly isolated macrophages were then double-stained with PE-conjugated anti-TRAIL and PerCP-conjugated anti-F4/80 antibodies for flow cytometry analysis. *, p < 0.02 versus solvent; #, p < 0.001 versus DMSO + PEDF. B, TRAIL-R2-Fc blocks PEDF-induced TC-1 cell apoptosis. C57BL/6 mice bearing TC-1 tumors were pretreated with or without GW9662 for 6 h and then injected around the tumor with PEDF or PEDF combined with TRAIL-R2-Fc or control IgG1. At day 2 post-treatment, tumors were harvested, and tumor sections were double-stained with TUNEL to identify apoptotic cells (green) and F4/80 to identify macrophages (red). Representative photographs revealed PEDF-induced tumor cell apoptosis in the peripheral tumor and in the vicinity of macrophages. Apoptotic TC-1 cells were quantified under a microscope (×400, 10 fields/tumor section) using a digital program. Data are representative of three individual experiments. *, p < 0.001 versus solvent. **, p < 0.001 versus PEDF. #, p < 0.005 versus PEDF + IgG1. Error bars, S.E.
Immunohistochemical analysis of solvent- and PEDF-injected tumors using the F4/80 antibody showed an accumulation of macrophages in the stromal capsule surrounding the tumor cell bulk, with only a few macrophages inside the tumor cell bulk (Fig. 4B, stained red) and the density of macrophage in PEDF-treated tumors to be not significantly different from that in solvent-treated tumors. This observation was consistent with a previous report (42). In the vicinity of macrophages, we observed prominent apoptosis of TC-1 cells, as determined by in situ TUNEL assays (stained green). In addition, co-injection with TRAIL-R2-Fc abolished PEDF-induced apoptosis, but co-injection with the isotype control IgG did not, indicating that the PEDF-induced cytotoxicity depended on TRAIL. The tumor cell apoptosis induced by PEDF was completely abrogated by GW9662 pretreatment. Cancer-associated fibroblasts are abundant in tumor stroma too (15, 16). To address the distributions of macrophages and cancer-associated fibroblasts in TC-1 tumor, we also performed immunohistochemical analysis and found that most of the fibroblasts, however, are further away from the TC-1 tumor core (supplemental Fig. S2), indicating that fibroblast-tumor cell contact is unlikely in the small TC-1 tumors. Our animal study indicated that PEDF induced TRAIL expression in tumor macrophages, and this resulted in tumor cell apoptosis in vivo.
PEDF Induces TRAIL Expression in Human Monocyte-derived Macrophages
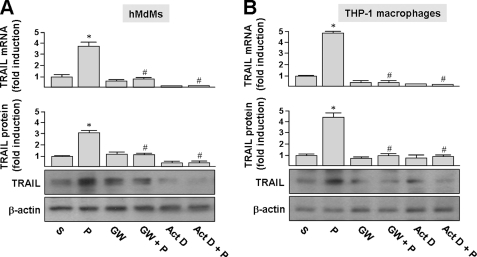
To determine if human macrophages also expressed TRAIL in response to PEDF, human macrophages derived from peripheral monocytes (monocyte-derived macrophages; hMDMs) and the THP-1 monocytic cell line were treated with PEDF, and TRAIL expression was assessed by quantitative real-time PCR and Western blot analysis. As shown in Fig. 5A, exposure of hMDMs to 200 ng/ml PEDF for 24 h induced a 3.7- and 2.9-fold induction of the TRAIL mRNA and protein, respectively, as compared with solvent-treated cells. Exposure of THP-1 macrophages to 200 ng/ml PEDF for 24 h increased the TRAIL mRNA and protein levels by 4.7- and 4.4-fold, respectively, as compared with solvent-treated cells (Fig. 5B). In addition, the TRAIL mRNA induced by PEDF was completely blocked by the general transcription inhibitor actinomycin D, suggesting that the effect of PEDF was transcription-dependent.
FIGURE 5.
PEDF induces TRAIL expression in human macrophages. A and B, hMdMs and THP-1 macrophages were treated with PEDF (P) or solvent (S) for 24 h or pretreated with 5 ng/ml actinomycin D (Act D) or 5 μm GW9662 (GW) for 1 h and then incubated with PEDF for an additional 24 h. Cells were then harvested and assayed by quantitative real-time PCR and Western blot analysis. TRAIL mRNA and protein expression were calculated as the -fold increase of TRAIL expression compared with the solvent-only control. Representative immunoblots and densitometric analysis with S.D. (error bars) are shown. *, p < 0.05 versus solvent-treated cells. #, p < 0.05 versus PEDF-treated cells.
GW9662 pretreatment (5 μm, 1 h) attenuated PEDF-induced TRAIL mRNA and protein levels in hMDMs and THP-1 macrophages (Fig. 5). This and our previous observation that PEDF induces the expression of the PPARγ transcription factor in THP-1 macrophages and hMDMs (35, 38) indicate a role for PPARγ in human macrophages during PEDF-induced TRAIL expression.
PPARγ Activates the Human TRAIL Promoter by Binding to a PPRE
The role of PPARγ in TRAIL induction raises the possibility that the PPARγ transcription factor activates TRAIL expression by binding to its promoter. To investigate this possibility, a plasmid construct containing 1.5 kb of the human TRAIL 5′-flanking sequence fused to a luciferase reporter gene (Fig. 6A; pGL3-TRAILp) was constructed, and a plasmid expressing the entire coding region of human PPARγ (pcDNA-Pγ) was employed. Cotransfection of pcDNA-Pγ with pGL3-TRAILp in HuH-7 cells resulted in up to a 7-fold increase in the transcriptional activity of the luciferase construct (Fig. 6B). The addition of the PPARγ antagonist GW9662 suppressed the PPARγ-induced TRAIL promoter activity by a factor of 3.5-fold.
FIGURE 6.
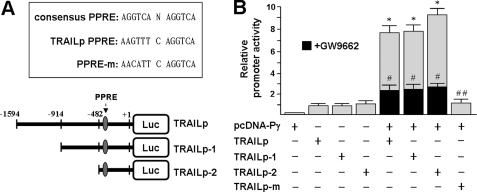
A, sequence of the consensus PPRE and sequence of the candidate PPRE contained within the human TRAIL promoter 5′-flanking region. The location of the candidate PPRE (nt −382/−370) is marked with an arrow. The candidate PPRE mutated from GT to CA is designated as PPRE-m. Numbers are relative to the transcription start site (+1). The region spanning 1594 bp of the promoter was progressively deleted from its 5′-end and fused to the pGL3 basic vector. B, PPARγ transactivates the human TRAIL promoter. HuH-7 cells were transiently transfected with the indicated reporter construct in the presence or absence of the PPARγ-expressing plasmid, pcDNA-Pγ. Black columns represent cells that were treated with 20 μm GW9662 during transfection. Values (mean ± S.D. (error bars)) represent firefly luciferase activity normalized relative to a Renilla luciferase internal control. Luciferase activities are shown relative to the activity of the TRAILp vector, which was arbitrarily set to 1. *, p < 0.05 versus. TRAILp. #, p < 0.05 versus pcDNA-Pγ + TRAILp. ##, p < 0.001 versus pcDNA-Pγ + TRAILp.
To delineate the regulatory region on the human TRAIL gene essential for this effect, transfection studies were performed using pGL3-TRAILp carrying a 5′-terminal deletion. PPARγ-mediated activation of the human TRAIL gene was retained even with the removal of nucleotides (nt) −1594 to −482 (Fig. 6, A and B), suggesting that a critical PPRE resides within nt −482 to +1. PPREs have been characterized as specific DNA-binding sites for the nuclear receptor superfamily of PPARs and generally consist of a direct repeat of the hexamer AGGTCA sequence, separated by one or two nucleotides (30). We analyzed the TRAIL promoter and found a putative hexanucleotide separated by a single nucleotide located at nt −382/−370 (Fig. 6A). The putative hexamer sequence contained three mismatches with respect to the consensus PPRE.
We next examined TRAIL promoter activity in the THP-1 macrophages using a transient transfection method. THP-1 macrophages were transfected with the pGL3-TRAILp and pGL3-TRAILp-2 (a plasmid retaining the putative PPRE) followed by stimulation with PEDF in the presence or absence of GW9662. Luciferase activity was significantly enhanced by PEDF but not by PEDF together with GW9662, as compared with vector-transfected controls (Fig. 7A).
FIGURE 7.
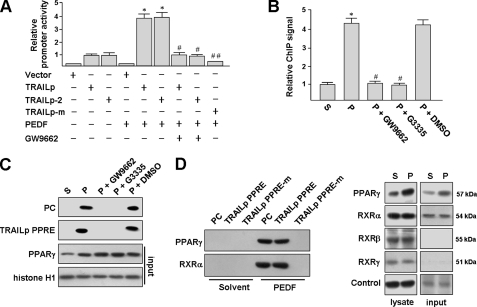
PPARγ binds to the TRAIL promoter. A, PEDF enhances human TRAIL promoter activity. THP-1 macrophages were transfected with the indicated reporter construct. After 48 h post-transfection, cells were stimulated with PEDF or PEDF combined with 10 μm GW9662 for a further 24 h prior to reporter gene activity assays as described above. *, p < 0.05 versus. TRAILp. #, p < 0.05 versus PEDF + TRAILp. ##, p < 0.05 versus PEDF + TRAILp. B, ChIP assay. THP-1 macrophages were treated with PEDF (P) or PEDF solvent (S) for 24 h or pretreated with 10 μm GW9662 for 1 h and then incubated with PEDF for an additional 24 h. PPARγ-DNA complexes were immunoprecipitated with the anti-PPARγ antibody or IgG. Data are normalized to IgG immunoprecipitated DNA and input DNA. The binding of PPARγ to the TRAIL promoter in PEDF-stimulated THP-1 macrophages was measured and quantified by real-time PCR. C and D, identification of PPARγ and PPRE association by DNA pull-down assay. The 5′-biotinylated double-stranded oligonucleotides containing a consensus PPRE (positive control (PC)) or TRAILp PPRE or its mutant, as indicated in Fig. 6A, were used to precipitate PPARγ and RXRα from nuclear protein extracts of THP-1 macrophages treated with PEDF or its solvent. The mixtures were mixed with or without PPARγ antagonists (reaching a final concentration of 20 μm) and then pulled down by streptavidin beads. The proteins in the complex were analyzed by Western blotting using antibodies against PPARγ or RXR isoforms. Aliquots of nuclear extracts were also analyzed before the DNA pull-down assay (input). Shown are representative experiments that were repeated four times with similar observations. Error bars, S.E.
To test whether PPARγ bound to the putative PPRE within the TRAIL promoter in vivo, ChIP was performed in THP-1 macrophages. The genomic DNA fragment between nt −422 and −252, encompassing the putative PPRE of the TRAIL promoter, was amplified by PCR after immunoprecipitation with a monoclonal anti-PPARγ antibody or a control IgG. Strikingly, PEDF stimulation caused an increase in PPARγ binding by 4.2-fold compared with solvent treatment (Fig. 7B). In the IgG control, no TRAIL promoter fragment was detected. Pretreatment of cells with the PPARγ antagonists abrogated this binding.
To confirm the physical interaction between PPARγ and the putative PPRE, we performed a DNA pull-down assay using a biotinylated PPRE probe (nt −385 to −367). This labeled PPRE was incubated with nuclear extracts from PEDF-induced THP-1 macrophages and then immobilized on streptavidin-agarose beads (Fig. 7C). A biotinylated consensus PPRE probe was used as a positive control. We found that PPARγ was pulled down by the consensus PPRE and the putative TRAIL PPRE probes, as assayed by immunoblotting using an anti-PPARγ antibody. The binding specificity of PPARγ to these PPRE sites was demonstrated by mixing nuclear extracts with a PPARγ antagonist (GW9662 or G3335). In addition, a basal level of PPARγ was detected in nuclear extract from solvent-treated THP-1 macrophages (input). However, we did not detect any positive bands by immunoblotting.
To determine the heterodimerization of PPARγ and RXR isoforms (α, β, and γ) on this putative TRAIL PPRE, immunoblot analysis was performed and revealed that three RXR isoforms are expressed in THP-1 macrophages (Fig. 7D). Among the three isoforms, only RXRα was detectable in the nuclear extracts of THP-1 macrophages; RXRβ and RXRγ were barely detectable. These results were consistent with immunocytochemical findings that RXRβ and RXRγ cannot be detected in nucleus, but PPARγ and RXRα can (supplemental Fig. S3). DNA pull-down assays were also performed to examine whether the PPARγ-RXRα heterodimer binds to this putative TRAIL PPRE. Incubation of labeled PPRE probes with nuclear extracts from PEDF-induced THP-1 macrophages resulted in the formation of this complex, as observed by immunoblotting (Fig. 7D). This suggests that RXRα is an important partner of PPARγ for induction of TRAIL promoter activity. Furthermore, a mutated PPRE oligonucleotide corresponding to TRAIL PPRE (PPRE-m; Fig. 6A) demonstrated significantly impaired PPARγ binding, confirming that the putative hexamer was functional for interaction with PPARγ. No significant binding of PPARγ or RXRα was observed when oligonucleotides were incubated with nuclear extracts from solvent-treated THP-1 macrophages. Next, we investigated the importance of the putative PPRE in the regulation of the human TRAIL gene by PEDF. The PPRE-m (Fig. 6A) was introduced into pGL3-TRAIL plasmid to substitute the TRAIL PPRE. This mutation of the TRAIL promoter substantially abolished the induction of the TRAIL promoter by pcDNA-Pγ transfection (Fig. 6B) or PEDF treatment (Fig. 7A), indicating that the hexamer is a functional PPRE. Taken together, our results indicated that the major transcriptional activator responsible for TRAIL gene expression induced by PEDF is PPARγ. The results also implied that PPARγ binds in a ligand-dependent manner to a putative PPRE within the TRAIL promoter in vivo.
DISCUSSION
PEDF has been shown to inhibit tumor growth (5–14). Because PEDF harbors anti-angiogenesis functions similar to anti-VEGF antibodies, angiostatin, and endostatin, the tumor inhibition function of PEDF has been previously associated with the inhibition of tumor vessel growth. Although direct tumor cell killing and inhibition of metastasis has also been reported on several occasions, the mechanism involved is not clear (8, 10–12, 14). Among the multiple functions of PEDF, recent reports of anti-inflammation properties suggest that PEDF modulates the innate immune system. This raises our interest in the possible involvement of the immune system in PEDF-induced tumor cell killing. Our observations in cell culture showed that PEDF exposure did not affect the survival of TC-1 or SK-Hep-1 cells and that PEDF only induced the apoptosis of tumor cells when cocultured with macrophages. This macrophage-mediated tumor cell killing was abolished by treatment with the soluble TRAIL receptor and by knockdown of TRAIL expression, indicating that macrophages killed tumor cells via TRAIL-mediated apoptosis. These observations indicated that the induction of the tumoricidal activity of macrophages could be a possible mechanism of the anticancer activity of PEDF.
Because of the anti-angiogenesis activity of PEDF, PEDF-induced tumor cell killing is expected to occur by the deprivation of nutrients. This may mask the effects of other tumor cell-killing mechanisms. One way to circumvent this is to study small tumors that depend less on vessel ingrowth for nutrient delivery. Under such conditions, tumor cell killing, particularly that occurring in the outer rim of the tumor, is less likely to be due to nutrient deprivation. In our study, very little vessel growth was observed in the TC-1 tumors, and PEDF did not induce central tumor necrosis. Only tumor cells at the tumor periphery that were in direct contact to the host immune cells were killed (Fig. 4B). This strongly indicated that the tumor cells were killed by an effector originating from the tumor capsule rather than nutrient deprivation. The induction of TRAIL in macrophages in the tumor capsule, which is in direct contact with apoptotic tumor cells, suggests that PEDF-induced TRAIL in tumor-associated macrophages has the potential to kill tumor cells in vivo. We propose that in advanced tumors where tumor-associated macrophages have penetrated deeply inside the tumor, the activation of TRAIL contributes to tumor cell killing and may be a novel tumor eradication mechanism.
A seemingly high macrophage to tumor cells ratio was employed in a cell culture experiment in this study as well as in a previous report of tumoricidal activity of BMDMs (43). The fraction of macrophage inside tumor varies from different reports. Macrophages can rise up to 60% of a tumor mass (44). It is still possible for the local ratio of macrophage to tumor cell to be high. Indeed, immunohistochemical staining of F4/80-positive macrophages in TC-1 tumor showed concentrated macrophages surrounding the tumor core (Fig. 4B and supplemental Fig. S2), and only tumor cells located at the tumor periphery that were in direct contact of macrophages were killed (Fig. 4B).
Several lines of evidence in this report sustained the notion that PPARγ mediates PEDF-induced TRAIL expression by activating the TRAIL promoter. This evidence includes the following. 1) PPARγ-expressing plasmids transiently transfected into HuH-7 cells caused ligand-dependent induction of a TRAIL promoter reporter. 2) Macrophages pretreated with PPARγ antagonists or specific siRNAs substantially blocked the ability of PEDF to induce TRAIL expression. 3) ChIP and DNA pull-down assays showed that a PPRE spanning nucleotides −382 to −370 in the proximal region of the human TRAIL promoter bound to PPARγ. Recent animal studies using PPARγ-deficient mice have proposed that PPARγ acts as a tumor suppressor gene to prevent carcinogenesis in the liver (45) and colon (46). Because TRAIL is expressed in the immune system and exerts a critical role in antitumor immunity (47), our study expanded the potential role of PPARγ and PEDF in tumor immunosurveillance mediated via macrophages.
We found that PPARγ played a crucial role in the PEDF-mediated induction of TRAIL in macrophages. To our knowledge, this is the first report of PPARγ as an activator of TRAIL expression in macrophages. This mechanism, if established, may also explain previous reports indicating that mature monocyte-derived dendritic cells and TCR-activated human CD4+ T cells both highly express PPARγ (48–50) and display cytotoxic effects that are at least partly dependent on TRAIL expression (51, 52). The promoter of the human TRAIL gene contains several potential responsive elements, which are associated with different transcription factors. For example, TRAIL is reportedly involved in activation-induced cell death of mature T lymphocytes. The mechanism invoked by NF-κB is activated and associated with a c-Rel binding site in the TRAIL promoter (53). TRAIL plays a role in virus-induced host cell apoptosis because Sendai virus infection causes host interferon regulatory factor-3 binding to IFN-stimulated response elements and subsequent induction of TRAIL-mediated cell apoptosis (39). Moreover, the involvement of other transcriptional activators, such as FOXO3A, in tumor cells has been demonstrated, which provides an additional level of complexity to the regulation of TRAIL expression (54). The involvement of these transcription mechanisms in PEDF-induced macrophages has not been investigated. However, from our inhibitor and knockdown experiments, the major signaling mechanism involved in TRAIL induction is through PPARγ.
Our study revealed that PEDF, via PPARγ, induces TRAIL expression in murine BMDMs and macrophages isolated from TC-1 tumors. This suggested that a PPRE also resided within the 5′-flanking sequence of the murine TRAIL promoter. A 0.5-kb murine TRAIL promoter cDNA (GenBankTM accession no. AB052771) has been isolated and contained an IFN-stimulated response element responsible for TRAIL promoter activation by IFN-β in NK cells (55), but no PPRE has been reported. Structurally, PPREs consists of a direct repeat of the hexamer AGGTCA (or TGACCT) sequence (30). By examining the sequence, we found a PPRE-like sequence separated by a single nucleotide at −60 to −72 in the murine TRAIL promoter (CTAACT G TGACCT; initiation codon ATG is designated +1) as similar to the PPRE of the human TRAIL gene (Fig. 6A), which contained a consensus hexamer and a putative hexanucleotide with less homology to the consensus hexamer. We are in the process of determining whether this is the PPRE within the murine TRAIL promoter.
PEDF is well known for its antiangiogenic activity. PEDF exerts this activity by direct induction of endothelial cell apoptosis (2). Our finding of macrophage-mediated apoptosis raised the possibility that macrophages participate in the antiangiogenesis process of PEDF in vivo. In support, a recent study indicated that TRAIL induces apoptosis of human brain endothelial cell hCMEC/D3 in vitro (56). This possibility awaits further investigation.
In summary, the antitumor activity of PEDF may involve multiple mechanisms. Our findings indicated a novel mechanism in which PEDF was able to enhance the antitumor efficacy of macrophages by inducing a TRAIL-mediated tumoricidal activity. In addition, we have defined a novel PPRE in the human TRAIL promoter that is crucial for up-regulation of TRAIL by PPARγ.
Supplementary Material
Acknowledgments
We thank Jin-Man Chen, Chu-Ping Ho, and Tzu-Hsiu Lo for excellent technical support.
This study was supported by National Science Council, Taiwan, Grant NSC 97-2314-B-195-014-MY3 and Mackay Memorial Hospital Grant MMH-E-100-006.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S3.
- PEDF
- pigment epithelium-derived factor
- BMDMs
- bone marrow-derived macrophages
- TRAIL
- TNF-related apoptosis-inducing ligand
- PPARγ
- peroxisome proliferator-activated receptor gamma
- PPRE
- peroxisome proliferator response elements
- TAMs
- tumor-associated macrophages
- RXR
- retinoic acid receptor
- hMDMs
- human monocyte-derived macrophages
- nt
- nucleotides.
REFERENCES
- 1. Steele F. R., Chader G. J., Johnson L. V., Tombran-Tink J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dawson D. W., Volpert O. V., Gillis P., Crawford S. E., Xu H., Benedict W., Bouck N. P. (1999) Science 285, 245–248 [DOI] [PubMed] [Google Scholar]
- 3. Zhang S. X., Wang J. J., Gao G., Shao C., Mott R., Ma J. X. (2006) FASEB J. 20, 323–335 [DOI] [PubMed] [Google Scholar]
- 4. Tombran-Tink J., Mazuruk K., Rodriguez I. R., Chung D., Linker T., Englander E., Chader G. J. (1996) Mol. Vis. 2, 11. [PubMed] [Google Scholar]
- 5. Guan M., Pang C. P., Yam H. F., Cheung K. F., Liu W. W., Lu Y. (2004) Cancer Gene Ther. 11, 325–332 [DOI] [PubMed] [Google Scholar]
- 6. Matsumoto K., Ishikawa H., Nishimura D., Hamasaki K., Nakao K., Eguchi K. (2004) Hepatology 40, 252–259 [DOI] [PubMed] [Google Scholar]
- 7. Cai J., Parr C., Watkins G., Jiang W. G., Boulton M. (2006) Clin. Cancer Res. 12, 3510–3517 [DOI] [PubMed] [Google Scholar]
- 8. Cheung L. W., Au S. C., Cheung A. N., Ngan H. Y., Tombran-Tink J., Auersperg N., Wong A. S. (2006) Endocrinology 147, 4179–4191 [DOI] [PubMed] [Google Scholar]
- 9. Zhang L., Chen J., Ke Y., Mansel R. E., Jiang W. G. (2006) Int. J. Mol. Med. 17, 937–944 [PubMed] [Google Scholar]
- 10. Orgaz J. L., Ladhani O., Hoek K. S., Fernández-Barral A., Mihic D., Aguilera O., Seftor E. A., Bernad A., Rodríguez-Peralto J. L., Hendrix M. J., Volpert O. V., Jiménez B. (2009) Oncogene 28, 4147–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hase R., Miyamoto M., Uehara H., Kadoya M., Ebihara Y., Murakami Y., Takahashi R., Mega S., Li L., Shichinohe T., Kawarada Y., Kondo S. (2005) Clin. Cancer Res. 11, 8737–8744 [DOI] [PubMed] [Google Scholar]
- 12. Halin S., Rudolfsson S. H., Doll J. A., Crawford S. E., Wikström P., Bergh A. (2010) Neoplasia 12, 336–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doll J. A., Stellmach V. M., Bouck N. P., Bergh A. R., Lee C., Abramson L. P., Cornwell M. L., Pins M. R., Borensztajn J., Crawford S. E. (2003) Nat. Med. 9, 774–780 [DOI] [PubMed] [Google Scholar]
- 14. Takenaka K., Yamagishi S., Jinnouchi Y., Nakamura K., Matsui T., Imaizumi T. (2005) Life Sci. 77, 3231–3241 [DOI] [PubMed] [Google Scholar]
- 15. Joyce J. A., Pollard J. W. (2009) Nat. Rev. Cancer 9, 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siveen K. S., Kuttan G. (2009) Immunol. Lett. 123, 97–102 [DOI] [PubMed] [Google Scholar]
- 17. Solis M., Goubau D., Romieu-Mourez R., Genin P., Civas A., Hiscott J. (2006) Biochem. Pharmacol. 72, 1469–1476 [DOI] [PubMed] [Google Scholar]
- 18. Taniguchi H., Shimada Y., Sawachi K., Hirota K., Inagawa H., Kohchi C., Soma G., Makino K., Terada H. (2010) Anticancer Res. 30, 3159–3165 [PubMed] [Google Scholar]
- 19. Vishvakarma N. K., Singh S. M. (2010) Immunol. Lett. 134, 83–92 [DOI] [PubMed] [Google Scholar]
- 20. Watkins S. K., Li B., Richardson K. S., Head K., Egilmez N. K., Zeng Q., Suttles J., Stout R. D. (2009) Eur. J. Immunol. 39, 2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galmbacher K., Heisig M., Hotz C., Wischhusen J., Galmiche A., Bergmann B., Gentschev I., Goebel W., Rapp U. R., Fensterle J. (2010) PLoS One 5, e9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang W., Zhu X. D., Sun H. C., Xiong Y. Q., Zhuang P. Y., Xu H. X., Kong L. Q., Wang L., Wu W. Z., Tang Z. Y. (2010) Clin. Cancer Res. 16, 3420–3430 [DOI] [PubMed] [Google Scholar]
- 23. Meng Y., Beckett M. A., Liang H., Mauceri H. J., van Rooijen N., Cohen K. S., Weichselbaum R. R. (2010) Cancer Res. 70, 1534–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finnberg N., Gruber J. J., Fei P., Rudolph D., Bric A., Kim S. H., Burns T. F., Ajuha H., Page R., Wu G. S., Chen Y., McKenna W. G., Bernhard E., Lowe S., Mak T., El-Deiry W. S. (2005) Mol. Cell Biol. 25, 2000–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pan G., O'Rourke K., Chinnaiyan A. M., Gentz R., Ebner R., Ni J., Dixit V. M. (1997) Science 276, 111–113 [DOI] [PubMed] [Google Scholar]
- 26. Cretney E., Takeda K., Yagita H., Glaccum M., Peschon J. J., Smyth M. J. (2002) J. Immunol. 168, 1356–1361 [DOI] [PubMed] [Google Scholar]
- 27. Griffith T. S., Wiley S. R., Kubin M. Z., Sedger L. M., Maliszewski C. R., Fanger N. A. (1999) J. Exp. Med. 189, 1343–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. (1998) Nature 395, 137–143 [DOI] [PubMed] [Google Scholar]
- 29. Hihi A. K., Michalik L., Wahli W. (2002) Cell Mol. Life Sci. 59, 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michalik L., Desvergne B., Wahli W. (2004) Nat. Rev. Cancer 4, 61–70 [DOI] [PubMed] [Google Scholar]
- 31. Tontonoz P., Nagy L., Alvarez J. G., Thomazy V. A., Evans R. M. (1998) Cell 93, 241–252 [DOI] [PubMed] [Google Scholar]
- 32. Majdalawieh A., Ro H. S. (2010) Nucl. Recept. Signal. 8, e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glass C. K., Saijo K. (2010) Nat. Rev. Immunol. 10, 365–376 [DOI] [PubMed] [Google Scholar]
- 34. Ho T. C., Chen S. L., Yang Y. C., Liao C. L., Cheng H. C., Tsao Y. P. (2007) Cardiovasc. Res. 76, 213–223 [DOI] [PubMed] [Google Scholar]
- 35. Yang S. L., Chen S. L., Wu J. Y., Ho T. C., Tsao Y. P. (2010) Life Sci. 87, 26–35 [DOI] [PubMed] [Google Scholar]
- 36. Van Ginderachter J. A., Meerschaut S., Liu Y., Brys L., De Groeve K., Hassanzadeh Ghassabeh G., Raes G., De Baetselier P. (2006) Blood 108, 525–535 [DOI] [PubMed] [Google Scholar]
- 37. Petersen S. V., Valnickova Z., Enghild J. J. (2003) Biochem. J. 374, 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ho T. C., Yang Y. C., Chen S. L., Kuo P. C., Sytwu H. K., Cheng H. C., Tsao Y. P. (2008) Mol. Immunol. 45, 898–909 [DOI] [PubMed] [Google Scholar]
- 39. Kirshner J. R., Karpova A. Y., Kops M., Howley P. M. (2005) J. Virol. 79, 9320–9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang Y. C., Ho T. C., Chen S. L., Lai H. Y., Wu J. Y., Tsao Y. P. (2007) BMC Cancer 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deng W. G., Zhu Y., Montero A., Wu K. K. (2003) Anal. Biochem. 323, 12–18 [DOI] [PubMed] [Google Scholar]
- 42. Gérard C. M., Baudson N., Kraemer K., Ledent C., Pardoll D., Bruck C. (2001) Clin. Cancer Res. 7, (suppl.) 838s–847s [PubMed] [Google Scholar]
- 43. Hagemann T., Lawrence T., McNeish I., Charles K. A., Kulbe H., Thompson R. G., Robinson S. C., Balkwill F. R. (2008) J. Exp. Med. 205, 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dong Z., Kumar R., Yang X., Fidler I. J. (1997) Cell 88, 801–810 [DOI] [PubMed] [Google Scholar]
- 45. Yu J., Shen B., Chu E. S., Teoh N., Cheung K. F., Wu C. W., Wang S., Lam C. N., Feng H., Zhao J., Cheng A. S., To K. F., Chan H. L., Sung J. J. (2010) Hepatology 51, 2008–2019 [DOI] [PubMed] [Google Scholar]
- 46. Evans N. P., Misyak S. A., Schmelz E. M., Guri A. J., Hontecillas R., Bassaganya-Riera J. (2010) J. Nutr. 140, 515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bellail A. C., Qi L., Mulligan P., Chhabra V., Hao C. (2009) Rev. Recent Clin. Trials 4, 34–41 [DOI] [PubMed] [Google Scholar]
- 48. Szatmari I., Vámosi G., Brazda P., Balint B. L., Benko S., Széles L., Jeney V., Ozvegy-Laczka C., Szántó A., Barta E., Balla J., Sarkadi B., Nagy L. (2006) J. Biol. Chem. 281, 23812–23823 [DOI] [PubMed] [Google Scholar]
- 49. Yang X. Y., Wang L. H., Chen T., Hodge D. R., Resau J. H., DaSilva L., Farrar W. L. (2000) J. Biol. Chem. 275, 4541–4544 [DOI] [PubMed] [Google Scholar]
- 50. Thompson P. W., Bayliffe A. I., Warren A. P., Lamb J. R. (2007) Cytokine 39, 184–191 [DOI] [PubMed] [Google Scholar]
- 51. Fanger N. A., Maliszewski C. R., Schooley K., Griffith T. S. (1999) J. Exp. Med. 190, 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kayagaki N., Yamaguchi N., Nakayama M., Kawasaki A., Akiba H., Okumura K., Yagita H. (1999) J. Immunol. 162, 2639–2647 [PubMed] [Google Scholar]
- 53. Baetu T. M., Kwon H., Sharma S., Grandvaux N., Hiscott J. (2001) J. Immunol. 167, 3164–3173 [DOI] [PubMed] [Google Scholar]
- 54. Sakoe Y., Sakoe K., Kirito K., Ozawa K., Komatsu N. (2010) Blood 115, 3787–3795 [DOI] [PubMed] [Google Scholar]
- 55. Sato K., Hida S., Takayanagi H., Yokochi T., Kayagaki N., Takeda K., Yagita H., Okumura K., Tanaka N., Taniguchi T., Ogasawara K. (2001) Eur. J. Immunol. 31, 3138–3146 [DOI] [PubMed] [Google Scholar]
- 56. Chen P. L., Easton A. S. (2010) Biochem. Biophys. Res. Commun. 391, 936–941 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.