Abstract
Acyl-coenzyme A (acyl-CoA) thioesterases play a crucial role in the metabolism of activated fatty acids, coenzyme A, and other metabolic precursor molecules including arachidonic acid and palmitic acid. These enzymes hydrolyze coenzyme A from acyl-CoA esters to mediate a range of cellular functions including β-oxidation, lipid biosynthesis, and signal transduction. Here, we present the crystal structure of a hexameric hot-dog domain-containing acyl-CoA thioesterase from Bacillus halodurans in the apo-form and provide structural and comparative analyses to the coenzyme A-bound form to identify key conformational changes induced upon ligand binding. We observed dramatic ligand-induced changes at both the hot-dog dimer and the trimer-of-dimer interfaces; the dimer interfaces in the apo-structure differ by over 20% and decrease to about half the size in the ligand-bound state. We also assessed the specificity of the enzyme against a range of fatty acyl-CoA substrates and have identified a preference for short-chain fatty acyl-CoAs. Coenzyme A was shown both to negatively regulate enzyme activity, representing a direct inhibitory feedback, and consistent with the structural data, to destabilize the quaternary structure of the enzyme. Coenzyme A-induced conformational changes in the C-terminal helices of enzyme were assessed through mutational analysis and shown to play a role in regulating enzyme activity. The conformational changes are likely to be conserved from bacteria through to humans and provide a greater understanding, particularly at a structural level, of thioesterase function and regulation.
Keywords: Enzyme Catalysis, Enzymes, Fatty Acid, Fatty Acid Metabolism, Protein Structure
Introduction
Acyl-coenzyme A (acyl-CoA) thioesterases play important metabolic and regulatory roles in all living systems through their ability to modulate intracellular concentrations of free coenzyme A (CoASH)4 and of esterified and non-esterified fatty acids. They catalyze the cleavage of the thioester bond present in a wide range of CoA-esters; the most well characterized substrates are fatty acyl-CoAs, which range from short- to long-chain fatty acyl-CoAs, saturated and polyunsaturated fatty acyl-CoAs, branched-chain fatty acyl-CoAs, and methyl-branched-chain fatty acyl-CoAs (1). Acyl-coenzyme A thioesterases (hereafter abbreviated as Acots) have been isolated from organisms ranging from prokaryotes through to humans, and in higher order eukaryotes, they are localized within the cytosol, mitochondria, peroxisomes, and microsomes (1). Acyl-CoAs are important in β-oxidation, lipid biosynthesis, regulation of ion channel opening, signal transduction, the budding and fusion of intracellular membranes, and the regulation of gene transcription via nuclear receptors (1). There is also a high demand for free CoASH in mitochondria as a critical intermediate in the citric acid cycle, β-oxidation, and other metabolic pathways. Non-esterified fatty acid products of Acot catalysis are also important in biological processes such as regulation of calcium channels in cells and induction of calcium fluxes and as second messengers in signaling cascades (1).
In support of their key roles in cell function, these enzymes are conserved throughout evolution, and aberrations in Acot function have been implicated in a number of diseases in higher eukaryotes. Two-dimensional electrophoresis in combination with mass spectrometry and immunoblotting identified a significant decrease in the expression of the gene encoding Acot7 in mesial lobe epilepsy (2), whereas other studies have identified roles for long-chain acyl-CoA esters in insulin secretion (3). More generally, inappropriately high levels of free fatty acids are symptomatic in conditions such as obesity (4), insulin resistance (5), cancer (6), and hyperlipidemia (7).
At present, the protein structures of Acots that have been deposited in the Protein Data Bank (PDB) fall within three classes: the β-sandwich + α/β-hydrolase fold Acots, which include human Acot2 (PDB ID 3HLK (8)) and human Acot4 (PDB ID 3K2I); the tetrameric hot-dog fold Acots, which include thioesterases from Campylobacter jejuni Cj0915 (PDB ID 3D6L (9) and Helicobacter pylori Hp0496 (PDB ID 2PZH); and finally, Acots with a hexameric hot-dog fold arrangement, with structures from Acot7 (PDB IDs 2QQ2, 2Q2B, and 2V1O (10, 11)), Acot12 (PDB ID 3B7K), and Xanthomonas campestris (PDB ID 2FUJ). In all but one of these structures, the enzyme contained one or more ligands, presumably due to the instability or reduced propensity to crystallize in the apo-form.
In this study, we have determined the structure of Bacillus halodurans Acot (BhAcot) in the apo-form and present a structural analysis of the apo- and CoASH-bound forms of this hexameric hot-dog fold thioesterase. We provide novel insights into the dramatic ligand-induced conformational flexibility within this family of enzymes and assess the specificity of the enzyme against a range of fatty acyl-CoA substrates.
MATERIALS AND METHODS
Protein Expression and Purification
The bacterial expression plasmid encoding the BhAcot was obtained from the Joint Centre for Structural Genomics (JCSG). Plasmid DNA was transformed into the Escherichia coli BL21(DE3) expression strain, and a single colony was used to inoculate a 5-ml starter culture consisting of Luria-Bertani (LB) medium and ampicillin (100 μg/ml). This was used to inoculate 2 liters of LB-ampicillin medium and grown at 37 °C until an A600 °of 0.4 was reached, whereby protein overexpression was induced by the addition of 0.2% arabinose, and bacterial cells were grown overnight at 25 °C. Bacterial cells were collected by centrifugation and resuspended in 1/20 volume in His Buffer A (50 mm phosphate buffer, pH 8.0, 300 mm NaCl, 20 mm imidazole). Cells were lysed with three freeze-thaw cycles. The DNA was digested by the addition of 1 mg of DNase I (Invitrogen), and cell debris were removed by centrifugation. The soluble cell extract was injected onto a 5-ml prewashed nickel-nitrilotriacetic acid affinity matrix column (Qiagen) and washed with 10 column volumes of His Buffer A. Elution of the recombinant protein was achieved by applying a linear gradient of increasing imidazole concentration until a maximum concentration of 500 mm imidazole (His Buffer B). Fractions containing the desired protein were collected and injected onto a pre-equilibrated size exclusion column S200 26/60 (Amersham Biosciences) containing 20 mm Tris, pH 8.0, and 50 mm NaCl. The eluted protein was concentrated using an Amicon ultracentrifugal device, and aliquots were stored at −80 °C. Analysis of the protein by SDS-PAGE revealed greater than 99% purity.
Crystallization
Conditions that induce crystallization were screened by the sparse matrix and hanging-drop vapor diffusion techniques using a range of commercially available crystallization screens (Crystal Screen and Crystal Screen 2, Hampton Research; PACT Premier, Molecular Dimensions). Briefly, a 1-μl protein sample was combined with 1 μl of reservoir solution and suspended above a 500-μl reservoir solution and incubated at 290 K. Diffraction quality crystals were obtained in similar conditions to the CoASH-bound form (22% PEG 3350 and 0.13–0.17 m sodium formate) after 2 days at 298 K. For data collection, crystals were transiently soaked in reservoir solution containing 20% glycerol and flash-cooled under a 100 K αnitrogen stream (Cryocool, Cryo Industries) Diffraction data were collected from a single crystal on an R-Axis IV++ image plate detector using copper Kα radiation produced by a Rigaku FR-E rotating anode generator (Rigaku/MSC, Spring, TX). Raw data were autoindexed, integrated, and scaled using the HKL-2000 package (collection statistics are summarized in Table 1). The crystals had the symmetry of the space group P212121 with unit cell dimensions of a = 71.18 Å, b = 78.54 Å, and c = 187.38 Å, and the asymmetric unit contained six thioesterase molecules. Initial phases were obtained by molecular replacement using Phaser (12) and a single thioesterase monomer from PDB ID 1VPM as a search model. Local rebuilding was undertaken using Coot (13) and refinement with Refmac (14) from the CCP4 program suite (15). The final model shows excellent overall stereochemistry (Table 1).
TABLE 1.
Crystallographic data for apo-BhAcot
| Data collection | |
|---|---|
| Space group | P212121 |
| Unit cell dimensions | a = 71.18 Å, b = 78.54 Å, c = 187.38 Å |
| Resolution range (Å) | 25–2.9 (2.99–2.90)a |
| Total reflections | 160, 257 (15, 413)a |
| Unique observations | 23, 985 (2, 348)a |
| Completeness (%) | 99.7 (98.2)a |
| Redundancy (%) | 6.61 (6.55)a |
| Rmerge (%)b | 11.7 (32.9)a |
| Average I/σ(I) | 9.1 (4.3)a |
| Refinement | |
| Rcryst/Rfree (%)c | 23.2/28.6 |
| Bond length r.m.s.d. (Å) | 0.007 |
| Bond angle r.m.s.d. (°) | 1.03 |
| Ramachandran plot (%)d | |
| Most favored | 92.7 |
| Allowed | 7.3 |
| Generously allowed | 0.0 |
| Disallowed | 0.0 |
a Numbers in parenthesis are for the highest resolution shell.
b Rmerge = Σhkl(Σi(|I hkl,I − 〈Ihkl 〉|))/Σhkl,i 〈Ihkl〉, where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and 〈Ihkl〉 is the mean intensity of that reflection. Calculated for I > −3σ(I).
c Rcryst = Σhkl(‖Fobshkl| − |Fcalchkl‖)/|Fobshkl|, where |Fobshkl| and |Fcalchkl| are the observed and calculated structure factor amplitudes. Rfree is equivalent to Rcryst but calculated with reflections (5%) omitted from the refinement process.
d Calculated with the program PROCHECK (19).
Thioesterase Activity Assay
The activity assay described by Yamada et al. (16) was utilized to measure the specificity of the enzyme against a range of fatty acyl-CoA substrates. The standard reaction mixture contained fatty acyl-CoA substrates ranging from 10 to 250 μm, 0.1 μg of protein sample, and 100 mm sodium phosphate (pH 7.4) in a final volume of 1 ml. The absorbance at 232 nm was monitored immediately after adding the substrate and followed for 3 min at 20-s intervals. The molar absorption coefficient, ϵ232 (4,250 m−1cm−1), was used to calculate cleavage of the thioester bond. Prism (GraphPad) was used to plot the data and calculate maximum velocity and Michaelis-Menten constants.
ThermoFluor Stability Assay
Wild-type BhAcot and the N149C mutant were diluted to 0.5 mg/ml in 20 mm Tris, 125 mm NaCl, pH 7.6, alone, or supplemented with 5 mm CoA. SYPRO Orange was added to the solution (5× final concentration; Invitrogen) and gradually heated over a temperature range of 25–95 °C. Fluorescence was monitored using a 7900 HT real-time PCR machine (Applied Biosystems), and curve fitting (Boltzmann Sigmoid) was performed by GraphPad (Prism) to obtain the melting temperature of the protein (Tm). Each assay was done in triplicate, and the values above represent the mean ± S.E.
Mutagenesis
A mutation within the BhAcot gene was generated by PCR mutagenesis with KOD Hot Start DNA polymerase (Merck). An N149C mutation was incorporated using mutagenic primers (5′-CGCGCCTGCACGAAAAGAATGCCGGAAAAAACGAGCAGC-3′ and 5′-GCTGCTCGTTTTTTCCGGCATTCTTTTCGTGCAGGCGCG-3′; Invitrogen), and the fidelity of the clone was confirmed by DNA sequencing. Expression and purification of the recombinant mutant were undertaken as described for WT BhAcot, with the addition of DTT to all of the purification buffers. The protein was finally oxidized by buffer exchange containing 20 mm Tris, pH 8.0, and 50 mm NaCl, and subjected to extensive air oxidation. Disulfide bond formation was confirmed by non-reducing SDS-PAGE analysis.
RESULTS AND DISCUSSION
Structural Characterization of the Hexameric Apo-acyl-CoA Thioesterase
The structure of the acyl-CoA thioesterase from B. halodurans (BhAcot) was determined by x-ray crystallography at 2.9 Å resolution. The enzyme was expressed recombinantly and purified to homogeneity using affinity and size exclusion chromatography. The molecular mass of the native enzyme was estimated to be ∼100 kDa from its size exclusion chromatography elution profile, suggestive of a hexameric quaternary structure (monomer molecular mass 17.3 kDa), and confirmed by the presence of six monomers within the asymmetric unit of the crystal. The crystals were grown in the absence of substrate, and inspection of electron density maps confirmed the absence of ligands at the active site.
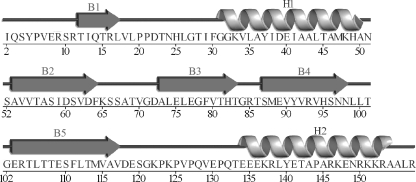
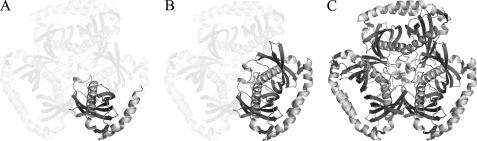
Each monomer is composed of a five-stranded antiparallel β-sheet that wraps around a central α-helix and packs against a C-terminal α-helix (Figs. 1 and 2A). All monomers within the asymmetric unit possess the same hot-dog fold tertiary structure, with the greatest root mean square distance (r.m.s.d.) between any two monomers being 0.39 Å. In forming a hexamer, each monomer associates into a dimer (Fig. 2B), which then further associates to form a trimer-of-dimers arrangement (Fig. 2C).
FIGURE 1.
Schematic representation of the secondary structure elements within each monomer of the hexameric BhAcot. The five β-strands (B1–B5) form an antiparallel β-sheet that surrounds helix H1. Helix H2 packs against the β-sheet on the opposite side to helix H1.
FIGURE 2.
Graphic representation of the monomer (A), dimer (B), and hexamer (C) of BhAcot. Each monomer forms a dimer (double hot-dog fold), which then associates with the two other double hot-dogs to form a hexamer essentially composed of a trimer-of-dimers.
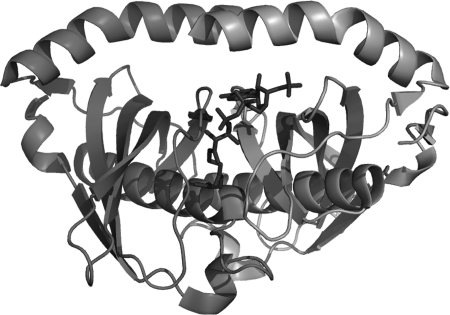
The enzyme contains six active sites, each formed by the juxtaposition of two monomers at the double hot-dog interface (Fig. 3). Residues involved in catalysis involve a strictly conserved Asn24 residue and polarizing Gly31 (positioned at the end of the central helix) from one monomer, together with a highly conserved Asp39 from the adjacent monomer. Although the mechanism of catalysis cannot be directly inferred from the structure, it is likely to involve a nucleophilic attack by the carboxyl group of Asp39 either directly on the substrate or through a water molecule (17).
FIGURE 3.
Graphic representation of the double hot-dog fold of the BhAcot dimer with CoA superimposed, illustrating the active site at the interface between two monomers.
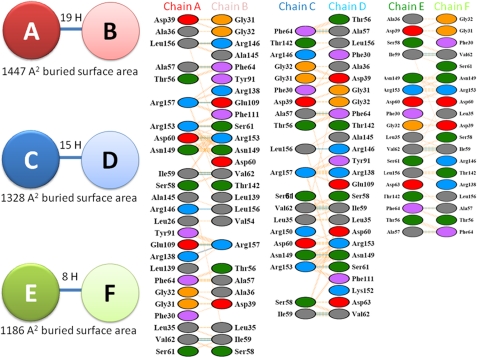
Interestingly, analyses of the interfaces that mediate association into the native hexameric quaternary structure revealed significant variability. Interfaces that mediate association of two hot-dog domains to form a double hot-dog dimer contribute not only the active sites within the enzyme but a large proportion of the overall stability of the hexamer (approximately half of the total buried surface area of the hexamer is found at the dimer interfaces). Given the importance of this interface to the catalytic activity and overall structural stability of the enzyme, it was striking to observe a significant variation at each of these interfaces. For example, the interface between chain A and B dimer contained 19 hydrogen bonds and a buried surface area of 1447 Å2 (determined using the program PISA (18)), whereas the interface at chains C:D contained 15 hydrogen bonds and a buried surface area of 1328 Å2, and the E:F dimer interface possessed only eight hydrogen bonds and a buried surface area of 1186 Å2. The interface areas therefore vary by more than 20%; the interfaces and the associated interactions are presented in Fig. 4. Inspection of the structure of a related hexameric Acot from Haemophilus influenzae deposited in the PDB (PDB ID 3BJK) revealed that this variation is possibly a general characteristic of the apo-form of hexameric thioesterases; this hexameric apo-acyl-CoA thioesterase exhibited similar variations at the dimer interfaces, with 777, 688, and 651 Å2 of buried surface area and 12, 10, and 9 hydrogen bonds at the dimer interfaces, respectively. As described below, analysis of the same interfaces of BhAcot in the CoASH-bound form reveals much greater uniformity, and it is therefore possible to speculate that ligand binding at this interface locks the dimers into position. In further support of this, the interfaces that mediate trimer association and which are not involved in ligand binding (i.e. chains A:F, B:C, and D:E) exhibit consistent interactions in both apo-bound (877, 867, and 867 A2 buried surface area) and CoASH-bound forms (see below) and are also consistent with the H. influenzae thioesterase structure (876, 878, and 855 A2).
FIGURE 4.
Variation in bonding at each of the double hot-dog fold BhAcot dimer interfaces.
Structural Comparison with the Coenzyme A-bound Acyl-CoA Thioesterase
An x-ray crystallographic model of BhAcot with bound CoASH ligand has been deposited to the PDB (ID 1VPM) by the Joint Center for Structural Genomics. However, a study describing the structure remains to be published. Here, we provide a brief analysis of the bound enzyme and draw comparisons with our apo-enzyme to reveal changes within the structure upon ligand binding.
The overall structure of the CoASH-bound BhAcot is similar to the apo-enzyme in terms of overall tertiary and quaternary structure; however, a number of distinct differences were observed. At the monomer level, each domain consists of the same hot-dog fold as observed in the apo-form (highest r.m.s.d. of the CoASH-bound monomers is 0.62 Å). However, comparison of the hexamer with the apo-form revealed greater variation (overall r.m.s.d. of 2.13 Å), with most of the variation occurring at the interfaces and at the C-terminal regions of each chain. In the CoASH-bound form, the enzyme appears to open up to accommodate the ligand, and as noted earlier, this is associated with greater uniformity at the dimer interfaces but significantly less buried surface area (ranging from 738 to 781 Å2): ∼60% of that observed in the apo-structure of the protein.
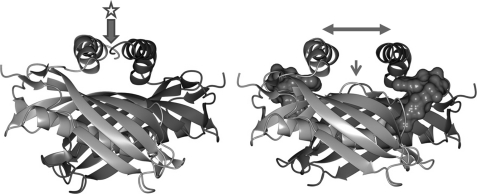
Upon ligand binding, significant variation is also observed at the C-terminal α-helices to accommodate the ligand (Fig. 5). In the apo-form, both helices within the double hot-dog are bound through interactions involving Asn149 and Arg153; Ala156 and Ala145; and Arg157 and Glu135. However, in the ligand-bound form, these interactions are disrupted due to favorable interactions with the ligand: in particular, Lys147 interaction with the 3′-phosphoryl group and Arg150 interaction with the adenine base.
FIGURE 5.
Conformational changes between the apo- (left) and CoASH-bound BhAcot. In the apo-form, the C-terminal helices from each monomer interact, and a continuous β-sheet is formed between the two monomers. In the CoASH-bound structure, the α-helices contact CoASH, and a β-bulge is formed at the interface to interrupt the β-sheet. The N149C mutation (discussed under “Interaction of the C-terminal Helices with Coenzyme A Regulates Catalytic Activity”) is located in the left panel in the position marked with a star.
We also observed a ligand-induced β-bulge (disruption of the regular hydrogen bonding of the β-sheet) at each of the double hot-dog interfaces that was not present at the apo-interfaces (Fig. 5). These conformational changes occur due to interactions between the adenine base of the CoASH ligand with both the Ser58 side chain of one monomer and the Asp63 side chain of the adjacent monomer. Interestingly, a similar observation was noted for the structurally related hexameric broad specificity acyl-coenzyme A thioesterase from H. influenzae (HIO827); however, the comparison was made with an inactive mutant enzyme, and therefore it was not certain whether these structural differences were the result of ligand binding or mutations within the active site (17). Inspection of other CoASH-bound thioesterases deposited in the PDB also display this bulge, including human Acot12 (PDB ID 3B7K) and Acot7 N-terminal domain, whereas in the apo-form, the β-sheet is not perturbed. This suggests that the observed ligand-induced conformational change may be an evolutionary conserved feature of hexameric acyl-CoA thioesterases and is likely to assist in maintaining the overall stability of the β-sheet, whereas the structure undergoes ligand-induced conformational changes at the dimer and trimer interfaces and the C-terminal helical regions.
Following the observed CoA-induced changes and reduction in buried surface area, we performed a thermal stability assay using the SYPRO Orange fluorescent dye to compare the stability of the apo- and CoA-bound forms of BhAcot. Consistent with the structural data, the melting temperature of the CoA ligand-bound form enzyme was significantly reduced when compared with that of the apo-enzyme (CoA ligand-bound form: Tm = 40.79 ± 0.1 °C; apo-BhAcot: Tm = 42.98 ± 0.05 °C).
Specificity for Short-chain Fatty Acyl-CoAs
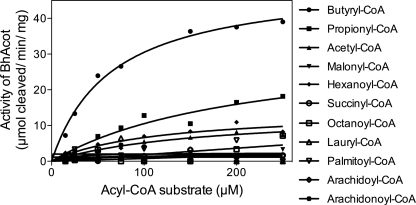
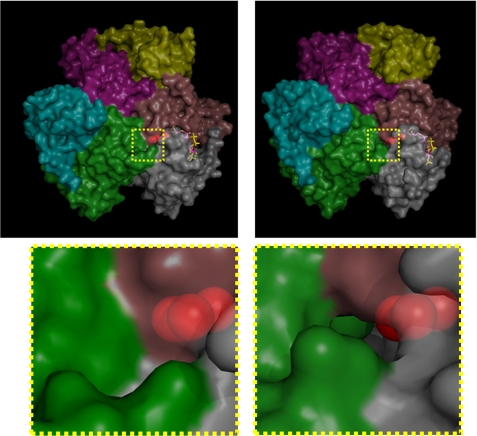
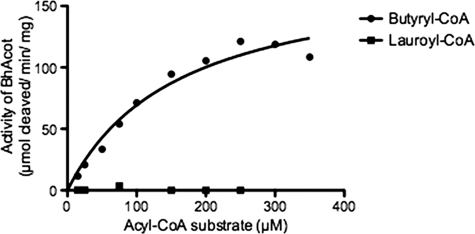
The structure of the enzyme revealed a large internal channel at the active site, which based on its hydrophobic nature and proximal location to coenzyme A, is likely to be the fatty acyl binding pocket. The channel could accommodate a short-chain fatty acid. We tested the thioesterase activity on a range of fatty acyl-CoA substrates to determine the optimal specificity of the enzyme. In accord with the structural observations, BhAcot showed the greatest activity for butyryl-CoA, with only negligible activity for longer chain fatty acids (Fig. 6; Table 2). Interestingly, molecules with a similar chain length to butyryl-CoA, such as succinyl- and malonyl-CoA, were poor substrates, presumably due to the carboxylate anion not favored in a strongly hydrophobic environment; the fatty acyl chain pocket is lined by hydrophobic amino acids: 42AALTA46 and 53AVVT56 from one monomer and 26LGTIF30 from another monomer. In the CoASH-bound form, the channel traverses to the exterior of the protein; however, in the apo-form, the channel is closed (Fig. 7), indicating that the active site undergoes dramatic conformational changes during the catalytic cycle.
FIGURE 6.
Michaelis-Menten plots of BhAcot thioesterase activity against a range of substrates.
TABLE 2.
Summary of kinetic values for acyl-CoA thioesterase activity of BhAcot against a range of fatty-acyl CoA substrates
ND, not determined due to low catalytic properties.
| Acyl-CoA substrate | Vmax | Km |
|---|---|---|
| μmol/min/mg | μmol | |
| Butyryl-CoA | 50 ± 2.5 | 64 ± 9 |
| Propionyl-CoA | 33 ± 9 | 213 ± 100 |
| Acetyl-CoA | 14 ± 2.5 | 163 ± 56 |
| Malonyl-CoA | <10 | ND |
| Succinyl-CoA | <10 | ND |
| Octanoyl-CoA | <10 | ND |
| Lauryl-CoA | <10 | ND |
| Palmitoyl-CoA | <10 | ND |
| Arachidonoyl-CoA | <10 | ND |
| Arachidoyl-CoA | <10 | ND |
FIGURE 7.
Surface representation of the BhAcot hexamer (each domain is colored differently) in the absence (left) and presence (right) of CoASH. In the CoASH-bound form, the enzyme opens up, and a channel is formed that extends from the active site, through to the surface of the protein.
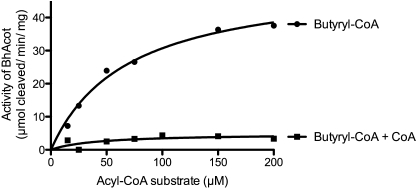
Based on our structural and ThermoFluor data indicating that CoA induces a less stable enzyme, we compared the activity of BhAcot against butyryl-CoA in the presence of CoA. We found that CoA dramatically inhibited the enzyme (Fig. 8). This likely provides a negative feedback mechanism, which in part, may be due to the conformational changes and instability of the enzyme caused by association of the ligand.
FIGURE 8.
Michaelis-Menten plots of BhAcot thioesterase activity against butyryl-CoA in the presence and absence of 50 μm CoA.
Interaction of the C-terminal Helices with Coenzyme A Regulates Catalytic Activity
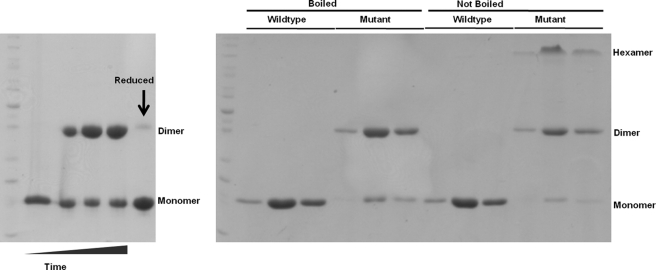
Our structural data show that in the presence of CoA, interactions between the two C-terminal helices are disrupted and the helices interact with two CoA ligands (Fig. 5). This is associated with further disruptions in the β-sheet and instability in the overall quaternary structure. To assess the role of interaction of the C-terminal helices with CoA, we attempted to prevent this interaction by incorporating a disulfide bond through mutation of the Asn149 to Cys (Fig. 5, left panel, star) to lock the enzyme in the apo-conformation. The mutant enzyme was recombinantly expressed and purified in the presence of reducing agent (DTT) and then oxidized through buffer exchange. As demonstrated in Fig. 9, the mutant formed a stable covalently linked dimer that could be reversed by the presence of reducing agent. Locking the C-terminal helices in the apo-form also appeared to have a further stabilizing effect on overall quaternary structure as shown in both the non-reducing electrophoretic gel analysis (Not Boiled panels), with an increase in the hexamer form, as well as in ThermoFluor stability assays, with the mutant exhibiting a Tm of 45.21 ± 0.2 (when compared with 42.98 ± 0.05 for WT). Interestingly, we also observed that addition of CoA to the mutant no longer destabilized the enzyme, but rather stabilized it (Tm: 48.3 ± 0.2). This is most likely due to CoA making contacts at the dimer interface, without the associated disruptions in the C-terminal helices and other regions in the enzyme. Consistent with this observation, the activity of the mutant was greater than that observed for WT because CoA, the product of the reaction, is no longer able to induce the same reduction in enzyme stability (Fig 10).
FIGURE 9.
SDS-PAGE analysis of WT and N149C mutant BhAcot. Left panel, oxidation of the disulfide linkage over time. Lane 1, time 0, prior to removal of reducing agent; lanes 2-4, 1, 3, and 5 days after removal of reducing agent, respectively; lane 5, the addition of reducing agent to the oxidized protein (day 5) shows reduction to monomeric mutant BhAcot. Right panel, analysis of fractions from size exclusion chromatography including boiling and non-boiling heat treatments.
FIGURE 10.
Michaelis-Menten plots of BhAcot N149C mutation in the presence of butyryl-and lauroyl-CoA. The introduced N149C mutation did not affect the substrate specificity of the enzyme.
Conclusion
The structure of the apo-form of BhAcot reveals a large variability between individual dimeric interfaces within the hexamer and dramatic conformational differences from the CoASH-bound form including disruptions in the C-terminal helices, a β-bulge in the adjacent sheets, and an overall reduction in the internal buried surface area. Coenzyme A binding induces reduced stability as determined by ThermoFluor stability assays and supported by the structural data and provides negative feedback inhibition. The role of the C-terminal helices was assessed, and we determined that their interaction with CoA is at least in part involved in destabilizing the enzyme and reducing enzyme activity because preventing the interaction by locking the helices together increased the stability and activity of the enzyme. An internal hydrophobic channel consistent with the size of the optimal substrate was identified, which traverses to the exterior in the CoASH-bound structure, highlighting the remarkable conformational changes Acots appear to undergo during the catalytic cycle.
Acknowledgments
We thank the Joint Center for Structural Genomics for donating the expression plasmid encoding BhAcot. We acknowledge the use of the University of Queensland Remote Operation Crystallization and X-ray (UQROCX) Diffraction Facility.
This work was funded in part by the Australian Research Council (ARC) and the National Health and Medical Research Council (NHMRC).
The atomic coordinates and structure factors (code 3SPS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CoASH
- coenzyme A
- Acot
- acyl-coenzyme A thioesterase
- BhAcot
- B. halodurans Acot
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Kirkby B., Roman N., Kobe B., Kellie S., Forwood J. K. (2010) Prog. Lipid Res. 49, 366–377 [DOI] [PubMed] [Google Scholar]
- 2. Dalby N. O., Mody I. (2001) Curr. Opin. Neurol. 14, 187–192 [DOI] [PubMed] [Google Scholar]
- 3. Gribble F. M., Proks P., Corkey B. E., Ashcroft F. M. (1998) J. Biol. Chem. 273, 26383–26387 [DOI] [PubMed] [Google Scholar]
- 4. Adams S. H., Chui C., Schilbach S. L., Yu X. X., Goddard A. D., Grimaldi J. C., Lee J., Dowd P., Colman S., Lewin D. A. (2001) Biochem. J. 360, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Storlien L. H., Kraegen E. W., Chisholm D. J., Ford G. L., Bruce D. G., Pascoe W. S. (1987) Science 237, 885–888 [DOI] [PubMed] [Google Scholar]
- 6. Welsch C. W. (1992) Cancer Res. 52, 2040s–2048s [PubMed] [Google Scholar]
- 7. Grundy S. M., Denke M. A. (1990) J. Lipid Res. 31, 1149–1172 [PubMed] [Google Scholar]
- 8. Mandel C. R., Tweel B., Tong L. (2009) Biochem. Biophys. Res. Commun. 385, 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yokoyama T., Choi K. J., Bosch A. M., Yeo H. J. (2009) Biochim. Biophys. Acta 1794, 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forwood J. K., Thakur A. S., Guncar G., Marfori M., Mouradov D., Meng W., Robinson J., Huber T., Kellie S., Martin J. L., Hume D. A., Kobe B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10382–10387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Serek R., Forwood J. K., Hume D. A., Martin J. L., Kobe B. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 133–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 13. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 15. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 16. Yamada J., Matsumoto I., Furihata T., Sakuma M., Suga T. (1994) Arch. Biochem. Biophys. 308, 118–125 [DOI] [PubMed] [Google Scholar]
- 17. Willis M. A., Zhuang Z., Song F., Howard A., Dunaway-Mariano D., Herzberg O. (2008) Biochemistry 47, 2797–2805 [DOI] [PubMed] [Google Scholar]
- 18. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 19. Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]