Abstract
There are seven linker histone variants in human somatic cells (H1.0 to H1.5 and H1X), and their prevalence varies as a function of cell type and differentiation stage, suggesting that the different variants may have distinct roles. We have revisited this notion by using new methodologies to study pluripotency and differentiation, including the in vitro differentiation of human embryonic stem (ES) and teratocarcinoma cells and the reprogramming of keratinocytes to induced pluripotent stem cells. Our results show that pluripotent cells (PCs) have decreased levels of H1.0 and increased levels of H1.1, H1.3, and H1.5 compared with differentiated cells. PCs have a more diverse repertoire of H1 variants, whereas in differentiated cells, H1.0 expression represents ∼80% of the H1 transcripts. In agreement with their prevalent expression in ES cells, the regulatory regions of H1.3 and H1.5 genes were found to be occupied by pluripotency factors. Moreover, the H1.0 gene promoter contains bivalent domains (H3K4me2 and H3K27me3) in PCs, suggesting that this variant is likely to have an important role during differentiation. Indeed, the knockdown of H1.0 in human ES did not affect self-renewal but impaired differentiation. Accordingly, H1.0 was recruited to the regulatory regions of differentiation and pluripotency genes during differentiation, confirming that this histone variant plays a critical role in the regulation of these genes. Thus, histone H1 variant expression is controlled by a variety of mechanisms that produce distinct but consistent H1 repertoires in pluripotent and differentiated cells that appear critical to maintain the functionality of such cells.
Keywords: Cell Differentiation, Embryonic Stem Cell, Histones, Induced Pluripotent Stem (iPS) Cell, Transcription Factors, Bivalent Domains, Histone Variants, Linker Histone H1
Introduction
Eukaryotic DNA is packaged into chromatin through its association with histone proteins. Chromatin is composed of nucleosomes, repetitive units of 146 base pairs of DNA wrapped around an octamer of four core histone proteins, and the linker histone H1 bound near the entry and exit sites of the core particle to stabilize two full turns of DNA. Histone H1 participates in nucleosome spacing and in the folding and stabilization of the 30-nm chromatin fiber (1, 2). Histone H1 is a lysine-rich protein with a short basic N-terminal tail region, a highly conserved central globular domain, and a long positively charged C-terminal tail. These tails are post-translationally modified, mostly by phosphorylation but also by acetylation and methylation (3–5).
Histone H1 in humans is a family of closely related, single-gene encoded proteins, including seven somatic subtypes (from H1.1 to H1.5, H1.0, and H1X), three testis-specific variants (H1t, H1T2m and HILS1), and one restricted to oocytes (H1oo) (6–8). Among the somatic histone H1 variants, H1.1 to H1.5 are expressed in a replication-dependent manner, whereas H1.0 and H1X are replication-independent. The H1.1 to H1.5-encoding genes are clustered in a region of chromosome 6 together with the core histone genes, whereas H1X and H1.0 genes are on chromosome 3 and 22, respectively. H1.2 to H1.5 and H1X are ubiquitously expressed, H1.1 is restricted to certain tissues, and H1.0 accumulates in terminally differentiated cells.
As it participates in the formation of higher order chromatin structures, H1 is seen as a structural component related to chromatin compaction and inaccessibility to transcription factors and to RNA polymerase. Nonetheless, it has also been suggested that histone H1 plays a more dynamic and gene-specific role, participating in the regulation of gene expression. Previous studies on the effect of H1 depletion on global gene expression have found no effect on the vast majority of genes but rather have detected up- or down-regulation of small groups of genes (9–13). It is not clear whether the different variants have specific roles or regulate specific promoters. In mice, single or double H1 variant knock-outs have no apparent phenotype because of compensatory up-regulation of other subtypes (14). These reports have favored the view that H1 variants are redundant. On the other hand, we have reported that depletion of single H1 subtypes by inducible RNA interference in breast cancer cells produced different phenotypic effects (13), suggesting differential functions for the various H1 variants in somatic cells. Gene-specific effects of H1 might result from interactions with specific regulatory factors or DNA-binding proteins. Such effects might also be the origin of reported specific functions for some H1 variants (15–18).
The pluripotency and self-renewal properties of embryonic stem (ES)7 cells are controlled at the molecular level by a tightly regulated network of transcription factors, including the master transcription factors OCT4, SOX2, and NANOG. These pluripotency factors occupy the regulatory regions of both self-renewal and developmental genes and have the dual ability to maintain the high levels of expression of genes needed for self-renewal and suppress the expression of differentiation genes (19).
ES cell chromatin has structural peculiarities with respect to differentiated cells. In particular, heterochromatin appears more relaxed, perhaps because the proteins involved in its formation, such as HP1 and linker histone H1, have hyperdynamic interactions with chromatin (20). ES cells also display unique histone modification patterns at the regulatory regions of developmental genes called “bivalent domains,” consisting of the simultaneous presence of histone H3 trimethylated at residue Lys-27 (H3K27me3) and di/trimethylated at residue Lys-4 (H3K4me2/me3) around the transcriptional start site of critical developmental genes. This coexistence of marks associated with transcriptional activation (H3K4me2/me3) and repression (H3K27me3) has been suggested to play a role in silencing developmental genes in ES cells while keeping them poised for activation upon initiation of specific developmental pathways (21, 22). Genes marked with bivalent domains commonly encode master regulators that are critical for differentiation. In the course of differentiation, the permissive chromatin structure of multipotent cells is progressively and selectively closed up. Accessible regulatory areas, such as bivalent domains, close up in a tissue specific manner and are no longer accessible to transcription factors, leading to a loss of regulatory potential (23).
Recently, it has been reported that adult somatic cells can be reprogrammed to pluripotency by the overexpression of critical transcription factors, most commonly OCT4, SOX2, c-MYC, and KLF4 (24, 25). Such induced pluripotent stem (iPS) cells have the same properties as embryonic stem cells regarding self-renewal and pluripotency and are thus of great importance for regenerative medicine.
Changes in the relative amounts of the somatic H1 variants and their potential roles during differentiation have been little investigated; among the few examples are in vitro in murine erythroleukemic cells (26) and in vivo in differentiating tissues of the young mouse and rat (27, 28). Recent advances in stem cell biology and reprogramming make it possible to investigate changes in the relative human H1 content upon differentiation and how the expression of the different H1 variants modulates self-renewal and differentiation. To address this questions, we have used two different models: 1) in vitro differentiation of established human embryonic stem cell lines and 2) reprogramming of juvenile human primary keratinocytes to iPS cells. Overall, we describe that human pluripotent cells (ES and iPS) express a wider repertoire of histone H1 variants and higher levels of H1.1, H1.3, and H1.5 than somatic cells. Accordingly, we detected the presence of pluripotency transcription factors at the promoters of H1.3 and H1.5 variants. However, the knockdown of H1.3 in human ES cells did not affect self-renewal. In contrast, H1.0 was found to be the main H1 variant in adult somatic cells. The H1.0 gene contains bivalent domains in human ES and iPS cells, and its expression is induced during differentiation. Knockdown of H1.0 did not affect the self-renewal abilities of human ES cells but affected the in vitro differentiation capacity of these cells. In agreement with this phenotype, we found that histone H1, and more specifically the H1.0 variant, is recruited to certain pluripotency and differentiation genes during the differentiation of human ES cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Culturing Conditions
The human embryonic stem cell lines ES[4] and ES[2] (29) and the induced pluripotent cell lines KiPS4F1 and KiPS4F4 (30) were grown in Matrigel-coated plates and in the presence of irradiated mouse embryonic fibroblast-conditioned medium supplemented with FGF and passed as clumps using trypsin. To assess differentiation, we produced embryoid bodies (EBs), aggregates in which cells differentiate into a heterogeneous mix of derivatives of the three germ layers. For EB generation, the cells were trypsinized to single cells and counted. Approximately 50,000 cells/well were placed in 96-well V-bottom microplates and centrifuged to facilitate the formation of aggregates. Two days later, the aggregates were moved to gelatin-coated plates and cultured in differentiation media containing 20% FBS and absence of FGF for 20 days.
Primary culture of human keratinocytes from foreskin was carried out as previously described (30). The foreskin fibroblast line HFF-1 (ATCC CRL-2429) was cultured in DMEM containing 10% FBS.
NT2-D1 cells were cultured in DMEM containing 10% FBS and 2 mm l-glutamine at 37 °C in 5% CO2. Differentiation was induced by 10−6 m retinoic acid treatment for up to 20 days.
Viral Transduction to Stably Deplete or Overexpress H1 Variants
Lentivirus for the expression of H1.0 and H1.3 shRNAs (in the pLVTHM vector) or HA-tagged H1 variants (in pEV833) were described elsewhere (13). Viral production and infection by spinoculation were also described. Because all of the vectors coexpressed the GFP marker, effectively infected cells were purified by cell sorting (FACS).
RNA Extraction and RT-qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen) or the HighPure RNA isolation kit (Roche Applied Science) according to the manufacturer's instructions. Quality assessment and quantification of RNA was performed in the Nanodrop ND1000 (Thermo Scientific) machine. cDNA was generated from 100 ng of total RNA using SuperScript VILO cDNA synthesis (Invitrogen). Quantification of gene products was performed by real time PCR with specific oligonucleotides using SYBR Green I Master mix (Roche) in a Roche 480 Lightcycler. Each value was corrected by human GAPDH and expressed as relative units. When comparison between H1 variants was required, each histone H1 cDNA value was normalized to the value of real time PCR amplification of genomic DNA extracted from the cell line analyzed, with the same primers set. Gene-specific oligonucleotide sequences are available on request.
Histone H1 Extraction, Gel Electrophoresis, and Immunoblotting
Histone H1 was purified by 5% perchloric acid lysis for 1 h at 4 °C. Soluble acid proteins were precipitated with 30% trichloroacetic acid overnight at 4 °C, washed twice with 0.5 ml of acetone, and reconstituted in water. Protein concentration was determined with the Micro BCA protein assay (Pierce). Purified histones were subjected to 10% SDS-PAGE, transferred to a PVDF membrane, blocked with Odyssey blocking buffer (LI-COR Biosciences) for 1 h, and incubated with primary antibodies overnight at 4 °C and with secondary antibodies conjugated to fluorescence (IRDye 680 goat anti-rabbit IgG or IRDye 800CW goat anti-mouse IgG, Li-Cor) for 1 h at room temperature. The bands were visualized and quantified in the Odyssey Infrared Imaging System. Antibodies specifically recognizing human H1 variants, including those generated in our laboratory (13), are available from Abcam: H1.2 (ab17677), H1.3 (ab24174), H1.5 (ab24175), H1X (ab31972), and H1.0 (ab11079).
Chromatin Immunoprecipitation
Immunoprecipitation was performed according to the Upstate-Millipore standard protocol. Briefly, the cells were fixed using 1% formaldehyde, harvested, and sonicated to generate chromatin fragments between 200 and 500 bp. Sheared chromatin was immunoprecipitated overnight using 2 μg of antibody against H3K4me2 (07-030; Millipore), H3K27me3 (07-449; Millipore), OCT4 (sc-8628; Santa Cruz), SOX2 (sc-17320; Santa Cruz), NANOG (AF1997; R & D), H1 (clone AE-4; Millipore), H1X (ab31972), H1.2 (ab4086), or HA tag (ab9110). Immunocomplexes were recovered using a mix of protein A and protein G-Sepharose, washed, and eluted. Cross-linking was reversed at 65 °C overnight, and immunoprecipitated DNA was recovered using the PCR purification kit from Qiagen or iPure kit from Diagenode. The presence of the genomic regions of interests was checked by real time PCR. Oligonucleotide sequences used for the amplifications are available on request. For re-ChIP experiments, chromatin was eluted in 10 mm DTT after immunoprecipitating with anti-H3K4me2 antibody and reimmunoprecipitated with anti-H3K27me3 antibody or control IgG.
RESULTS
Changes of Histone H1 Variant Gene Expression during the Differentiation of Human Embryonic Stem and Teratocarcinoma Cells
To gain insights into the role of histone H1 variants during differentiation, we analyzed the mRNA levels of H1.0, H1.1, H1.2, H1.3, H1.4, H1.5, and H1X at different time points during the differentiation of human ES cell lines. Over this period, the expression of the pluripotency factors NANOG, SOX2, and OCT4 encoding genes was strongly silenced (Fig. 1A), and most cells acquired a differentiated morphology. Adjustment of the expression data obtained from RT-qPCR by GAPDH expression and by the data from qPCR amplification of genomic DNA with the same sets of primers used on cDNA enabled the expression of the different histone variants to be compared in self-renewing cells (Fig. 1B) and during differentiation (Fig. 1C).
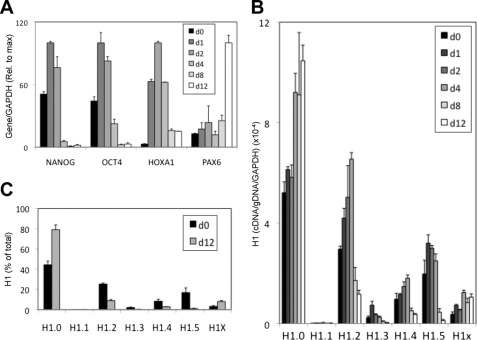
FIGURE 1.
Changes of histone H1 variant gene expression during the differentiation of human embryonic stem cells. A, silencing of NANOG, SOX2, and OCT4 genes during the differentiation of ES[4] stem cells. ES[4] cells were cultured in differentiation media as described under “Experimental Procedures” for 4, 8, 15, or 20 days. At the indicated time points, the cells were processed for RNA extraction. Expression of NANOG, SOX2, and OCT4 genes was determined by RT-qPCR with specific oligonucleotide pairs. GAPDH expression was measured for normalization, and the data are shown relative to the maximal level of each gene. B, comparison of H1 variant gene expression in ES cells. RNA extracted from ES[4] cells was used to measure histone H1 expression by RT-qPCR with variant-specific oligonucleotide pairs, as well as FOXA2 and SOX2 expression with oligonucleotide pairs in the same exon. GAPDH was used for normalization. To allow comparison between genes, each cDNA value was normalized to the value of qPCR amplification of genomic DNA (gDNA) extracted from ES[4], with the same primer set. C, histone H1 variant gene expression during the differentiation of ES[4]. The RNA samples of the ES[4] differentiation curve described in A were used to measure histone H1 expression by RT-qPCR with variant-specific oligonucleotide pairs. GAPDH was used for normalization. To allow comparison between H1 variants, each histone H1 cDNA value was normalized to the value of qPCR amplification of genomic DNA as in B. D, proportion of histone H1 variant transcripts in differentiated and undifferentiated cells. The relative proportion of each H1 variant expression in the initial and final time points along the differentiation kinetic was calculated from the data in C and expressed as percentage of the total H1 content. In addition to ES[4], data for ES[2] differentiation are also shown calculated from a kinetic shown in supplemental Fig. S1. For the ES[2] experiment, day 4 is the first time point available. The means and S.D. of a representative experiment quantified in triplicate are shown.
In undifferentiated ES cells, the variants expressed at higher rates were H1.2, H1.0, H1.5, H1.3, and H1X, whereas H1.1 and H1.4 were lower but still significantly expressed compared with a gene that is not expressed in ES cells, namely FOXA2 (Fig. 1B). Upon differentiation of ES[4] cells, H1.2, H1X, H1.4, and, more importantly, H1.0 gene expression increased, H1.3 and H1.1 expression decreased, and H1.5 was initially induced but fell from day 8 to day 15. The total level of H1 expression was higher upon differentiation (Fig. 1C). In ES[2] cells, H1.0 expression was also increased upon differentiation, and the other H1 variants were, overall, down-regulated, except for H1X, which remained unaltered. Nonetheless, for H1.5, H1.3, H1.4, and, more importantly, H1.2, expression at day 20 was higher than at day 15 (supplemental Fig. S1). At the end of the differentiation process of both ES cell lines, cells expressed H1.0 predominantly, but also H1.2 and H1X. Comparison of the relative contribution of each variant with the total H1 content, at the mRNA level, in the initial ES cells and the resulting differentiated cells at day 20, showed that the contribution of H1.0 doubled upon differentiation (up to 40–45% of the total H1 mRNA), whereas the contributions of H1.2 and H1X remained similar (20–25% of total H1), and those of H1.3 and H1.5 decreased by ∼2-fold (Fig. 1D).
To extend these observations, we investigated changes in expression of H1 variants and key genes during the retinoic acid (RA)-induced differentiation of a human pluripotent embryonal carcinoma cell line, NT2, into neural lineages. As expected, OCT4 and NANOG were down-regulated along differentiation, and PAX6, a neuroectodermal marker gene, and the homeobox HOXA1 gene were up-regulated (Fig. 2A). Initially, all H1 variants except H1.1 were up-regulated to some extent, but later H1.2, H1.3, H1.4, and H1.5 were clearly down-regulated to below base-line levels (Fig. 2B). Only H1.0 and H1X variants increased expression upon differentiation and continued accumulating upon RA treatment beyond the 12-day experiment shown. Similar to ES cells, upon differentiation, the expression of a more diverse repertoire of H1 variants in pluripotent cells was converted into H1.0 mRNA prevalence (Fig. 2C).
FIGURE 2.
Histone H1 variant expression in the course of the RA-induced differentiation of NT2 cells. A and B, NT2 cells were incubated with RA 10−6 m for the indicated days, and the cells were processed for RNA extraction. Expression of several pluripotency, differentiation (A), and H1 genes (B) was determined by RT-qPCR with specific oligonucleotide pairs. Histone H1 variant cDNA amplification was normalized with the values of NT2-derived genomic DNA amplified with the same primers set and GAPDH expression, as described for Fig. 1B. C, proportions of histone H1 variant transcripts in undifferentiated and differentiated NT2 cells. The relative proportions of each H1 variant expression at days 0 and 12 of RA treatment were calculated from the data in B and expressed as percentages of the total H1 content. The means and S.D. of a representative experiment quantified in triplicate are shown.
Diversification of the Histone H1 Variant Content upon Reprogramming of Human Keratinocytes to Induced Pluripotent Stem Cells
We have investigated changes in mRNA expression levels of H1 variants in human keratinocytes (K1 and K2), in iPS cell lines generated from human keratinocytes KiPS4F1 (F1) and KiPS4F4 (F4) (30), and in human fibroblasts (HF) as a control. As expected, FOXA2 was only detected in differentiated cells and absent from iPS cells. The reverse was true for SOX2, OCT4, and NANOG expression, namely, they were absent in differentiated cells and present in iPS cells, similar to the ES cells tested as a control (Fig. 3A). Because NANOG was not included in the transduction to induce reprogramming, its expression was indicative of acquired pluripotency in the iPS lines.
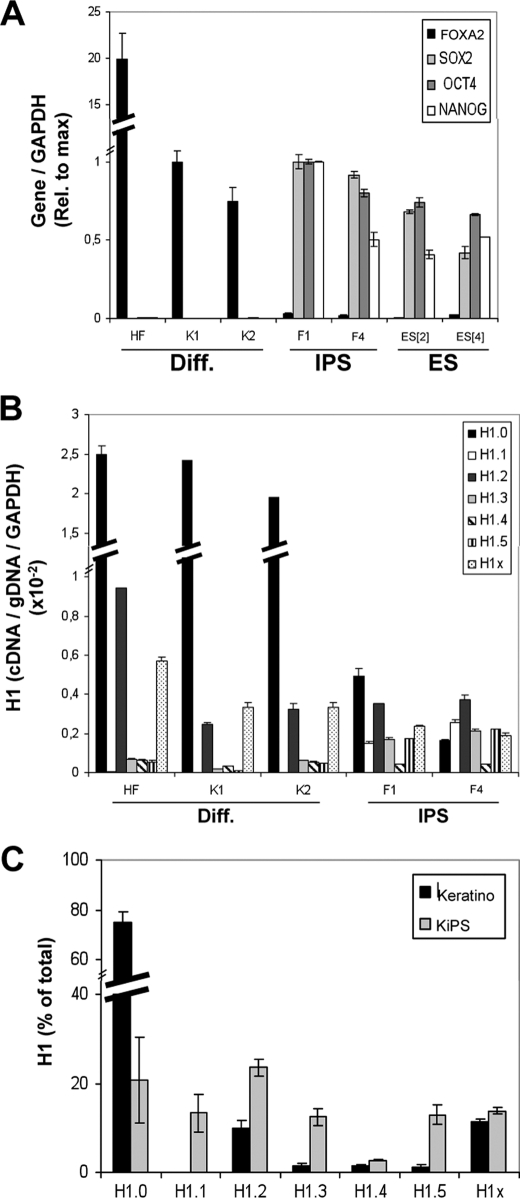
FIGURE 3.
Changes of histone H1 variant gene expression upon reprogramming of human keratinocytes to iPS cells. A, expression of pluripotency or differentiation markers (Diff.) in normal and reprogrammed keratinocytes. RNA was extracted from human cell lines derived from HF, keratinocytes (K1 and K2), iPS cells obtained by reprogramming of keratinocytes (KiPS4F1 and KiPS4F4; named F1 and F4 in the figures), and hES cells (ES[2] and ES[4]) and used to determine expression of FOXA2, SOX2, OCT4, and NANOG by RT-qPCR with specific primer pairs. GAPDH expression was measured for normalization. Expression of each gene relative to the maximal value in keratinocytes (FOXA2) or iPS cells (SOX2, OCT4, and NANOG) is represented. B, histone H1 variant gene expression in normal and reprogrammed keratinocytes. The RNA samples of HF, keratinocytes (K1 and K2) and iPS cells (KiPS4F1 and KiPS4F4) were used to measure histone H1 expression by RT-qPCR with variant-specific oligonucleotide pairs. Histone H1 variant cDNA amplification was normalized with the values of HF or K1-derived genomic DNA amplified with the same primer set and GAPDH expression, as described in Fig. 1B. C, proportions of histone H1 variant transcripts in normal and reprogrammed keratinocytes. The relative proportion of each H1 variant expression in keratinocytes and iPS cells was calculated from the data in B and expressed as a percentage of the total H1 content. The data are expressed as the mean values for K1 and K2 (keratinocytes) or KiPS4F1 and KiPS4F4 (KiPS), respectively. The means and S.D. of a representative experiment quantified in triplicate are shown.
Comparison of H1 variant expression in keratinocytes and iPS cells showed that the expression of H1.0 and H1X decreased upon reprogramming; H1.1, H1.3, and H1.5 were up-regulated; and H1.2 and H1.4 remained virtually unaltered (Fig. 3B and supplemental Fig. S2). Comparing the relative contribution of each variant to the total H1 transcript content, in keratinocytes and in the resulting reprogrammed iPS cells, revealed that H1.0 represented almost 80% of the H1 content in keratinocytes but was reduced to ∼20% upon reprogramming (Fig. 3C). Further, in keratinocytes, H1.2 and H1X represented ∼10% of total H1 transcripts, and there were only traces of the other variants (<2%). By contrast, in reprogrammed cells all variants (except H1.4) had similar levels of expression (12–23%), H1.2 being the most abundant variant.
Comparing these results with data in Fig. 1D, it can be seen that the relative abundance of H1 variants in iPS cells was similar to that in ES cells, except that H1.1 was less abundant in ES. Furthermore, H1 variant expression in differentiated ES cells resembled the H1 distribution in keratinocytes and fibroblasts, except that H1.0 made a greater contribution in adult somatic cells. In summary, pluripotent cells have lower levels of H1.0 and higher levels of H1.1, H1.3, and H1.5, compared with adult differentiated cells. Overall, pluripotent cells have a more diverse repertoire of histone H1 variants. However, total H1 expression appears lower in pluripotent cells than in differentiated cells, because of the great reduction in H1.0.
Pluripotent Cells Have Lower Levels of Histone H1.0 and Higher Levels of H1.3 and H1.5 Compared with Differentiated Cells
To confirm the results obtained by measuring H1 variant mRNA accumulation by RT-qPCR, H1 histones were extracted from the cell lines used in the aforementioned experiments, and the abundance of the different H1 variants was assessed by immunohybridization with specific antibodies (Fig. 4A and supplemental Fig. S3). Because the level of H1.2 expression was found to be relatively similar across the cell lines by RT-qPCR, the protein levels of this variant were expected to serve as loading control for normalization. Although H1.2 levels were very homogeneous, H1.0 was overrepresented in differentiated cells, and H1.3 and H1.5 were more abundant in iPS and ES cells. Because H1X hybridization showed two products, the observed increase in fibroblasts has to be taken cautiously (Fig. 4A). When the values from several samples (supplemental Fig. S3) were pooled and normalized with respect to H1.2 abundance, the differences between differentiated and pluripotent (iPS +ES) cells were clear (Fig. 4B).
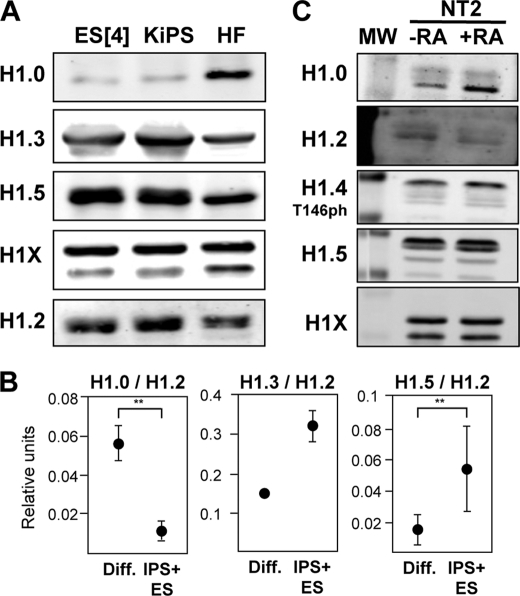
FIGURE 4.
Histone H1 variant content analyzed by immunohybridization with specific antibodies. A, immunoblot of H1 variant abundance in differentiated (Diff.) and pluripotent cells. Histone H1 was extracted from hES, iPS cells obtained by reprogramming of keratinocytes and HF, resolved in SDS-PAGE, and immunoblotted with antibodies specifically recognizing human H1.0, H1.2, H1.3, H1.5, or H1X. Equivalent amounts of protein were loaded for each sample. B, relative abundance of H1.0, H1.3, and H1.5 normalized by H1.2 abundance. Histone H1 was extracted from several differentiated (n = 4) and pluripotent (n = 4) cell lines and analyzed by immunoblot as in A. The immunoblot (supplemental Fig. S3) was quantified, and the relative light intensity units for all differentiated (HF, K1, and K2) or pluripotent (KiPS4F1, KiPS4F4, ES[2], and ES[4]) samples were pooled. Graphics represent means and S.D. of the abundance of each tested H1 variant normalized to H1.2 units. The differences were interrogated with the Student's t test for statistical significance when possible. **, p < 0.05. C, H1 variant abundance in undifferentiated and differentiating NT2 cells. H1 was extracted from untreated or 10-day RA-treated NT2 cells and immunoblotted with the indicated antibodies as in A. The molecular mass marker (MW) bands shown correspond to 34 and 26 kDa.
Analysis of H1 variants accumulation in NT2 cells showed a significant increase of H1.0 in the course of retinoic acid treatment for 10 days but limited or no reduction of other variants, in contrast with the detected decrease in mRNA levels (Fig. 4C). Because of the relatively high stability of H1 proteins, mRNA decays may need a longer time to translate into H1 protein reduction.
In conclusion, the determination of the abundance of H1 variants by immunohybridization confirmed and validated the measurements of H1 gene expression by RT-qPCR with specific oligonucleotides, indicating that H1 variant accumulation greatly depends on the rate of expression of their encoding genes. Additionally, relative proportions of H1 variants are not randomly established but rather follow precise cellular programs, some variants being more common in differentiated cells (H1.0 and to a less extend H1X), whereas others are more closely linked to pluripotency (H1.1, H1.3, and H1.5), supporting the hypothesis that these variants may be functionally different.
The H1.0 Promoter Contains a Bivalent Domain in Pluripotent Cells
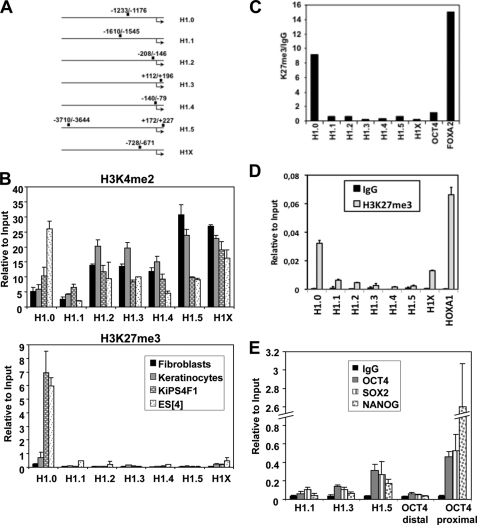
To investigate how expression of the different H1 variant-encoding genes is controlled in the transition between pluripotent and differentiated cells, we used ChIP to explore the presence of histone marks related to transcriptional activation (H3K4me2) and repression (H3K27me3), as well as the presence of pluripotency transcription factors, at their regulatory regions (Fig. 5A). H3K4me2 was detected at all H1 variant genomic regions and in all cell lines analyzed. In contrast, H3K27me3 was only detected at the H1.0 promoter and only in pluripotent cells (ES[4] and KiPS4F1) (Fig. 5B). To confirm that these two histone marks were present simultaneously at the H1.0 promoter in ES cells, chromatin immunoprecipitated with the anti-H3K4me2 antibody was used for reimmunoprecipitation with the anti-H3K27me3 antibody (re-ChIP), and a positive result was obtained for the H1.0 promoter as well as for the bivalent gene FOXA2 (Fig. 5C). Analysis of histone H3 methylation marks at the H1 promoters in undifferentiated NT2 cells revealed that in these cells the H1.0 promoter is also marked with H3K27me3 (Fig. 5D). The presence of bivalent marks at the promoter of the H1.0 variant suggest that this gene is poised for activation during the differentiation of human ES cells.
FIGURE 5.
Detection of H3K4me2 and H3K27me3 and pluripotency transcription factors binding at the promoters of histone H1 variants. A, representation of the regulatory regions of the H1 variants and the amplified areas in ChIP coupled to real time PCR. The numbers refer to the positions from the transcriptional start site. B, detection of histone H3 Lys-4 and Lys-27 methylation at the promoters of the somatic human histone H1 variant genes in differentiated and pluripotent cells. Cross-linked chromatin from human fibroblasts, human keratinocytes, iPS cells (KiPS4F1), and ES[4] cells was used for ChIP experiments with specific antibodies for H3K4me2 (upper panel) and H3K27me3 (lower panel). The precipitated DNA fragments were subjected to qPCR with primers for the regulatory regions of the indicated H1 variants. For variant H1.5, oligonucleotides covering +172/+227 were used. Amplification of input DNA (representing 1% of immunoprecipitated DNA) was used for normalization. The means and S.D. of at least two independent quantifications are shown. C, bivalent domains are found at the histone H1.0 promoter in ES cells. DNA precipitated with the H3K4me2 antibody from ES[4] cells was submitted to a second immunoprecipitation (re-ChIP) with antibody against H3K27me3 or with unrelated serum (rabbit IgG) as a control. The precipitated DNA fragments were subjected to qPCR with specific primers for the indicated H1 variants and for the promoter regions of OCT4 (−2464/−2399) and FOXA2 (+173/+229) genes as controls. D, detection of H3K27me3 at the regulatory region of H1 variants in undifferentiated NT2 cells. NT2 cells were submitted to ChIP analysis with an antibody for H3K27me3 or rabbit IgG as a control, and qPCR was performed for the H1 variant regulatory regions as described in Fig. 5B and for the HOXA1 promoter. The data are normalized to amplified input DNA. The means and S.D. of a representative experiment quantified in triplicate are shown. E, pluripotency factors are found at the promoter of histone H1 variants expressed in ES cells. Chromatin from ES[4] cells was immunoprecipitated with antibodies against OCT4, SOX2, or NANOG pluripotency transcription factors or IgG as a control. The precipitated DNA fragments were subjected to qPCR with specific primers for H1.1, H1.3, and H1.5 (−3710/−3644) and for two promoter regions of OCT4 (distal, −3399/−3338; and proximal, −1615/−1553) as controls. The data are normalized to amplified input DNA. The means and S.D. of at least two independent quantifications are shown.
Pluripotency Transcription Factors Are Present at the Promoter of Certain Histone H1 Variants
H1.1, H1.3, and H1.5 variants might have a prevalent role in pluripotent cells, because they are up-regulated in these cells. Moreover, genome-wide occupancy studies in human ES cells describe binding regions for OCT4, SOX2, and NANOG at the regulatory regions of H1.1, H1.3, and H1.5 variants (19). To verify this data, we investigated the presence of these factors at the promoters of these variants in ES cells, using the OCT4 promoter as a positive control (Fig. 5E). The three factors were unequivocally found at H1.5 promoter, at a lower level in H1.3, and only at marginal levels in H1.1. Therefore, H1.3 and H1.5 seem to be direct targets of the pluripotency network, in agreement with the fact that these variants are expressed at higher levels in pluripotent cells than somatic cells.
H1.0 Knockdown Impairs the Differentiation Program of Human ES Cells
Next, we asked whether those H1 variants that were differentially expressed in ES cells and somatic cells could be involved in self-renewal or differentiation. For that, stable ES[4] cell lines were obtained after transduction with a previously characterized shRNA-expressing lentiviral vector against the H1.3, H1.5, or H1.0 variants or a control random shRNA. Unfortunately, we were unable to significantly reduce the levels of H1.5 in human ES cells, but the levels of H1.3 and H1.0 could be efficiently knocked down (supplemental Fig. S4). The knockdown of H1.3 or H1.0 did not affect the self-renewal properties of ES[4] as judged by the absence of phenotypical and gene expression changes compared with the control cell line (supplemental Fig. S4). However, in vitro differentiation experiments showed that the silencing of pluripotency factors was delayed in cells knockdown for H1.0 compared with the control line, and the differentiation-related genes HNF4, FOXA2, and SOX17 were not induced, indicating that differentiation was impaired in cells lacking this H1 variant (Fig. 6). In the course of differentiation, the expression of all H1 variants in the H1.0 knocked down cells followed the same pattern as in control cells, including H1.3 and H1.5 down-regulation, except that H1.0 was not accumulated in the course of this process because of the ongoing action of the specific shRNA (Fig. 6 and supplemental Fig. S5).
FIGURE 6.
Interference of H1.0 expression in differentiating hES cells. ES[4] H1.0 knockdown (shH1.0) or control (shRD) cells were cultured in differentiation media for the indicated days and processed for RNA extraction. The expression of several pluripotency, differentiation, and H1 genes was determined by RT-qPCR with specific oligonucleotide pairs. GAPDH expression was measured for normalization. The means and S.D. of a representative experiment quantified in triplicate are shown.
Histone H1 Accumulates at the Promoter of Pluripotency Factors in Differentiated Cells and during Differentiation
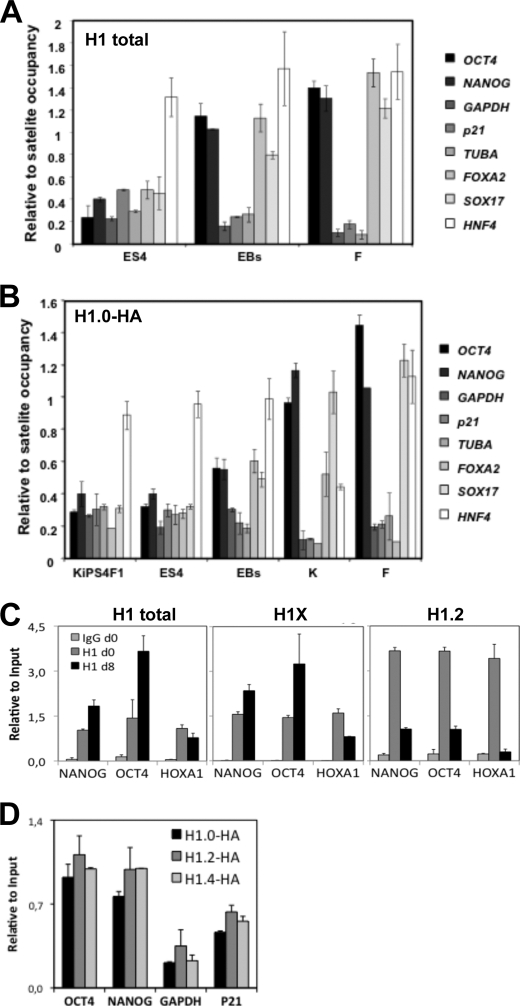
The effects of the H1.0 knockdown in the differentiation of human ES cells prompted us to investigate whether changes in H1 variant expression during differentiation could have a regulatory role in the expression of pluripotency or differentiation genes. For that, we performed ChIP assays with H1 antibodies in undifferentiated and differentiated ES cells, as well as iPS cells, keratinocytes and fibroblasts. First, H1 occupancy at several promoters was compared in undifferentiated ES[4], ES[4]-derived EBs at day 15 of differentiation, and fibroblasts using a pan-H1 antibody (Fig. 7A). H1 was accumulated at OCT4 and NANOG promoters upon differentiation, as well as at the FOXA2 and SOX17 regulatory regions, but was displaced from very actively transcribed genes such as GAPDH, TUBA, and p21-encoding gene promoters. In somatic cell types, the differences in occupancy between repressed pluripotency genes and actively transcribed genes became even more conspicuous.
FIGURE 7.
Recruitment of H1 to the regulatory region of pluripotency and differentiation genes. A, recruitment of total H1 in undifferentiated ES[4], ES-derived EBs at day 15 of differentiation and human fibroblasts (F). ChIP with a pan-H1 antibody (AE-4) was performed, and H1 occupancy at several promoters was interrogated with specific oligonucleotides by qPCR. Amplification of input DNA (representing 1% of immunoprecipitated DNA) was used for normalization. To allow proper comparison of the H1 occupancy between different cell lines, the data were normalized to the mean of H1 occupancy at two satellite regions (SATAB and SATXL) that may represent a highly invariable portion of the genome in terms of H1 occupancy. B, recruitment of overexpressed HA-tagged H1.0 in KiPS4F1, ES[4], and ES-derived EBs at day 15 of differentiation, keratinocytes (K) and fibroblasts (F). The cells were transduced with an HA-tagged H1.0 expressing lentivirus and submitted to ChIP analysis with an anti-HA antibody, and H1.0 occupancy at several gene promoters was interrogated as in A. To overcome potential differences in the expression of HA-H1.0 in each transduced cell line, the data were normalized to the mean of HA-H1.0 occupancy at two satellite regions (SATAB and SATXL). C, recruitment of H1 in undifferentiated and differentiated NT2 cells. ChIP with antibodies against total H1 (AE-4), H1X, or H1.2 was performed, and occupancy at the indicated promoters was interrogated with specific oligonucleotides by qPCR. Amplification of input DNA was used for normalization. D, H1 occupancy of NANOG and OCT4 promoters in breast cancer cells. T47D cells stably expressing HA-tagged H1.0, H1.2, or H1.4 were submitted to ChIP analysis with an anti-HA antibody, and H1 occupancy at several promoters was interrogated with specific oligonucleotides by qPCR. The data are normalized to amplified input DNA. The means and S.D. of at least two independent quantifications of representative experiments are shown throughout the figure.
Because H1.0 accumulated upon differentiation becoming the predominant H1 variant and its knockdown affects differentiation, we hypothesized that this histone may have a key role in repressing the expression of pluripotency factors. Because ChIP grade anti-H1.0 antibodies are not available, to specifically assess the presence of H1.0 at promoters by ChIP, we transduced ES[4], iPS cells, keratinocytes, and fibroblasts with an HA epitope-tagged H1.0 expression lentivirus. ChIP with an anti-HA antibody confirmed that H1.0 accumulates at OCT4, NANOG, FOXA2, and SOX17 promoters upon differentiation but not at the promoters of actively transcribed genes (Fig. 7B). Overall, occupancy of total H1 and H1.0 showed similar patterns in the tested cell lines.
A recent report suggests that the H1X variant is up-regulated and recruited to the NANOG promoter during the differentiation of NT2 cells (31). This prompted us to investigate whether other H1 variants different from H1.0 could bind the regulatory regions of pluripotency genes. This was assessed in NT2 cells using specific antibodies against H1.2 and H1X and in previously described T47D stable cell lines expressing HA-tagged versions of the H1 variants for which specific ChIP grade antibodies are not available (13). Our results confirm that H1X is recruited to these promoters during NT2 differentiation but displaced from the HOXA1 promoter, in agreement with the induction of this gene. H1.2 was displaced from the tested promoters, according to its down-regulation during differentiation (Fig. 7C). The data in T47D breast cancer cells where pluripotency genes are repressed show that these promoters can be occupied by any available HA-tagged variant (Fig. 7D). Taken together, these findings indicate that repressed pluripotency gene promoters are not exclusively occupied by H1.0 and further suggest that the dynamic changes in H1 variant expression during differentiation could contribute to regulate the expression of pluripotency factor-encoding genes.
DISCUSSION
Using human ES, iPS, and teratocarcinoma cell lines, we have investigated the changes in the proportions of H1 variants in the course of differentiation and as a consequence of the reprogramming of adult somatic cells to pluripotency. The relative levels of H1 variants in pluripotent and differentiated cells were different, mainly attributable to H1.0 being prevalent in differentiated cells and higher levels of expression of H1.1, H1.3, and H1.5 in pluripotent than in differentiated cells. Interestingly, the proportions of H1 variants were similar in all pluripotent cells, irrespective of their embryonic or somatic origin. Similarly, proportions of H1 histones were consistent when comparing keratinocytes or fibroblasts and in vitro differentiated ES cells, except that H1.0 represented a higher proportion of the total H1 content in adult somatic cells. The expression of H1.2 and H1.4 was not characteristically altered upon reprogramming or differentiation, but the relative levels were very different, with H1.2 representing ∼20% of the H1 content in all cell lines tested here and H1.4 accounting for less than 5%. H1.2 and H1.4 are the only somatic linker histone variants that have been found in all of the cell lines analyzed to date, accounting for at least 20% of the H1 content (32, 33), so it may be speculated that these two isoforms play an important role in cell homeostasis. This was also suggested by Sancho et al. (13) as knocking down H1.2 and H1.4, but not H1.0, H1.3, or H1.5, affected the proliferation of a breast cancer cell line. Despite the fact that it has been considered that H1X is ubiquitously expressed (34, 35), our results suggests that H1X may have a more prevalent role in differentiated cells than in pluripotent cells because H1X expression was diminished upon reprogramming and slightly up-regulated during differentiation. Overall, the histone H1 changes in expression that we report here follow the same trend as those reported by Helliger et al. (26) in the course of in vitro induced differentiation of erythroleukemic cells, in which the murine homologs of H1.1, H1.3, and H1.5 were down-regulated, whereas H1.0 was strongly up-regulated, and levels of H1.2 and H1.4 increased by more or less, depending on which inducer treatment was used.
The changes in the expression of particular H1 variants during differentiation and reprogramming suggest that these genes are controlled by the master regulators of self-renewal or differentiation. H1.1 was absent from keratinocytes and fibroblasts but was up-regulated upon reprogramming and also detected in human ES cell lines. Therefore, in humans this variant appears to be specific to pluripotent cells, although it represents a small percentage of the total H1 content in these cells. Indeed the H1.1 expression is restricted to a few tissues in the adult mouse but has also been detected in mouse oocytes and early embryos, in male germ cells during early stages of spermatogenesis, and in several tissues shortly after birth (36, 37). Despite the fact that the occupancy of the H1.1 promoter by pluripotency factors has been described in genome wide approaches (19), we did not detect their presence at the reported site. These factors could, however, still be located at other regions of the H1.1 gene and contribute to maintaining the expression of this variant in pluripotent cells. Importantly, the detection of the pluripotency transcription factors OCT4, SOX2, and NANOG at the promoters of H1.3 and H1.5 encoding genes in human ES cells and the fact that these two H1 variants are more highly expressed in pluripotent cells than in differentiated cells indicate that these factors may be acting as positive regulators of these genes and suggest that H1.3 and H1.5 could have a role in maintaining pluripotency/self-renewal, either by producing a more relaxed chromatin structure or by specifically contributing to the regulation of a subset of genes, such as repressing certain tissue-specific genes. However, we did not observe significant effects after knocking down H1.3 in human ES cells, suggesting that the functions of this variant can be assumed by other variants in pluripotent cells.
Several key genes critical for the process of differentiation are kept silenced in pluripotent cells but poised for activation upon initiation of specific developmental programs by harboring bivalent domains at their regulatory regions (21, 22). Our finding that the H1.0 promoter contains bivalent domains in pluripotent cells suggests that it has a key role as a master regulator of differentiation and helps explain how its expression is controlled in pluripotent cells and rapidly activated as soon as cells enter into a developmental program. It is likely that the recruitment of the specific H3K27 demethylases UTX and JMJD3 (38) during differentiation triggers the rapid removal of the H3K27 methylation mark and the transcriptional activation of this gene. Interestingly, we have observed the re-establishment of the bivalent domain at the H1.0 promoter upon reprogramming of keratinocytes to iPS cells, through the regaining of H3K27 methylation, and without altering the H3K4 methylation levels. This fact indicates that the H3K27 is actively methylated during the reprogramming process and that regaining of this mark is needed to down-regulate H1.0 expression. OCT4, SOX2, and NANOG occupy the promoters of many genes containing bivalent domains, pointing to these factors having a role in the regulation of these domains; however, genome wide studies performed in human ES were not able to confirm the presence of the pluripotency factors at the regulatory regions of the H1.0 variant (19).
Pluripotent cells have notable levels of fluid chromatin that likely allow access to transcription factors that maintain the programs for self-renewal and pluripotency and eventually the binding of pioneer transcription factors during early stages of differentiation. During differentiation, chromatin becomes more condensed and refractory to stimulation. Concomitantly, the affinity of H1 for chromatin increases and its dynamic exchange slows down (20, 39). As a consequence, the average number of H1 molecules per nucleosome increases (40). These reported facts are in agreement with our observations. In ES cells, the occupancy of total H1, and more specifically H1.0, is lower in highly expressed (OCT4, NANOG, GADPH, and TUBA) and poised bivalent genes (SOX17, FOXA2, and p21) compared with tissue-specific repressed genes that do not contain bivalent domains (HNF4). During differentiation and in differentiated cells, the regions of euchromatin and heterochromatin become more defined regarding H1 content, being repressed genes heavily loaded with H1. This could be caused by an overall increase in the H1 content in differentiating cells and/or by the reported changes in the proportions of H1 variants. The fact that H1.0 is among the strongest condensers of histone H1 subtypes (41) suggests that changes in its level of expression may have structural consequences with respect to the compaction of chromatin. In agreement with this, we observed that the knockdown of H1.0 in human ES cells have dramatic effects in differentiation, causing a delay in the silencing of pluripotency genes and defects in the activation of differentiation genes. The proper differentiation of human ES requires the silencing of pluripotency genes as well as the coordinated activation and repression of genes involved in the specification of the three embryonic layers. Our results point to a role for H1.0 in the silencing of pluripotency genes during differentiation; however, the direct effects on differentiation genes are more difficult to dissect. Although we cannot rule out the possibility that H1.0 is involved in the transcriptional activation of these genes, it seems more likely that the effects of the H1.0 knockdown on the induction of differentiation genes are at the level of repression of bivalent domain containing genes. Although the endoderm-related bivalent genes SOX17 and FOXA2 are strongly induced during differentiation, immunostaining reveals that only 10–20% of cells stain positive for these markers in the EBs (data not shown), suggesting that in most of the cells, these genes are becoming repressed and likely recruiting H1.0 to their promoters. Moreover, H1.0 is displaced from the bivalent gene p21 during differentiation in correlation with its induction in all three embryonic layers. Despite the fact that most H1 variants can occupy the promoters of pluripotency genes in differentiated cells, the effects of the H1.0 knockdown can be explained by the prevalent expression of H1.0 during differentiation, although the relative abundance of the different variants at these promoters remains to be determined.
In conclusion, we have shown that specific changes in the expression and accumulation of histone H1 variants occur in the course of differentiation of human ES cells or reprogramming of adult somatic cells to pluripotency. Using the two reverse systems, we have found nearly complementary changes in H1 expression that support the view that there are functional differences between H1 variants. More specifically, we have shown that the H1.0 variant plays a critical role in the differentiation of human ES cells. Overall, our data suggest that changes in the expression of H1 variants are key determinants of the potency of a cell and regulators of the differentiation process.
Supplementary Material
Acknowledgments
We thank M. Carrió and L. Casano for establishment of primary cell lines and M. Sancho for construction of H1 expression and knockdown vectors. We also thank M. A. González at Hospital del Mar for providing tissues for the establishment of primary cultures, and L. di Croce (Centro de Regulació Genomica) for providing reagents.
This work was supported by the Ministerio de Ciencia e Innovación of Spain (MICINN); Fondo Europeo Desarrollo Regional Grants 200820I090 and BFU2008-00359/BMC (to A. J.) and RYC-2007-01510 and SAF2009-08588 (to M. J. B.); and Generalitat de Catalunya Grant 2009-SGR-1222 (to A. J.). Work at the laboratory of J.C.I.B. is supported by grants from the G. Harold and Leila Y. Mathers Charitable Foundation, Sanofi-Aventis, MICINN, Centro Investigación Biomédica en Red de Bioingeniería, and Fundacion Cellex.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- ES
- embryonic stem
- iPS
- induced pluripotent stem
- EB
- embryoid body
- qPCR
- quantitative PCR
- RA
- retinoic acid
- HF
- human fibroblast.
REFERENCES
- 1. Brown D. T. (2003) Biochem. Cell Biol. 81, 221–227 [DOI] [PubMed] [Google Scholar]
- 2. Bustin M., Catez F., Lim J. H. (2005) Mol. Cell 17, 617–620 [DOI] [PubMed] [Google Scholar]
- 3. Garcia B. A., Busby S. A., Barber C. M., Shabanowitz J., Allis C. D., Hunt D. F. (2004) J. Proteome Res. 3, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 4. Wisniewski J. R., Zougman A., Krüger S., Mann M. (2007) Mol. Cell Proteomics 6, 72–87 [DOI] [PubMed] [Google Scholar]
- 5. Wood C., Snijders A., Williamson J., Reynolds C., Baldwin J., Dickman M. (2009) FEBS J. 276, 3685–3697 [DOI] [PubMed] [Google Scholar]
- 6. Parseghian M. H., Newcomb R. L., Hamkalo B. A. (2001) J. Cell Biochem. 83, 643–659 [DOI] [PubMed] [Google Scholar]
- 7. Happel N., Doenecke D. (2009) Gene 431, 1–12 [DOI] [PubMed] [Google Scholar]
- 8. Izzo A., Kamieniarz K., Schneider R. (2008) Biol. Chem. 389, 333–343 [DOI] [PubMed] [Google Scholar]
- 9. Fan Y., Nikitina T., Zhao J., Fleury T. J., Bhattacharyya R., Bouhassira E. E., Stein A., Woodcock C. L., Skoultchi A. I. (2005) Cell 123, 1199–1212 [DOI] [PubMed] [Google Scholar]
- 10. Shen X., Gorovsky M. A. (1996) Cell 86, 475–483 [DOI] [PubMed] [Google Scholar]
- 11. Lin Q., Inselman A., Han X., Xu H., Zhang W., Handel M. A., Skoultchi A. I. (2004) J. Biol. Chem. 279, 23525–23535 [DOI] [PubMed] [Google Scholar]
- 12. Hellauer K., Sirard E., Turcotte B. (2001) J. Biol. Chem. 276, 13587–13592 [DOI] [PubMed] [Google Scholar]
- 13. Sancho M., Diani E., Beato M., Jordan A. (2008) PLoS Genet. 4, e1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan Y., Sirotkin A., Russell R. G., Ayala J., Skoultchi A. I. (2001) Mol. Cell Biol. 21, 7933–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee H., Habas R., Abate-Shen C. (2004) Science 304, 1675–1678 [DOI] [PubMed] [Google Scholar]
- 16. Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. (2004) Mol. Cell 16, 93–105 [DOI] [PubMed] [Google Scholar]
- 17. Daujat S., Zeissler U., Waldmann T., Happel N., Schneider R. (2005) J. Biol. Chem. 280, 38090–38095 [DOI] [PubMed] [Google Scholar]
- 18. Kim K., Choi J., Heo K., Kim H., Levens D., Kohno K., Johnson E. M., Brock H. W., An W. (2008) J. Biol. Chem. 283, 9113–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., Misteli T. (2006) Dev. Cell 10, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 22. Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A. G. (2006) Nat. Cell Biol. 8, 532–538 [DOI] [PubMed] [Google Scholar]
- 23. Gargiulo G., Levy S., Bucci G., Romanenghi M., Fornasari L., Beeson K. Y., Goldberg S. M., Cesaroni M., Ballarini M., Santoro F., Bezman N., Frigè G., Gregory P. D., Holmes M. C., Strausberg R. L., Pelicci P. G., Urnov F. D., Minucci S. (2009) Dev. Cell 16, 466–481 [DOI] [PubMed] [Google Scholar]
- 24. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 25. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 26. Helliger W., Lindner H., Grübl-Knosp O., Puschendorf B. (1992) Biochem. J. 288, 747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lennox R. W., Cohen L. H. (1983) J. Biol. Chem. 258, 262–268 [PubMed] [Google Scholar]
- 28. Piña B., Martínez P., Suau P. (1987) Eur. J. Biochem. 164, 71–76 [DOI] [PubMed] [Google Scholar]
- 29. Raya A., Rodríguez-Pizà I., Arán B., Consiglio A., Barri P. N., Veiga A., Izpisúa Belmonte J. C. (2008) Cold Spring Harbor Symp. Quant. Biol. 73, 127–135 [DOI] [PubMed] [Google Scholar]
- 30. Aasen T., Raya A., Barrero M. J., Garreta E., Consiglio A., Gonzalez F., Vassena R., Bilić J., Pekarik V., Tiscornia G., Edel M., Boué S., Izpisúa Belmonte J. C. (2008) Nat. Biotechnol. 26, 1276–1284 [DOI] [PubMed] [Google Scholar]
- 31. Shahhoseini M., Favaedi R., Baharvand H., Sharma V., Stunnenberg H. G. (2010) FEBS Lett. 584, 4661–4664 [DOI] [PubMed] [Google Scholar]
- 32. Meergans T., Albig W., Doenecke D. (1997) DNA Cell Biol. 16, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 33. Kratzmeier M., Albig W., Meergans T., Doenecke D. (1999) Biochem. J. 337, 319–327 [PMC free article] [PubMed] [Google Scholar]
- 34. Yamamoto T., Horikoshi M. (1996) Gene 173, 281–285 [DOI] [PubMed] [Google Scholar]
- 35. Happel N., Schulze E., Doenecke D. (2005) Biol. Chem. 386, 541–551 [DOI] [PubMed] [Google Scholar]
- 36. Franke K., Drabent B., Doenecke D. (1998) Biochim. Biophys. Acta 1398, 232–242 [DOI] [PubMed] [Google Scholar]
- 37. Fu G., Ghadam P., Sirotkin A., Khochbin S., Skoultchi A. I., Clarke H. J. (2003) Biol. Reprod. 68, 1569–1576 [DOI] [PubMed] [Google Scholar]
- 38. Swigut T., Wysocka J. (2007) Cell 131, 29–32 [DOI] [PubMed] [Google Scholar]
- 39. Yellajoshyula D., Brown D. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18568–18573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Woodcock C. L., Skoultchi A. I., Fan Y. (2006) Chromosome Res. 14, 17–25 [DOI] [PubMed] [Google Scholar]
- 41. Clausell J., Happel N., Hale T. K., Doenecke D., Beato M. (2009) PLoS One 4, e0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.