Background: Protein deacylation by sirtuins yields O-acyl-ADP-ribose molecules, which are implicated in cell signaling and metabolism.
Results: Structural and functional analysis of a human orphan macrodomain protein reveals a common core structure and ability to hydrolyze O-acyl-ADP-ribose.
Conclusion: Diverse macrodomains are capable of hydrolyzing O-acyl-ADP-ribose but utilize a different set of catalytic amino acids.
Significance: Macrodomain-containing proteins provide a biochemical and biological link to sirtuin-dependent deacylation.
Keywords: Histone Deacetylase, Hydrolases, Metabolism, Protein Acylation, Sirtuins, NAD, O-Acetyl-ADP-ribose, Deacylation, Macrodomains, sir2
Abstract
Post-translational modification of proteins/histones by lysine acylation has profound effects on the physiological function of modified proteins. Deacylation by NAD+-dependent sirtuin reactions yields as a product O-acyl-ADP-ribose, which has been implicated as a signaling molecule in modulating cellular processes. Macrodomain-containing proteins are reported to bind NAD+-derived metabolites. Here, we describe the structure and function of an orphan macrodomain protein, human C6orf130. This unique 17-kDa protein is a stand-alone macrodomain protein that occupies a distinct branch in the phylogenic tree. We demonstrate that C6orf130 catalyzes the efficient deacylation of O-acetyl-ADP-ribose, O-propionyl-ADP-ribose, and O-butyryl-ADP-ribose to produce ADP-ribose (ADPr) and acetate, propionate, and butyrate, respectively. Using NMR spectroscopy, we solved the structure of C6orf130 in the presence and absence of ADPr. The structures showed a canonical fold with a deep ligand (ADPr)-binding cleft. Structural comparisons of apo-C6orf130 and the ADPr-C6orf130 complex revealed fluctuations of the β5-α4 loop that covers the bound ADPr, suggesting that the β5-α4 loop functions as a gate to sequester substrate and offer flexibility to accommodate alternative substrates. The ADPr-C6orf130 complex identified amino acid residues involved in substrate binding and suggested residues that function in catalysis. Site-specific mutagenesis and steady-state kinetic analyses revealed two critical catalytic residues, Ser-35 and Asp-125. We propose a catalytic mechanism for deacylation of O-acyl-ADP-ribose by C6orf130 and discuss the biological implications in the context of reversible protein acylation at lysine residues.
Introduction
The macrodomain is an evolutionarily conserved protein fold found in isolation or embedded within larger polypeptides in the genomes of bacteria, archaea, and eukaryotes (1). Named for their presence in the C terminus of the large variant core histone macroH2A (2), macrodomains adopt a globular α/β-fold of ∼140 residues that contain a deep cleft that serves as a ligand-binding site (3, 4). Macrodomain-containing proteins participate in a diverse array of cellular functions (4, 5), and recent reports have shown that several macrodomains can bind NAD+ metabolites including ADP-ribose (ADPr),4 poly(ADPr), and 2-O-acetyl-ADP-ribose (OAADPr) (6–9). Eleven macrodomain proteins encoded by 10 genes (7, 10, 11) have been identified in humans. Structural studies of the ADPr-binding core of histone variant macroH2A1.1 in complex with ADPr have shed light on macroH2a1.1-dependent chromatin rearrangements upon PARP1 activation (12). In addition, structural genomics research has generated the structures of two of the three macrodomain-containing poly(ADPr) polymerases (PARP14 and PARP15) in complex with ADPr (PDB codes 3Q71 and 3KH6, respectively). No structural information is available for the human macrodomain proteins GDAP2 and CHD1L (ALC1).
Our research has focused on the function of the novel metabolite OAADPr. OAADPr is produced in protein deacetylation reactions catalyzed by NAD+-dependent protein deacetylases (13, 14), which regulate gene silencing, metabolic enzymes, and life span (15–18). OAADPr has been implicated as a signaling molecule that modulates cellular processes and harnesses the regulation of metabolism and gene activity (19–21). Recently we have shown that a group of macrodomain proteins, i.e. MacroD-like proteins, including human MacroD1, human MacroD2, Escherichia coli YmdB, and the sirtuin-linked SAV0325 protein from Staphylococcus aureus, function as OAADPr deacetylases, catalyzing the deacetylation of OAADPr to form ADP-ribose and acetate (9). Mutagenesis and modeling of ADPr into the putative active site of MacroD1 suggested important roles for Asn-171, Asn-174, Asp-184, and His-188. Unlike other human macrodomain proteins, MacroD1 and MacroD2 contain no other known functional domains. The OAADPr deacetylation activity functionally links the MacroD-like proteins with the NAD+-dependent protein deacetylases, connecting metabolic regulation with chromatin structure and gene activity (5–7, 22).
C6orf130 is a unique stand-alone 17-kDa human macrodomain protein of unknown function. The 152-amino acid protein is encoded by open reading frame 130 of chromosome 6 (23). It shares the least amino acid sequence similarity to the other MacroD-like proteins, among which human MacroD1 and MacroD2 are closely related and share more similarity to E. coli YmdB and the sirtuin-linked SAV0325 protein from S. aureus. The divergent amino acid sequence of C6orf130 makes it a distinct branch on the phylogenetic tree (9). C6orf130 transcription and expression are enriched in chronic lymphocytic leukemia cells and in B-cells of the chronic lymphocytic leukemia patients when effective graft-versus-leukemia responses are achieved after donor lymphocyte infusion (24). C6orf130 can therefore be used as a biomarker for chronic lymphocytic leukemia and for monitoring the effectiveness of adoptive immunotherapy for this disease. The high level expression of C6orf130 in chronic lymphocytic leukemia suggests the possibility of using C6orf130 as a potential immunogen for developing chronic lymphocytic leukemia-specific vaccines. A molecular understanding of the association of C6orf130 with chronic lymphocytic leukemia is not understood. In a recent proteomic analysis, the putative C6orf130 ortholog from Torpedo californica was identified in the electric organ (25). To understand the molecular function of this distinct member of the macrodomain family, we investigated the catalytic and structural properties of human C6orf130.
Here we report the solution structures of C6orf130 in the apo and ADPr bound states using NMR spectroscopy as well as the biochemical characterization of its catalytic properties. In this study we show that C6orf130 is a new member of the OAADPr deacetylase family that efficiently catalyzes the hydrolysis of OAADPr to produce ADP-ribose and free acetate. We also show that C6orf130 can catalyze the deacylation of ADPr derivatives with longer acyl chains, including O-propionyl-ADP-ribose (OPADPr) and O-butyryl-ADP-ribose (OBADPr), to produce ADPr and the corresponding acyl acids. To our knowledge, the C6orf130-ADPr NMR structure is the first of a binary macrodomain complex that harbors O-acyl-ADP-ribose deacylase activity, revealing key residues for substrate binding and catalysis. Structural analyses of C6orf130 in the apo and ADPr bound states indicate that the β5-α4 loop in the core macrodomain fold functions as a gate to sequester ADPr in the ligand-binding cleft. Based on structural, mutagenic, and catalytic studies, we propose a minimal catalytic mechanism for the deacylation reaction of O-acyl-ADP-ribose by C6orf130. Comparisons with the MacroD-like proteins indicate that the sequence variation within the macrodomain family permits a different set of catalytic residues to perform O-acyl-ADPr hydrolysis.
EXPERIMENTAL PROCEDURES
Materials
Tritium-labeled acetyl-CoA was purchased from Moravek Biochemicals (Brea, CA). Synthetic peptide and acyl-peptides corresponding to the 11 amino acids surrounding lysine 14 of histone H3, H2N-KSTGGKAPRKQ-CONH2 (11-mer H3 peptide), H2N-KSTGGK(-acetyl)APRKQ-CONH2, H2N-KSTGGK(-propionyl)APRKQ-CONH2, H2N-KSTGGK(-n-butyryl)APRKQ-CONH2, were synthesized in the University of Wisconsin-Madison Biotechnology Center Peptide Synthesis Facility. Tritium-labeled acetyl H3 peptide, H2N-KSTGGK(3H-acetyl)APRKQ-CONH2, was synthesized enzymatically from the 11-mer H3 peptide and purified as described previously (26). OAADPr, OPADPr, OBADPr, and O-3H-acetyl-ADP-ribose (3H-OAADPr) were synthesized enzymatically from ADPr and the acyl-peptides using yeast deacetylase HST2 and nicotinamidase from Salmonella enterica fused to maltose-binding protein (MBP-PncA) following procedures described previously (9, 14). All other chemicals used were of the highest purity available commercially and were purchased from Sigma-Aldrich or Fisher Scientific.
Protein Expression and Purification for Enzymatic Assays
Expression and purification of yeast HST2 (14, 27) and nicotinamidase from S. enterica fused to maltose-binding protein (28) were performed as described previously (9). Expression and purification of the His-tagged C6orf130 protein for enzymatic studies was achieved by transforming E. coli BL21(DE3) cells with the pDEST17 plasmid containing the C6orf130 gene insert and induction of mid-log phase cells (A600 = 0.7) with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside at 23 °C. Specific point mutations were created using the QuikChange II site-directed mutagenesis kit (Stratagene). Mutated genes were sequenced at the University of Wisconsin Biotechnology Center DNA Sequencing Facility to ensure that only the desired mutations were introduced. All protein purification was performed at 4 °C or on ice as described previously. The purified protein aliquots were reconcentrated, flash-frozen in liquid nitrogen, and stored at −80 °C.
Enzyme Activity and Inhibition Assays
Enzyme activity assays for the deacylation of OAADPr, OPADPr, and OBADPr to ADPr and the corresponding acyl acids were performed by radioactive and HPLC methods using conditions as described (9). Inhibition of C6orf130 activity by ADPr and the non-hydrolysable OAADPr analog N-acetyl-ADP-ribose (NAADPr) were assayed in reactions containing from 20 to 1600 μm O-3H-acetyl-ADP-ribose and 0.5 μm C6orf130 enzyme and ADPr concentrations ranging from 0 to 400 μm or NAADPr concentrations from 0 to 500 μm in 50 mm Tris, pH 7.3, at 23 °C. Reactions were initiated by the addition of enzyme and terminated by mixing an aliquot of the reaction mixtures with 1.5 volumes of activated charcoal slurry at the desired time points. Inhibition was analyzed by measuring the initial forward rate of 3H-acetate formation as described (9). The initial velocity data were fitted and displayed using Kaleidagraph (Synergy Software, Reading, PA).
C6orf130 Expression and Purification for Structural Studies
NMR samples of the C6orf130 gene product from Homo sapiens were initially prepared in a uniformly 13C,15N-labeled form according to wheat germ cell-free protocols described previously (29). Briefly, the protein was expressed with an N-terminal His6 fusion tag in wheat germ extract supplemented with uniformly 13C,15N-labeled amino acids (Cambridge Isotope Laboratories) and purified by metal affinity chromatography followed by size-exclusion chromatography as described previously (30). Samples prepared using cell-free protocols were used to solve the structure of apo-C6orf130.
Additional NMR samples for ADPr titrations and structure determination of the ADPr-C6orf130 complex were prepared recombinantly in E. coil strain SG13009[pRPEP4] (Qiagen) using the pQE308HT vector as described previously (31). Briefly, cells were grown at 37 °C in M9 minimal broth containing 150 μg/ml ampicillin and 50 μg/ml kanamycin until reaching a cell density of A600 = 0.7. Protein expression was then induced by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 1 mm. Cells were grown for another 5 h, harvested by centrifugation, and stored at −80 °C until processed further. Isotopically labeled proteins were prepared for NMR by supplementing the M9 medium with 15N-ammonium chloride and/or 13C-glucose as the sole nitrogen and carbon source, respectively. Cells harvested from a 1-liter culture were lysed with a French pressure cell and purified by metal affinity chromatography using a previously published protocol with one modification (32). In place of a second round of metal affinity chromatography, the proteolytically cleaved His8 affinity tag was separated from C6orf130 using a Sephacryl S-100 size-exclusion chromatography column equilibrated with 50 mm bis-Tris, pH 6.5, and 200 mm sodium chloride.
NMR Spectroscopy
NMR samples were prepared in buffer containing 50 mm bis-Tris, pH 6.5, 200 mm sodium chloride, 2 mm dithiothreitol, and 5–10% 2H2O. All NMR data were acquired at 25 °C on a Bruker 600 MHz spectrometer equipped with a triple resonance CryoProbeTM and processed with NMRPipe software (33). ADPr (Sigma) was titrated incrementally into a sample containing 250 μm 15N-labeled C6orf130 and monitored by two-dimensional 15N,1H HSQC to confirm binding. Backbone 1H, 15N, and 13C chemical shift assignments for apo-C6orf130 were obtained automatically as described previously using a 550 μm sample of 15N,13C-labeled apo-C6orf130 (34, 35). Side chain assignments were completed manually from three-dimensional HBHACONH, HCCONH, and HCCH total correlation spectroscopy and 13C (aromatic)-edited NOESY-HSQC spectra. Backbone and side chain assignments were transferred to the ADPr-C6orf130 complex by inspection and confirmed with three-dimensional HNCACB and HCCH total correlation spectroscopy acquired using a 800 μm sample of 15N,13C-labeled ADPr-C6orf130. Chemical shift assignments were ∼91 and ∼93% complete for the apo- and holoproteins, respectively. Heteronuclear NOE values were measured from an interleaved pair of two-dimensional 15N-1H sensitivity-enhanced correlation spectra recorded with and without a 5-s proton saturation period. The combined chemical shift difference for each residue was calculated as [((ΔδN/5)2 + (ΔδH)2)/2]1/2, where ΔδN and ΔδH are the 15N and 1HN chemical shift differences (ppm), respectively, between the two conditions.
Structure Calculations of apo-C6orf130 and ADPr-C6orf130
Distance constraints were obtained from three-dimensional 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC spectra (τmix = 80 ms). The intermolecular distance constraints between C6orf130 and ADPr were obtained from a three-dimensional F1-13C-filtered/F3-13C-edited NOESY-HSQC spectrum (τmix = 120 ms) (36). Backbone φ and ψ dihedral angle constraints were generated from secondary shifts of the 1H, 13Cα, 13Cβ, 13C′, and 15N nuclei using the program TALOS (37). A CYANA library file was generated for the ADPr ligand as we had described previously for small molecules (38). Structure calculations were performed using the torsion angle dynamics program CYANA (39) followed by iterative rounds of manual refinement to eliminate constraint violations. Of the 100 CYANA structures calculated, the 20 conformers with the lowest target function were subjected to a molecular dynamics protocol in explicit solvent (40) using XPLOR-NIH (41). The appropriate topology and parameter files for ADPr were generated in a semi-automated manner using the PRODRG server and edited manually to add the omitted protons (38). The apo-C6orf130 and ADPr-C6orf130 structures have been deposited in the Protein Data Bank (PDB codes 2LGR and 2L8R, respectively) and the Biological Magnetic Resonance Bank (BMRB IDs 15593 and 17421, respectively).
RESULTS
C6orf130 Deacetylase Activity
Eleven macrodomain proteins encoded by 10 genes (7, 10, 11) have been identified in humans. Two of the human macrodomain proteins, MacroD1 and MacroD2, are structurally and functionally homologous and are closely related to bacterial YmdB proteins and to the sirtuin-linked protein SAV0325 from pathogenic bacterium S. aureus (supplemental Table S1). As a member of the macrodomain superfamily, C6orf130 shares minimal homology with the MacroD-like proteins including human MacroD1 and MacroD2 (Fig. 1a and supplemental Table S1). C6orf130 transcription and expression are enriched in chronic lymphocytic leukemia cells, suggesting that this protein might be a promising serological marker for this disease (24). To explore the molecular and biological functions of this unique macrodomain protein, we examined whether C6orf130 was capable of hydrolyzing OAADPr and other NAD+ metabolites derived from the sirtuin reaction.
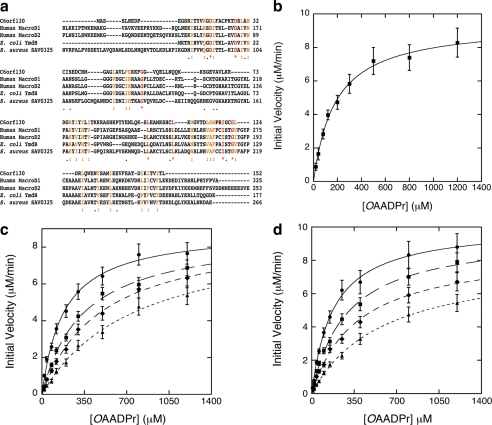
FIGURE 1.
C6orf130 deacetylates OAADPr. a, sequence alignment C6orf130 with other MacroD-like proteins human MacroD1, human MacroD2, E. coli YmdB, and S. aureus SAV0325. b, steady-state kinetic analysis of C6orf130. The deacetylation reaction of OAADPr catalyzed by C6orf130 follows saturation kinetics. The steady-state kinetic parameters are determined by radioactive assays for acetate formation and HPLC assays for ADP-ribose formation. Apparent Km as measured was 182 ± 17 μm; the kcat as measured was 0.31 ± 0.03 s−1. The reaction mixtures contained 0.5 μm C6orf130. c, inhibition of C6orf130 deacetylase activity by ADPr. The deacetylation reactions were carried out in 50 mm Tris-HCl, pH 7.3, containing 0.5 μm C6orf130. The reactions were initiated in the presence of 0 μm (circles), 100 μm (squares), 200 μm (diamonds), and 400 μm (triangles) ADPr, respectively. The Km′ values measured were 183.3, 340.6, 480.1, and 782.2 μm in the presence of 0, 100, 200, and 400 μm initial ADPr, respectively. The Vmax values were 0.31, 0.29, 0.29, and 0.30 s−1. The Ki was determined to be 119.3 ± 4.5 μm. The error bars represent standard deviations calculated from the measured initial velocities at each substrate concentration from three separate experiments. d, inhibition of C6orf130 deacetylase activity by NAADPr. The deacetylation reactions were carried out under the same conditions as described in c in the presence of 0 μm (circles), 125 μm (squares), 250 μm (diamonds), and 500 μm (triangles) NAADPr, respectively. The Km′ values measured were 183.3, 340.6, 480.1, and 782.2 μm in the presence of 0, 125, 250, and 500 μm initial NAADPr, respectively. The Vmax values were 0.31, 0.29, 0.29, and 0.30 s−1. The Ki was determined to be 223.3 ± 11.4 μm. The error bars represent standard deviations calculated from the initial velocities at each substrate concentration from three separate experiments.
By incubating radiolabeled O-3H-acetyl-ADP-ribose with purified C6orf130 and analyzing the incubation mixture using radioactive charcoal binding and HPLC (9), we found that C6orf130 carries a robust catalytic activity toward deacetylation of OAADPr, forming ADPr and acetate. Fig. 1b shows that the initial rates of OAADPr deacetylation catalyzed by C6orf130 depend on the concentration of OAADPr and exhibit saturation kinetics. An apparent Km of 182 ± 17 μm and a Vmax of 0.31 ± 0.03 s−1 at 23 °C were determined (Table 1). To investigate the substrate specificity and whether C6orf130 also possesses catalytic activity toward other sirtuin-derived NAD+ metabolites, we synthesized longer acyl derivatives of ADPr and used them as alternative substrates. Catalytic assays using the alternative substrates show that C6orf130 efficiently catalyzed the deacylation of OPADPr and OBADPr to form ADPr and propionate and butyrate, respectively. The deacylation of OPADPr and OBADPr by C6orf130 followed similar saturation kinetics. Increasing the acyl chains of O-acyl-ADPr only slightly increased the Km and decreased catalytic turnover kcat. The steady-state kinetic parameters for deacylation of OPADPr, OBADPr, and OAADPr are summarized in Table 1. The data indicate that C6orf130 has a slight preference for ADPr derivatives with shorter acyl chains.
TABLE 1.
Comparison of kinetic parameters using OAADPr, OPADPr, and OBADPr as substrates
| Substrate | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | μm | m−1s−1 | |
| OAADPr | 0.31 ± 0.03 | 182.3 ± 17.4 | (1.70 ± 0.23) × 103 |
| OPADPr | 0.24 ± 0.03 | 220.7 ± 26.4 | (1.09 ± 0.19) × 103 |
| OBADPr | 0.18 ± 0.02 | 295.6 ± 52.6 | (6.10 ± 1.27) × 102 |
Inhibition of C6orf130 by ADPr and an OAADPr Analog
Previous x-ray crystallography and biochemical studies revealed that human macroH2A1.1 functions as a binding module for ADPr, OAADPr, and other analogs (6, 22). Human MacroD1 and MacroD2 bind ADPr with high affinity (7), and ADPr serves as a competitive inhibitor for OAADPr deacetylation catalyzed by MacroD1 (9). Here, we examined the ability of ADPr and a non-hydrolyzable substrate analog, NAADPr, to function as inhibitors for the deacetylation of OAADPr by C6orf130. Deacetylation reactions of OAADPr were initiated by addition of C6orf130 in the presence of 0–400 μm ADPr or 0–500 μm NAADPr, and the initial rates of acetate formation were measured under steady-state conditions. As shown in Fig. 1, c and d, increasing the concentration of ADPr or NAADPr resulted in an increase of the apparent Michaelis constant, Km, whereas the maximum catalytic rates were minimally affected. These data are consistent with reversible competitive inhibition of C6orf130 by ADPr and NAADPr. Inhibition constants (Ki) of 119.3 ± 4.5 μm and 223.5 ± 11.3 μm were determined at 23 °C for ADPr and NAADPr, respectively. These results suggest that C6orf130 contains a specific binding site for ADPr and NAADPr, which is identical to the active site for OAADPr deacetylation. Inhibition of OAADPr deacetylation by ADPr indicates that structural information from a C6orf130-ADPr binary complex can be used to investigate the functions of active site residues. Inhibition by ADPr also suggests that ADPr could act as an in vivo inhibitor of macrodomain-dependent OAADPr deacetylation, providing feedback regulation of cellular OAADPr concentration. The relatively larger inhibition constant of NAADPr suggests that the rigidity of the amide bond of NAADPr is less favored at the substrate-binding site.
Structure of the apo-C6orf130 Macrodomain
To investigate whether C6orf130 retains the canonical macrodomain fold, we determined its solution structure using standard NMR methods (35). The final ensemble of 20 conformers is shown in Fig. 2a, and the structural statistics are presented in Table 2. Like other macrodomains, C6orf130 exhibits the canonical three-layered α-β-α sandwich with a central six-stranded β-sheet containing a mixture of anti-parallel (β3-β4-β2) and parallel (β2-β5-β6-β1) strands and a deep ligand-binding cleft (Fig. 2b). Structural alignment of C6orf130 with the macroD1 and macroH2A1.1 macrodomains reveals that C6orf130 contains only the core domain and lacks the extended N- and C-terminal structural elements commonly found in other macrodomains (Fig. 2c) (9, 42).
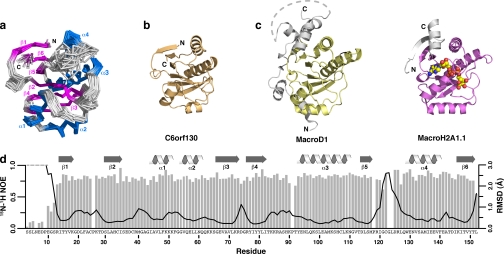
FIGURE 2.
NMR structure of the C6orf130 macrodomain. a, ensemble of the final 20 NMR structures (Cα trace). α-Helices and β-sheets are shown in blue and magenta, respectively. Residues 3–12 are unstructured and have been omitted for clarity. b, ribbon view of C6orf130 showing the core α-β-α sandwich fold typical of macrodomains. c, the MacroD1 (yellow; PDB code 2X47) and MacroH2A1.1 (magenta; PDB code 3IID) macrodomains contain the central α-β-α sandwich but also contain extended structural features at the N and C termini (gray). d, 15N-1H heteronuclear NOE values and global backbone atomic r.m.s.d. values plotted as a function of residue number. Values are missing for Gly-123 and Leu-124. Global r.m.s.d. values were calculated with the ensemble superimposed using residues 14–120 and 126–152.
TABLE 2.
Refinement statistics for the C6orf130 NMR ensembles
| Statistics | Apo state | ADPr-bound |
|---|---|---|
| Experimental constraints | ||
| Distance constraints | ||
| Long | 557 | 630 |
| Medium [1< (i − j) ≤ 5] | 238 | 303 |
| Sequential [(i − j) = 1] | 418 | 416 |
| Intra-residue [i = j] | 758 | 632 |
| Total | 1971 | 2020 |
| Dihedral angle constraints (φ and ψ) | 180 | 211 |
| Intermolecular constraints | 0 | 39 |
| Average atomic r.m.s.d. to the mean structure (Å) | ||
| Residues | 14-120, 126-152 | 14-152 |
| Backbone (Cα, C′, N) | 0.62 ± 0.07 | 0.56 ± 0.07 |
| Heavy atoms | 1.08 ± 0.09 | 0.97 ± 0.07 |
| Deviations from idealized covalent geometry | ||
| Bond lengths, r.m.s.d. (Å) | 0.017 | 0.016 |
| Torsion angle violations, r.m.s.d. (°) | 1.3 | 1.3 |
| Constraint violations | ||
| NOE distance, number > 0.5 Åa | 0.0 ± 0 | 0.0 ± 0 |
| NOE distance, r.m.s.d. (Å) | 0.026 ± 0.001 | 0.018 ± 0.001 |
| Torsion angle violations, number > 5°b | 0.0 ± 0 | 0.0 ± 0 |
| Torsion angle violations, r.m.s.d. (°) | 0.661 ± 0.086 | 0.571 ± 0.067 |
| WHATCHECK quality indicators | ||
| Z-score | −0.54 ± 0.19 | −1.01 ± 0.21 |
| r.m.s. Z-score | ||
| Bond lengths | 0.77 ± 0.02 | 0.74 ± 0.02 |
| Bond angles | 0.75 ± 0.01 | 0.75 ± 0.02 |
| Bumps | 0.0 ± 0 | 0.0 ± 0 |
| Lennard-Jones energyc (kJ mol−1) | −3719 ± 78 | −4156 ± 83 |
| Ramachandran statistics (% of all residues) | ||
| Most favored | 85.2 ± 2.0 | 87.2 ± 2.5 |
| Additionally allowed | 13.0 ± 2.4 | 11.8 ± 2.6 |
| Generously allowed | 1.1 ± 1.0 | 0.9 ± 0.8 |
| Disallowed | 0.7 ± 0.7 | 0 |
a The largest NOE violation in the ensemble of structures was 0.49 Å.
b The largest torsion angle violation in the ensemble of structures was 4.9°.
c Nonbonded energy was calculated in XPLOR-NIH.
A Dali search (43) using the apo-C6orf130 structure revealed the presence of numerous structural homologs. The top scoring homologs are: a hypothetical protein BT1257 from Bacteroides thetaiotaomicron (PDB codes 2FG1 and 2AFC; Z-scores of 17.0 for both; r.m.s.d. values of 2.3 and 2.1 Å for 140 and 137 superimposed Cα atoms, respectively); a hypothetical protein, TTHA0132, from Thermus thermophilus (PDB code 2DX6; Z-score of 14.9; r.m.s.d. of 2.5 Å for 134 superimposed Cα atoms); the nsP3 macrodomain of Chikungunya virus in complex with ADP-ribose (PDB code 3GPO; Z-score of 13.2; r.m.s.d. of 2.6 Å for 131 superimposed Cα atoms); and core histone macroH2A1.1 (PDB code 1YD9; Z-score of 12.6; r.m.s.d. of 2.7 Å for 138 superimposed Cα atoms).
Low backbone r.m.s.d. and high 15N-1H heteronuclear NOE values (Fig. 2d) over the length of C6orf130 suggest that the protein is well ordered, with two exceptions. The sharp increase in r.m.s.d. values and concurrent decrease in 15N-1H heteronuclear NOE values for the first 14 residues indicates that the N terminus is unstructured and dynamically disordered. This conclusion is supported by the lack of long range NOEs for these residues in NOESY spectra (data not shown). A similar increase in r.m.s.d. values and lack of long range NOEs is observed for residues 119–129 in the loop connecting β-strand 5 to α-helix 4 (β5-α4 loop), but in contrast to the N terminus, 15N-1H heteronuclear NOE values remain uniformly high. Taken together, the disparity between the r.m.s.d. and 15N-1H heteronuclear NOE values and the line broadening observed for β5-α4 15N-1H HSQC signals indicates that this loop undergoes conformational exchange on an intermediate (μs–ms) time scale.
NMR Analysis of ADPr Binding to C6orf130
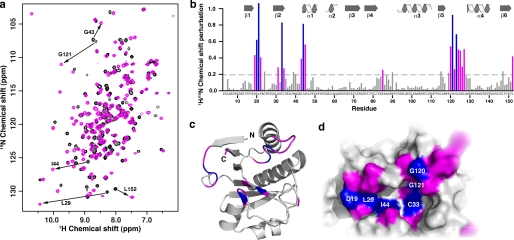
Our enzymatic studies demonstrate that C6orf130 catalyzes the deacylation of O-acylated-ADPr to yield ADPr and the corresponding organic acid. Inhibition studies indicate that ADPr serves as a competitive inhibitor for OAADPr binding (Fig. 1c). To determine whether the conserved binding cleft functions as the substrate recognition site, we titrated 15N-labeled C6orf130 incrementally with ADPr and monitored the changes by NMR. Inspection of the resulting 15N-1H HSQC spectra identified ∼20 resonances that respond in a dose-dependent manner to ADPr (Fig. 3a). Movement of only a subset of peaks indicated that the interaction with ADPr is specific and utilizes a defined binding site. The ADPr-induced chemical shift perturbations localized to the loop connecting β-strands 1 and 2, β-strand 2, the N-terminal end of α-helix 2, and the β5-α4 loop (Fig. 3b). Mapping these perturbations to the three-dimensional structure of apo-C6orf130 confirmed that the deep cleft conserved in the core macrodomain fold serves as the ADPr-binding pocket, and residues that exhibited the largest perturbations (>0.6) lined the floor of the cleft (Fig. 3, c and d). The majority of residues in the β5-α4 loop exhibit perturbations above the threshold value (≥0.25). The sensitivity of the β5-α4 loop to ADPr binding, its proximity to the ligand-binding groove, and the conformational fluctuation on the intermediate time scale suggest that the β5-α4 loop may serve as a gate that limits substrate binding or product release from the ligand-binding cleft. Depending on its conformation in the substrate-bound state, specific β5-α4 residues may also participate in O-acyl-ADPr hydrolysis.
FIGURE 3.
C6orf130 binds ADPr specifically. a, 15N-1H HSQC of C6orf130 in the apo (black) and ADPr bound states (magenta). b, combined 1H/15N chemical shift perturbations in response to ADPr binding were calculated and plotted as a function of the C6orf130 residue number. Residues with chemical shift perturbations of ≥0.25 were mapped onto the C6orf130 apostructure and are shown in ribbon (c) and surface (d) views. The perturbations localize to one face of the molecule and identify the conserved binding cleft as the ADPr-binding site. Chemical shift perturbations ranging from 0.25 to 0.59 and those ≥0.6 are shown in magenta and blue, respectively.
Structure of ADPr-C6orf130 Complex
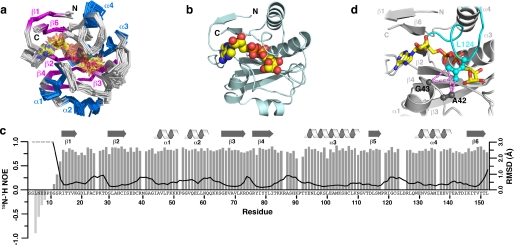
To further investigate the role of the β5-α4 loop in substrate binding and catalysis, we solved the NMR solution structure of the ADPr-C6orf130 complex. The final ensemble of 20 conformers is shown in Fig. 4a, and the structural statistics are presented in Table 2. ADPr binds in the deep cleft of C6orf130 and is covered by the β5-α4 loop (Fig. 4b and Fig. S1), consistent with the chemical shift perturbation data (Fig. 3b). The r.m.s.d. and 15N-1H heteronuclear NOE values for the ADPr-C6orf130 complex (Fig. 4c) are largely unchanged when compared with those for apo-C6orf130 (Fig. 2d) except in the β5-α4 loop. In the complex, r.m.s.d. values for residues 119–129 in the β5-α4 loop decreased to values consistent with all other well ordered residues. The low r.m.s.d. values and high 15N-1H heteronuclear NOE values indicate that the β5-α4 loop fluctuations present in the apo state were dampened upon ADPr binding. The appearance of long range NOEs between the methyl groups of Leu-124 and the side chains of Ala-42 and Gly-43 (Fig. 4d) reinforces this conclusion and confirms that the β5-α4 loop encloses ADPr or other ligands within the C6orf130 active site.
FIGURE 4.
NMR structure of the C6orf130-ADPr complex. a, ensemble of the final 20 NMR structures (Cα trace). α-Helices and β-sheets are shown in blue and magenta, respectively. Residues 3–12 are unstructured and have been omitted for clarity. b, ribbon view of C6orf130-ADPr complex with ADPr shown in yellow. c, 15N-1H heteronuclear NOE values and global backbone atomic r.m.s.d. values plotted as a function of residue number. 15N-1H heteronuclear NOE values are missing or excluded for residues Met-40, Phe-52, Arg-86, Leu-124, Asp-125, and Ile-136. Global r.m.s.d. values were calculated with the ensemble superimposed using residues 14–152. d, NOEs between the methyl groups of Leu-124 in the β5-α4 loop (cyan) and residues Ala-42 and Gly-43 (dark gray) are present only in the ADPr-C6orf130 complex. NOEs are shown in magenta as dashed lines.
Mutational Analysis of the C6orf130 Active Site
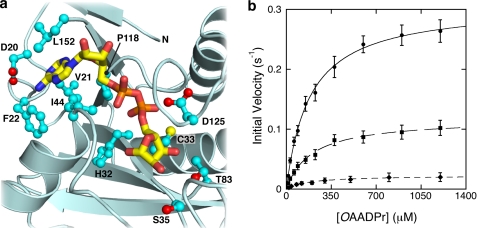
The structure of the C6orf130-ADPr binary complex revealed amino acid residues interacting with the bound ADPr (Fig. 5a) and suggested residues important for catalysis. The side chain carboxyl group of Asp-20 interacts with the adenine primary amine of the bound ADPr. Leu-21 and Phe-22 on the β1-β2 loop, Ile-44 and Leu-47 on the β2-α1 loop, and Pro-118, Tyr-150, and the C-terminal Leu-152 form a hydrophobic pocket in which the adenosine moiety is embedded. The electrostatic and hydrophobic interactions between the adenosine moiety and the protein-binding pocket seem to contribute the majority of the binding energy, as indicated by the higher coordinate precision of the adenosine side compared with the distal ribose side of the bound ADPr (Fig. 4a). The GXG sequence in the β5-α4 loop is positioned over the pyrophosphate group and is a common phosphate-binding element found in numerous nucleotide binding proteins (44). The side chains of Ser-35, Thr-83, and Asp-125 are located in the vicinity of the distal ribose and potentially interact with the C2- and C3-hydroxyl groups. His-32 and Cys-33 are also located nearby, and their main chain amides form part of the substrate-binding pocket.
FIGURE 5.
Kinetic comparison of wild type C6orf130 and its variants. a, key amino acid residues interacting with the bound ADPr. Side chains are shown in ball-and-stick form, and oxygen atoms are highlighted in red. b, Thr-83 and Ser-35 located in the vicinity of the 2- and 3-hydroxyl groups of the bound ADPr in the MacroD1 are mutated to alanine. A steady-state kinetic comparison of the wild type C6orf130 (circles) and the T83A (squares) and S35A (diamonds) mutants shows that these residues are important to catalysis. Substituting Thr-83 and Ser-35 with Ala mainly affects the Vmax. The Km values are only minimally affected. The error bars represent standard deviations calculated from the measured initial velocities at each substrate concentration from three separate experiments. Data points of mutant partial activities are the mean values from three separate measurements.
To verify that the ADPr-binding pocket constitutes the substrate-binding site and to identify important residues for catalysis, we performed site-directed mutagenesis on potentially important residues for substrate binding and catalysis. Gly-123 was substituted with Glu, and His-32, Cys-33, Ser-35, Thr-83, and Asp-125 were substituted with Ala. The purified mutant C6orf130 proteins were assayed for OAADPr deacetylation activity. Mutants retaining deacetylation activity were further analyzed under steady-state conditions. During the purification of these C6orf130 variants, no abnormal elution behaviors were observed in the Sephadex G-75 gel filtration chromatography, suggesting that amino acid substitution at these positions did not cause global structural perturbations. Substituting Cys-33 with Ala affected neither substrate binding nor catalytic efficiency. Substituting His-32, Gly-123, and Asp-125 with Ala resulted in enzymes for which the measured activities were within the rates observed for background hydrolysis. From the C6orf130-ADPr structure, the main chain NH of Gly-123 interacts with the pyrophosphate group, and the β5-α4 loop bearing Gly-123 interacts with the ribose hydroxyls of adenosine. Introducing a large side chain with a negative charge in the G123E mutant presumably disrupts the substrate binding and renders the variant inactive. The imidazole ring of His-32 is buried in a hydrophobic pocket formed by the side chains of Phe-48 and Tyr-80. Although one edge of the imidazole ring comes within ∼3 Å of the pyrophosphate, the main role of His-32 may be to maintain the integrity of the substrate-binding pocket. The carboxyl of Asp-125 is within H-bonding distance of the C2- and C3-hydroxyl groups of the distal ribose (i.e. ADP-ribose-C2(OH) and C3(OH)). The D125A mutant showed no detectable deacetylation activity after subtracting background activity obtained with rates in the absence of enzyme. No OAADPr deacetylation activity was detected using enzyme concentrations as high as 5 μm with prolonged incubation. The S35A point mutation resulted in an ∼20-fold decrease in kcat without significantly affecting Km (kcat = 0.023 ± 0.002 s−1, Km = 177.7 ± 12.6 (μm), and V/K = (1.29 ± 0.15) × 102 m−1s−1). Thus, substituting Ser-35 with Ala lowers the first-order rate constant for the chemical conversion of the enzyme-substrate complex but does not affect substrate binding as assessed by an unaffected Km value. The T83A substitution yielded an ∼3-fold decrease in kcat without affecting the Km. Steady-state kinetic parameters of the T83A and S35A mutants were determined (Fig. 5b) and were compared with those of the wild type enzyme (Table 3). Taken together the data indicate that Ser-35 and Asp-125 function as critical catalytic residues. A proposed catalytic mechanism is discussed below.
TABLE 3.
Steady-state kinetic parameters of wild type and selected mutant C6orf130 proteins
Catalytic turnover below 0.005 s−1 is considered background or inactive under the assay conditions.
| Proteins | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | μm | m−1s−1 | |
| C6orf130-WT | 0.31 ± 0.03 | 182 ± 17 | (1.7 ± 0.2) × 103 |
| C6orf130-C33A | 0.32 ± 0.02 | 181 ± 16 | (1.8 ± 0.2) × 103 |
| C6orf130-D125A | ∼0.002 | ||
| C6orf130-G123E | ∼0.005 | ||
| C6orf130-H32A | ∼0.004 | ||
| C6orf130-S35A | 0.023 ± 0.002 | 178 ± 13 | (1.3 ± 0.2) × 102 |
| C6orf130-T83A | 0.12 ± 0.01 | 189 ± 12 | (6.3 ± 0.7) × 102 |
DISCUSSION
Comparison of C6orf130 with Other Macrodomain Proteins
C6orf130 occupies a distinct branch in the phylogenic tree of macrodomain proteins (9). Despite its low sequence homology to MacroD-like proteins (Fig. 1a and supplemental Table S1), C6orf130 exhibits a canonical core fold similar to other macrodomain proteins, consisting of a three-layered α-β-α sandwich with a central six-stranded β-sheet containing a mixture of anti-parallel (β3-β4-β2) and parallel (β2-β5-β6-β1) strands and a ligand-binding cleft (Fig. 2b). Structural alignment of C6orf130 with other macrodomains reveals that C6orf130 contains only the core domain and lacks the extended N- and C-terminal structural elements found in MacroD1 and MacroH2A1.1 (Fig. 2c) (9, 42). In the apostructure of human MacroD1, an ADPr molecule was modeled in the ligand-binding cleft (9) with the diphosphate ribose moiety and distal 2- and 3-hydroxyl groups positioned at the surface of the protein in a highly solvent-accessible location. In contrast, the C6orf130-ADPr co-structure solved in this study reveals that ADPr binds deep within the macrodomain cleft, with the diphosphate ribose portion completely buried under the β2-α1, β4-α3, and β5-α4 loops. The canonical core fold and similar deacetylation activity toward OAADPr suggest a convergent evolution of the catalytic function between C6orf130 and the MacroD-like proteins.
Catalytic Mechanism of C6orf130
ADPr acts as a competitive inhibitor of C6orf130 deacylation, suggesting that the substrate O-acyl-ADPr binds in the same fashion as ADPr. The mobile β5-α4 loop appears to function as a gate that combines with the β2-α1 and β4-α3 loops to sequester substrate from the solvent environment. On these loops, Ser-35 (β2-α1), Thr-83 (β4-α3), and Asp-125 (β5-α4) are located close to the C2- and C3-hydroxyls of the distal ribose, serving potentially important roles in catalysis. The present mutagenic studies revealed that the T83A mutant retains nearly all of its deacetylation activity relative to wild type enzyme; therefore, the possibility of Thr-83 functioning as the key catalytic residue can be ruled out. Substituting Ser-35 with Ala severely impaired the catalytic capability with an ∼20-fold decrease in kcat. However, the Km of the S35A mutant was minimally affected (Table 3). The most dramatic substitution was the Asp-125 to Ala mutant, which resulted in a C6orf130 variant with no detectable deacetylation activity. Thus, Ser-35 and Asp-125 are candidate catalytic residues. Steady-state kinetic analyses of these mutant enzymes and the structural information from both apo-C6orf130 and C6orf130-ADPr complex led us to propose a minimal mechanism for the C6orf130-catalyzed deacylation reaction (Fig. 6c), bearing in mind that S35A retains ∼5% kcat activity of the wild type enzyme and there are no other functional groups in the vicinity of the C2- and C3-hydroxyls of the distal ribose (ADP-ribose-C2(OH) and C3(OH)). In this mechanism the hydroxyl of Ser-35 is H-bonded to the ester carbonyl oxygen to polarize the carbonyl bond. With Asp-125 acting as a general base, a water molecule is activated, which functions as a nucleophile to attack the carbonyl carbon and form an oxyanion tetrahedral intermediate. Protonation of the scissile ester oxygen atom and rearrangement result in the breakdown of the tetrahedral intermediate to form ADPr and the corresponding acyl acids (Fig. 6c).
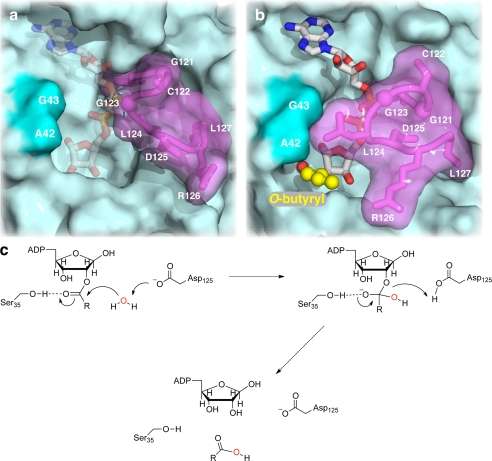
FIGURE 6.
Substrate binding and catalytic mechanism of C6orf130. a and b, C6orf130 accommodates longer acyl chains. A model of OBADPr bound in the active site of C6orf130 was constructed by addition of the butyryl moiety (yellow ball and stick) to the 2-hydroxyl of the distal ribose. a, in apo-C6orf130, residues 120–127 of the β5-α4 loop (magenta) are in the open position, exposing the ligand-binding cleft (indicated by ADPr). b, ADPr binding dampens the fluctuations of the β5-α4 loop (magenta) closing the gate that sequesters ADPr or other ligands in the C6orf130 active site. Lid closure forms a pliable pore through which longer acyl chains, such as the butyryl moiety modeled here (yellow ball and stick), can be accommodated in the C6orf130 active site. c, proposed catalytic mechanism of C6orf130. This mechanism is based on structural analyses of both the apo-C6orf130 and the C6orf130-ADPr complex, mutagenesis of key residues in the vicinity of the distal ribose of the bound ADPr, and kinetic studies of the mutant enzymes.
Previous kinetic analysis of MacroD1 mutants suggested that Asn-171, Asn-174, Asp-184, and His-188 are important for catalysis. Modeling of ADPr into the solved MacroD1 structure located these residues near the C2- and C3-hydroxyls of the distal ribose (9). Asn-171 and Asn-174 are widely conserved among many different macrodomains but not in C6orf130. Among MacroD-like proteins, Asp-184 and His-188 are conserved. In MacroD1, Asn-174 was proposed to help position the ribose and polarize the ester bond for hydrolysis. Asn-171 and His-188 interact with Asp-184 and might facilitate deprotonation, allowing Asp-184 to function as a general base by activating a water molecule for nucleophilic attack on the carbonyl carbon. In contrast to Ser-35 and Asp-125 from C6orf130, the four catalytic residues in MacroD1 individually contribute little to hydrolytic efficiency. The observation that the catalytic residues between C6orf130 and MacroD-like proteins are not conserved indicates sequence variation within the macrodomain family permits a different set of catalytic residues to perform O-acyl-ADPr hydrolysis.
Macrodomains as General Deacylases of O-Acyl-ADP-ribose
Protein lysine acetylation can have profound effects on the physiological function of modified proteins. Propionylation, butyrylation, and succinylation of lysine represent newly discovered protein post-translational modifications (45–47). Although it is unclear which enzymes are responsible for the addition of these groups, human acetyltransferases p300 and CBP (CREB-binding protein) catalyzed in vitro lysine propionylation and butyrylation of histones (45, 46). Berndsen et al. (48) showed that the yeast histone acetyltransferase Esa1 can readily perform propionyl transfer from propionyl-CoA to histones. In S. enterica, propionylation of propionyl-CoA synthetase is catalyzed by prokaryotic Gcn-5-related N-acetyltransferases (28). Thus, known acetyltransferases might catalyze these reactions in vivo, utilizing cellular propionyl-CoA, butyryl-CoA, and succinyl-CoA as the co-substrates. The propionyl-lysine and butyryl-lysine in the modified proteins can be deacylated in vitro by NAD+-dependent reactions catalyzed by sirtuins, producing O-acyl-ADPr as the co-product (19, 28, 46, 49). These observations suggest that propionyl-, butyryl-, and succinyl-lysine proteins might be natural substrates of sirtuins in vivo, although desuccinylation has yet to be reported. Acetyl-CoA, propionyl-CoA, butyryl-CoA, and succinyl-CoA are important intermediates in cellular energy metabolism, fatty acid β-oxidation, and the degradation of leucine, lysine, isoleucine, valine, and methionine. The reversible acylation of proteins and histones by acyl-CoA-dependent acyltransferases and NAD+-dependent protein deacylases connects energy metabolism with both the functional (de)acylation of targeted proteins and the production of a diverse set of O-acyl-ADPr metabolites. O-Acetyl-ADP-ribose has been implicated in metabolic regulation, gene expression, cellular redox control, and cation channel activity (5–7, 19–22, 50, 51). Thus, the O-acyl-ADPr deacylation activity of C6orf130 and the recently characterized MacroD-like proteins place these enzymes in a pivotal position to regulate the functions of various O-acyl-ADPr metabolites. The ability of C6orf130 to deacylate O-acyl-ADPr with varying acyl chains suggests that C6orf130 might function as an in vivo modulator of cellular processes regulated by the reversible acylation of proteins and histones (catalyzed by the acyl-CoA-dependent acyl transferases and the NAD+-dependent deacylases).
The ability of C6orf130 to accept acyl-ADPr substrates with longer acyl chains can be explained from the structures solved in this study. In the C6orf130-ADPr binary complex, the substrate is buried by flexible loops carrying the catalytic residues. ADPr binding dampens the fluctuations of the β5-α4 loop closing the “gate” sequestering ADPr or other ligands in the C6orf130 active site (Fig. 6a). Gate closure forms a pliable pore through which longer acyl chains, such as the modeled butyryl moiety, can be accommodated in the C6orf130 (Fig. 6b). Together, our structural studies of apo-C6orf130, the C6orf130-ADPr complex, and modeling of longer acyl chain ADPr esters are consistent with the observed catalytic properties using OAADPr, OPADPr, and OBADPr as substrates.
Supplementary Material
Acknowledgment
We thank Mark K. Devries of the Department of Biomolecular Chemistry, University of Wisconsin-Madison, for assisting with the synthesis and purification of O-butyryl-ADP-ribose.
This work was supported, in whole or in part, by National Institutes of Health Grants U54 GM064598 and P50 GM064598 from the NIGMS Protein Structure Initiative.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
The atomic coordinates and structure factors (codes 2LGR and 2L8R) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ADPr
- ADP-ribose
- OAADPr
- O-acetyl-ADP-ribose
- OPADPr
- O-propionyl-ADP-ribose
- OBADPr
- O-butyryl-ADP-ribose
- NAADPr
- N-acetyl-ADP-ribose
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1. Anantharaman V., Aravind L. (2002) Genome Biol. 3, RESEARCH0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pehrson J. R., Fuji R. N. (1998) Nucleic acids Res. 26, 2837–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allen M. D., Buckle A. M., Cordell S. C., Löwe J., Bycroft M. (2003) J. Mol. Biol. 330, 503–511 [DOI] [PubMed] [Google Scholar]
- 4. Ladurner A. G. (2003) Mol. Cell 12, 1–3 [DOI] [PubMed] [Google Scholar]
- 5. Till S., Ladurner A. G. (2009) Front. Biosci. 14, 3246–3258 [DOI] [PubMed] [Google Scholar]
- 6. Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A. G. (2005) Nat. Struct. Mol. Biol. 12, 624–625 [DOI] [PubMed] [Google Scholar]
- 7. Neuvonen M., Ahola T. (2009) J. Mol. Biol. 385, 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahel D., Horejsí Z., Wiechens N., Polo S. E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S. C., Jackson S. P., Owen-Hughes T., Boulton S. J. (2009) Science 325, 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen D., Vollmar M., Rossi M. N., Phillips C., Kraehenbuehl R., Slade D., Mehrotra P. V., von Delft F., Crosthwaite S. K., Gileadi O., Denu J. M., Ahel I. (2011) J. Biol. Chem. 286, 13261–13271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kraus W. L. (2009) Nat. Struct. Mol. Biol. 16, 904–907 [DOI] [PubMed] [Google Scholar]
- 11. Han W., Li X., Fu X. (2011) Mutat. Res. 727, 86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Timinszky G., Till S., Hassa P. O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E. H., Scheffzek K., Hottiger M. O., Ladurner A. G. (2009) Nat. Struct. Mol. Biol. 16, 923–929 [DOI] [PubMed] [Google Scholar]
- 13. Tanner K. G., Landry J., Sternglanz R., Denu J. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14178–14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jackson M. D., Denu J. M. (2002) J. Biol. Chem. 277, 18535–18544 [DOI] [PubMed] [Google Scholar]
- 15. Smith B. C., Hallows W. C., Denu J. M. (2008) Chem. Biol. 15, 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oberdoerffer P., Michan S., McVay M., Mostoslavsky R., Vann J., Park S. K., Hartlerode A., Stegmuller J., Hafner A., Loerch P., Wright S. M., Mills K. D., Bonni A., Yankner B. A., Scully R., Prolla T. A., Alt F. W., Sinclair D. A. (2008) Cell 135, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hallows W. C., Smith B. C., Lee S., Denu J. M. (2009) Cell 137, 404–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoff K. G., Wolberger C. (2005) Nat. Struct. Mol. Biol. 12, 560–561 [DOI] [PubMed] [Google Scholar]
- 20. Tong L., Lee S., Denu J. M. (2009) J. Biol. Chem. 284, 11256–11266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tong L., Denu J. M. (2010) Biochim. Biophys. Acta 1804, 1617–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Comstock L. R., Denu J. M. (2007) Org. Biomol. Chem. 5, 3087–3091 [DOI] [PubMed] [Google Scholar]
- 23. Mungall A. J., Palmer S. A., Sims S. K., Edwards C. A., Ashurst J. L., Wilming L., Jones M. C., Horton R., Hunt S. E., Scott C. E., Gilbert J. G., Clamp M. E., Bethel G., Milne S., Ainscough R., Almeida J. P., Ambrose K. D., Andrews T. D., Ashwell R. I., Babbage A. K., Bagguley C. L., Bailey J., Banerjee R., Barker D. J., Barlow K. F., Bates K., Beare D. M., Beasley H., Beasley O., Bird C. P., Blakey S., Bray-Allen S., Brook J., Brown A. J., Brown J. Y., Burford D. C., Burrill W., Burton J., Carder C., Carter N. P., Chapman J. C., Clark S. Y., Clark G., Clee C. M., Clegg S., Cobley V., Collier R. E., Collins J. E., Colman L. K., Corby N. R., Coville G. J., Culley K. M., Dhami P., Davies J., Dunn M., Earthrowl M. E., Ellington A. E., Evans K. A., Faulkner L., Francis M. D., Frankish A., Frankland J., French L., Garner P., Garnett J., Ghori M. J., Gilby L. M., Gillson C. J., Glithero R. J., Grafham D. V., Grant M., Gribble S., Griffiths C., Griffiths M., Hall R., Halls K. S., Hammond S., Harley J. L., Hart E. A., Heath P. D., Heathcott R., Holmes S. J., Howden P. J., Howe K. L., Howell G. R., Huckle E., Humphray S. J., Humphries M. D., Hunt A. R., Johnson C. M., Joy A. A., Kay M., Keenan S. J., Kimberley A. M., King A., Laird G. K., Langford C., Lawlor S., Leongamornlert D. A., Leversha M., Lloyd C. R., Lloyd D. M., Loveland J. E., Lovell J., Martin S., Mashreghi-Mohammadi M., Maslen G. L., Matthews L., McCann O. T., McLaren S. J., McLay K., McMurray A., Moore M. J., Mullikin J. C., Niblett D., Nickerson T., Novik K. L., Oliver K., Overton-Larty E. K., Parker A., Patel R., Pearce A. V., Peck A. I., Phillimore B., Phillips S., Plumb R. W., Porter K. M., Ramsey Y., Ranby S. A., Rice C. M., Ross M. T., Searle S. M., Sehra H. K., Sheridan E., Skuce C. D., Smith S., Smith M., Spraggon L., Squares S. L., Steward C. A., Sycamore N., Tamlyn-Hall G., Tester J., Theaker A. J., Thomas D. W., Thorpe A., Tracey A., Tromans A., Tubby B., Wall M., Wallis J. M., West A. P., White S. S., Whitehead S. L., Whittaker H., Wild A., Willey D. J., Wilmer T. E., Wood J. M., Wray P. W., Wyatt J. C., Young L., Younger R. M., Bentley D. R., Coulson A., Durbin R., Hubbard T., Sulston J. E., Dunham I., Rogers J., Beck S. (2003) Nature 425, 805–811 [DOI] [PubMed] [Google Scholar]
- 24. Marina O., Hainz U., Biernacki M. A., Zhang W., Cai A., Duke-Cohan J. S., Liu F., Brusic V., Neuberg D., Kutok J. L., Alyea E. P., Canning C. M., Soiffer R. J., Ritz J., Wu C. J. (2010) Cancer Res. 70, 1344–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mate S. E., Brown K. J., Hoffman E. P. (2011) Skelet. Muscle 1, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borra M. T., O'Neill F. J., Jackson M. D., Marshall B., Verdin E., Foltz K. R., Denu J. M. (2002) J. Biol. Chem. 277, 12632–12641 [DOI] [PubMed] [Google Scholar]
- 27. Jackson M. D., Schmidt M. T., Oppenheimer N. J., Denu J. M. (2003) J. Biol. Chem. 278, 50985–50998 [DOI] [PubMed] [Google Scholar]
- 28. Garrity J., Gardner J. G., Hawse W., Wolberger C., Escalante-Semerena J. C. (2007) J. Biol. Chem. 282, 30239–30245 [DOI] [PubMed] [Google Scholar]
- 29. Vinarov D. A., Lytle B. L., Peterson F. C., Tyler E. M., Volkman B. F., Markley J. L. (2004) Nat. Methods 1, 149–153 [DOI] [PubMed] [Google Scholar]
- 30. Tyler R. C., Sreenath H. K., Singh S., Aceti D. J., Bingman C. A., Markley J. L., Fox B. G. (2005) Protein Expr. Purif. 40, 268–278 [DOI] [PubMed] [Google Scholar]
- 31. Waltner J. K., Peterson F. C., Lytle B. L., Volkman B. F. (2005) Protein Sci. 14, 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lytle B. L., Peterson F. C., Kjer K. L., Frederick R. O., Zhao Q., Thao S., Bingman C., Johnson K. A., Phillips G. N., Jr., Volkman B. F. (2004) J. Biomol. NMR 28, 397–400 [DOI] [PubMed] [Google Scholar]
- 33. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 34. Wang L., Markley J. L. (2009) J. Biomol. NMR 44, 95–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Markley J. L., Bahrami A., Eghbalnia H. R., Peterson F. C., Ulrich E. L., Westler W. M., Volkman B. F. (2009) in Structural Bioinformatics (Gu J., Bourne P. E. eds) pp. 93–142, John Wiley and Sons, Inc., New York [Google Scholar]
- 36. Stuart A. C., Borzilleri K. A., Withka J. M., Palmer A. G., 3rd (1999) J. Am. Chem. Soc. 121, 5346–5347 [Google Scholar]
- 37. Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 38. Ziarek J. J., Peterson F. C., Lytle B. L., Volkman B. F. (2011) Methods Enzymol. 493, 241–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Güntert P. (2004) Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 40. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 41. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 42. Kuschert G. S., Hoogewerf A. J., Proudfoot A. E., Chung C. W., Cooke R. M., Hubbard R. E., Wells T. N., Sanderson P. N. (1998) Biochemistry 37, 11193–11201 [DOI] [PubMed] [Google Scholar]
- 43. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saraste M., Sibbald P. R., Wittinghofer A. (1990) Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 45. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., Zhao Y. (2007) Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu B., Lin Y., Darwanto A., Song X., Xu G., Zhang K. (2009) J. Biol. Chem. 284, 32288–32295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011) Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berndsen C. E., Albaugh B. N., Tan S., Denu J. M. (2007) Biochemistry 46, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith B. C., Denu J. M. (2007) J. Biol. Chem. 282, 37256–37265 [DOI] [PubMed] [Google Scholar]
- 50. Grubisha O., Rafty L. A., Takanishi C. L., Xu X., Tong L., Perraud A. L., Scharenberg A. M., Denu J. M. (2006) J. Biol. Chem. 281, 14057–14065 [DOI] [PubMed] [Google Scholar]
- 51. Lee S., Tong L., Denu J. M. (2008) Anal. Biochem. 383, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.