Background: Sphingomyelin synthase (SMS) catalyzes the synthesis of sphingomyelin.
Results: Sphingomyelin-deficient cells failed to proliferate in response to transferrin. Transfection of SMS1 enabled these cells to generate sphingomyelin, promoting clathrin-dependent uptake of transferrin and its dependent proliferation.
Conclusion: SMS1 is indispensable for transferrin internalization and cell proliferation.
Significance: Our findings provide new insights into the role of SMS1 in transferrin biology.
Keywords: Lipids, Membrane Trafficking, Sphingolipid, Trafficking, Vesicles, Cell Proliferation, Clathrin, Sphingomyelin, Sphingomyelin Synthase, Transferrin
Abstract
Transferrin (Tf) endocytosis and recycling are essential for iron uptake and the regulation of cell proliferation. Tf and Tf receptor (TfR) complexes are internalized via clathrin-coated pits composed of a variety of proteins and lipids and pass through early endosomes to recycling endosomes. We investigated the role of sphingomyelin (SM) synthases (SMS1 and SMS2) in clathrin-dependent trafficking of Tf and cell proliferation. We employed SM-deficient lymphoma cells that lacked SMSs and that failed to proliferate in response to Tf. Transfection of SMS1, but not SMS2, enabled these cells to incorporate SM into the plasma membrane, restoring Tf-mediated proliferation. SM-deficient cells showed a significant reduction in clathrin-dependent Tf uptake compared with the parental SM-producing cells. Both SMS1 gene transfection and exogenous short-chain SM treatment increased clathrin-dependent Tf uptake in SM-deficient cells, with the Tf being subsequently sorted to Rab11-positive recycling endosomes. We observed trafficking of the internalized Tf to late/endolysosomal compartments, and this was not dependent on the clathrin pathway in SM-deficient cells. Thus, SMS1-mediated SM synthesis directs Tf-TfR to undergo clathrin-dependent endocytosis and recycling, promoting the proliferation of lymphoma cells.
Introduction
Transferrin (Tf)4 is an important molecule regulating cell proliferation; it is the major iron-transport protein in serum and provides iron to cells by interacting with membrane Tf receptors (TfRs), such as TfR1 and TfR2 (1). Malignant cells highly express TfR and display a hyperactive Tf recycling system, reflecting their heightened need for iron for proliferation and DNA synthesis (2–4). Several studies have suggested that the TfR itself plays a role in cell proliferation (5–7). Thus, the Tf-TfR system appears to be crucial for cell proliferation.
The internalization of plasma membrane receptors and lipids occurs through the clathrin pathway and/or the lipid-raft endocytic pathway (8). Protein-lipid and protein-protein interactions are involved in the targeting of signaling molecules to specialized compartments in those pathways. Numerous studies have uncovered the proteins that participate in this machinery, including GTPases such as dynamins, coat components, and different adaptors (9–12). However, the role of lipid molecules remained relatively underappreciated by researchers until it was found that membrane lipids such as phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 4-phosphate are important for this process (13–15). The importance of lipid molecules was further demonstrated by the finding that endocytosis is regulated by the catalytic action of phospholipid metabolizing enzymes, such as phospholipase D (16–18). Additional evidence for the vital role of lipids is the existence of lipid binding domains in numerous vesicle-forming proteins (19).
Sphingolipids such as sphingomyelin (SM) and ceramide have emerged as vital components of cellular membranes and are involved in diverse cell functions, including cell proliferation, death, and immunity (20). In contrast to phosphatidylinositols, their role in cellular trafficking has not been thoroughly investigated. We previously described the role of the SM cycle in cell differentiation and death, demonstrating a physiologically important role for the mutual conversion of SM and ceramide in cell proliferation (21, 22). SM is produced by the catalytic action of SM synthase (SMS), which is thought to be the sole enzyme responsible for SM synthesis in mammals (23). SMS catalyzes the transfer of phosphocholine from phosphatidylcholine to ceramide, resulting in the generation of SM and diacylglycerol. In mammalians, two SMS isoforms (SMS1 and SMS2) have been shown to account for the synthesis of SM in the lumen of the trans-Golgi (24, 25). The human genome also contains an SMS-related gene (SMSr) whose product does not have the ability to synthesize SM but catalyzes the formation of the SM analog ceramide phosphoethanolamine (26, 27). Several lines of evidence indicate that SM formation by SMS is critical for cell proliferation and survival (25, 28, 29).
The SM recycling system utilizes the clathrin pathway like the Tf-TfR trafficking system (30). A small percentage of SM is trafficked to lysosomes where it is degraded to ceramide by acid sphingomyelinase (SMase) (31). Hydrolysis of plasma membrane SM by bacterial SMase (bSMase) rapidly induces the formation of numerous vesicles, probably containing ceramide, that pinch off from the plasma membrane in ATP-depleted macrophages and fibroblasts (32). Alterations in the sphingolipid composition of trafficking vesicles have the potential to affect the regulation of endocytosis and trafficking. In this study, we propose that SM formation by SMS1 on the plasma membrane regulates clathrin-dependent endocytosis of Tf and, consequently, cell proliferation.
EXPERIMENTAL PROCEDURES
Materials
Alexa Fluor 488-conjugated Tf, LysoTracker Red DND99, and Alexa Fluor-conjugated secondary antibody were purchased from Molecular Probes (Eugene, OR). C6-NBD-SM, C6-NBD-ceramide, and C6-SM were from Matreya Inc. (Pleasant Gap, PA). Hoechst33342 were obtained from Sigma. Anti-clathrin heavy chain IgG monoclonal antibody (MA1–065 clone X22) was from Affinity BioReagents (Golden, CO). Defatted bovine serum albumin (BSA) and human Tf were from Nacalai Tesque (Kyoto, Japan). bSMase was from Higeta Shoyu (Tokyo, Japan), and anti-MBP monoclonal antibody was from Upstate Biotechnology (Waltham, MA). Monoclonal mouse antibodies against Rab11 and the anti-TfR1 mouse monoclonal antibody were purchased from Zymed Laboratories Inc. (San Francisco, CA). Horseradish peroxidase-conjugated anti-Tf antibody was from Abcam (Cambridge, UK). Anti-actin polyclonal antibody was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Horseradish peroxidase-conjugated secondary antibody was purchased from Promega (Madison, WI). Cell culture and SM-deficient WR19L/Fas-SM(−) cells were generated in our laboratory (25). WR19L/Fas-SMS1 and WR19L/Fas-SMS2 cells were established by retroviral transfection of WR19L/Fas-SM(−) cells using pDON-hSMS1 and pDON-hSMS2, respectively. WR19L/Fas-SM(−) cells transfected with empty vector, pDON-AI, and the parental WR19L/Fas-SM(−) cells had the same characteristics, so we used WR19L/Fas-pDON-AI cells as a substitute for WR19L/Fas-SM(−) cells. Cells were cultured at 37 °C in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 50 μm β-mercaptoethanol in a 5% CO2 atmosphere.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
5 μg of total RNA, isolated using a RNA purification kit (RNeasy, Qiagen, Hilden, Germany), was used in reverse transcription reactions using a cDNA synthesis kit (TaKaRa Shuzo, Kyoto, Japan). The resulting cDNA was amplified using Premix TaqPCR kit (TaKaRa) in a PCR thermal cycler. The following primer sets were used: for glyceraldehyde-3-phosphate dehydrogenase, forward 5′-AAGGCTGTGGGCAAGGTCAT-3′ and reverse 5′-CACCACCCTGTTGCTGTAGC-3′; for human and mouse SMS1, forward 5′-TTAGCGCATGACCACTACAC-3′ and human reverse 5′-CAAAGTGGGGATCCGACAAA-3′ or mouse reverse 5′-CAAAGTGGGGATCCGACAAA-3′; for mouse SMS2, forward 5′-ATTCACCTCGTCACTTCTGG-3′ and reverse 5′-TTGCAGCACGGACTCTTTTG-3′; for human SMS2, forward 5′-ACTACTCTACCTGTGCCTGG-3′ and reverse 5′-AGCAGCCAGCAGATTAAATG-3′. PCR products were separated using 5% PAGE and detected with ethidium bromide staining.
SMS Activity
Cells (2 × 106/ml) were homogenized in ice-cold buffer (20 mm Tris-HCl (pH 7.4), 2 mm EDTA, 10 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 2.5 μg/ml leupeptin, and 2.5 μg/ml aprotinin). 100 μg of protein was mixed with the reaction solution (10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 20 μm C6-NBD-ceramide, 120 μm phosphatidylcholine) and incubated for 1 h at 37 °C. Lipids were extracted (33) and separated by thin-layer chromatography on silica gel G plates using chloroform, methanol, 12 mm MgCl2 (65:25:4, v/v/v). For in vivo SMS activity, 1 × 107 cells were treated with 5 μm C6-NBD-ceramide on ice, washed twice with ice-cold PBS, and incubated for 1 h at 37 °C. Cell homogenization and lipid extraction was performed as described for in vitro SMS activity. Fluorescent lipids were visualized by the LAS-1000 system (Fujifilm, Tokyo, Japan).
Lipid Measurement by High Performance Liquid Chromatography-Tandem Mass Spectrometry (LC/MS)
Analysis of SM and ceramide species in lipid extracts was performed using high performance liquid chromatography-tandem mass spectrometry as described by Bielawski et al. (34). Amounts of ceramide were expressed as the total of species with various carbon chains (C14, C16, C18, C18:1, C20, C20:1, C20:4, C22, C22:1, C24, C24:1, C26, and C26:1). Amounts of SM were similarly expressed as the total of species with various carbon-chains (C14, C16, C18, C18:1, C20, C20:1, C22, C22:1, C24, C24:1, C26, and C26:1).
Cell Viability and Proliferation
Cells were washed twice with serum-free RPMI 1640 medium and seeded at 2 × 105 cells/ml. Cell viability was counted by the trypan blue exclusion method.
Kinetics of Tf Uptake and Recycling
For the Tf transport assay, cells were incubated in serum-free medium for 60 min at 37 °C to deplete intracellular Tf. The cells were then incubated with 1 μg/ml [125I]Tf at 37 °C for the indicated time periods. Free and plasma membrane-associated Tf were extracted by washing with PBS and acidic buffer (20 mm MES (pH 5.0), 137 mm NaCl, 50 μm desferal, and 0.1% BSA) at 4 °C. The amount of internalized [125I]Tf was determined using a gamma counter. For measurement of Tf uptake, cells were preincubated in serum-free medium for 60 min and treated with 20 μg/ml Alexa Fluor 488-conjugated Tf for 10 min. Cells were washed twice with ice-cold PBS and treated with acidic buffer for 10 min at 4 °C. Then cells were washed with ice-cold PBS and fixed with 1% paraformaldehyde for 20 min at 4 °C. Fluorescence was measured with a fluorescence-activated cell sorter (FACS).
Immunofluorescence
For the detection of plasma membrane SM, cells were washed twice and kept for 1 h in serum-free RPMI 1640 medium supplemented with 0.2% defatted BSA at 37 °C to eliminate any lipids acquired from the serum. Cells were cytospun onto slides and fixed with 2% formaldehyde. They were probed with lysenin-MBP for 30 min at room temperature and stained with anti-MBP monoclonal IgG followed by Alexa Fluor 546 conjugated anti-mouse IgG antibodies. For determining Tf colocalization with clathrin heavy chain, Rab11, and Lamp2 (lysosomal-associated membrane protein 2), cells were treated with Alexa Fluor 488-conjugated Tf. Fixed cells were cytospinned onto glass slides and permeabilized with PBS containing 0.1% Triton X-100 for 10 min and incubated with PBS containing 1% BSA for 30 min at room temperature. Cells were washed with PBS and then incubated with primary antibodies for 2 h at room temperature. After washing with PBS, Alexa Fluor 546-conjugated anti-IgG antibodies were applied for 1 h. For studies of Tf colocalization with lysosomes in living cells, cells were treated with 200 nm LysoTracker Red before 1 h of Tf treatment and subsequently treated with Alexa Fluor 488-conjugated Tf and Hoechst33342 for 5 min. Confocal laser microscopy was performed using a TCS SP2 microscope (Leica). Colocalization of proteins was quantified with CoLocalizer Pro, Version 2.2, using the overlap coefficient according to Manders (CoLocalization Research Software, Boise, ID).
Western Blot Analysis
Cells were harvested, washed with PBS, and dispersed in hypotonic buffer containing 10 mm Tris-HCl (pH 7.4), 10 mm KCl, 1.5 mm MgCl2, 0.1% SDS, 1 mm PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. To detect intracellular Tf, cells (2 × 106 cells) were treated with or without 20 mm NH4Cl for 10 min and then incubated with 50 μg/ml Tf for 10 min at 37 °C. Cells were then washed with ice-cold PBS and treated with acidic buffer for 10 min, washed again with ice-cold PBS, and lysed. The lysates (50 μg of protein) were subjected to SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA) and blocked with PBS, 0.1% Tween 20 (PBS-T) containing 5% nonfat dried milk. Membranes were incubated with primary antibodies and then secondary antibodies in PBS-T containing 5% nonfat dried milk. Immunoreactive protein bands were visualized using an ECL-peroxidase detection system (Amersham Biosciences) according to the manufacturer's instruction. The bands were quantified with Image J 1.43.
Statistical Analysis
Comparison between two groups was carried out using the unpaired Student's t test.
RESULTS
Characterization of WR19L/Fas Cell Variants
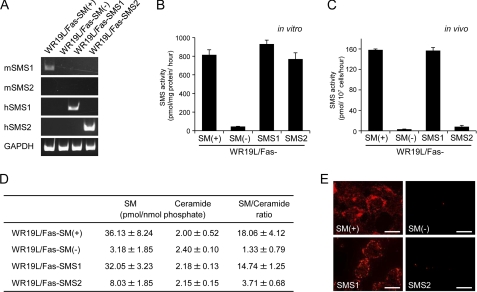
To examine the role of SMSs in Tf trafficking and cell proliferation, we employed SM-defective murine lymphoma WR19L/Fas-SM(−) cells that were established from SM-rich WR19L/Fas-SM(+) cells (35). The parent cells, but not WR19L/Fas-SM(−) cells, constitutively expressed mRNA for SMS1 (Fig. 1A). WR19L/Fas-SM(−) cells were stably transfected with human SMS1 or SMS2 genes to generate WR19L/Fas-SMS1 or WR19L/Fas-SMS2 cells, respectively. The respective mRNAs were detected in these cells (Fig. 1A). Next, we determined SMS activities in vitro and in vivo. WR19L/Fas-SM(−) cells were defective in in vitro (Fig. 1B) and in vivo (Fig. 1C) SMS activities, and cells overexpressing hSMS1 or hSMS2 showed in vitro SMS activities to a similar extent (Fig. 1B). The in vivo activity in WR19L/Fas-SMS1 cells was 150 pmol/107 cells/h, similar to the parental WR19L/Fas-SM(+) cells. SMS2-overexpressing WR19L/Fas-SMS2 cells showed only a slight increase in activity compared with WR19L/Fas-SM(−) cells (Fig. 1C). Furthermore, we determined the cellular content of the SMS substrate, ceramide, and the product, SM, by LC/MS. Introduction of the SMS1 gene dramatically augmented SM content to 32.05 ± 3.23 pmol/nmol of lipid phosphate from 3.18 ± 1.85 pmol/nmol of lipid phosphate in WR19L/Fas-SM(−) cells, but SMS2-overexpressing WR19L/Fas-SMS2 cells displayed only modest increases in SM. The modest elevation in in vivo activity by SMS2 gene introduction and the robust increase in in vitro activity was closely correlated to SM levels in WR19L/Fas-SMS2 cells. However, there was no significant difference in cellular ceramide levels among these cells (Fig. 1D). Immunocytochemical detection of cell surface SM by lysenin showed that plasma membrane SM was detectable only in SMS1-expressing cells (WR19L/Fas-SM(+) and WR19L/Fas-SMS1 cells; Fig. 1E). Thus, SMS1 appears to play an important role in forming SM, particularly at the plasma membrane.
FIGURE 1.
Characterization of WR19L/Fas cell variants for SM synthesis. A, mouse lymphoma cells were kept for 1 h in serum-free medium and washed twice with PBS, and then mRNA levels of mouse (m) and human (h) isoforms were detected by RT-PCR. The in vitro (B) and in vivo (C) SMS activities were measured by using C6-NBD-ceramide as a substrate. Data were obtained from at least three independent experiments. Error bars, S.D. D, ceramide (Cer) and SM levels were determined by LC/MS and normalized by organic phosphates. Values represent the means ± S.D. from four experiments. E, cells were fixed, stained with MBP-conjugated lysenin, and analyzed by confocal microscopy. Scale bars, 10 mm.
SMS1 Involvement in Tf-dependent Cell Proliferation
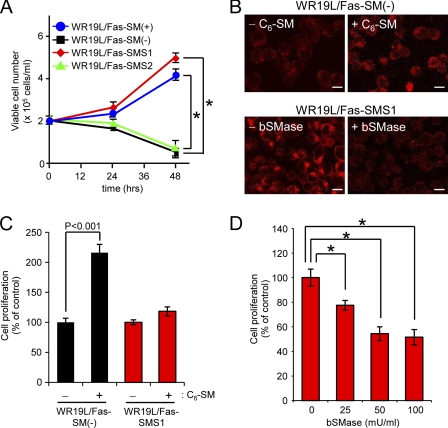
The Tf-TfR system plays an important role in cell proliferation, and we previously proposed that SMS1-generated SM is required for cell proliferation by insulin and Tf (25). However which SMS isoenzymes contribute to Tf-dependent cell proliferation remained unknown. As shown in Fig. 2A, SM-deficient WR19L/Fas-SM(−) cells failed to proliferate in response to Tf. Notably, only SMS1-expressing and SM-rich cells (WR19L/Fas-SMS1 and WR19L/Fas-SM(+) cells) displayed Tf-dependent proliferation, indicating an important role for SMS1-generated SM in Tf-dependent cell proliferation. Moreover, we examined the effects of exogenous short chain C6-SM or bSMase on Tf-dependent cell proliferation. In WR19L/Fas-SMS1 cells, bSMase treatment reduced cell surface SM, and C6-SM-treated WR19L/Fas-SM(−) cells had increased amounts of SM on the cell surface (Fig. 2B). As shown in Fig. 2C, C6-SM treatment imparted on WR19L/Fas-SM(−) cells the ability to proliferate in response to Tf. Conversely, the degradation of cell surface SM by bSMase suppressed Tf-induced proliferation of WR19L/Fas-SMS1 cells (Fig. 2D). These results suggest that Tf-induced cell proliferation requires SM at the cell surface and is dependent on the catalytic activity of SMS1.
FIGURE 2.
SMS1-mediated SM synthesis promotes Tf-mediated cell proliferation. A, cells (1 × 105 cells/ml) were kept in serum-free medium supplemented with 5 μg/ml Tf. After 24 or 48 h, viable cell numbers were counted by the trypan blue exclusion method. *, p < 0.001 versus WR19L/Fas-SM(−) cells. B, WR19L/Fas-SM(−) (black-filled column) or WR19L/Fas-SMS1 cells (red-filled column) were treated with 10 μm C6-SM or 10 milliunits/ml bSMase, respectively. Cells were fixed, stained with MBP-conjugated lysenin, and analyzed by confocal microscopy. Scale bars, 10 μm. C, WR19L/Fas-SM(−) and WR19L/Fas-SMS1 cells were treated with or without 10 μm C6-SM and then stimulated with Tf for 24 h. Viable cell numbers were counted by the trypan blue exclusion method. D, WR19L/Fas-SMS1 cells were treated with the indicated concentrations of bSMase and then incubated with Tf for 24 h. *, p < 0.001. Data were obtained from at least three independent experiments. Error bars, S.D.
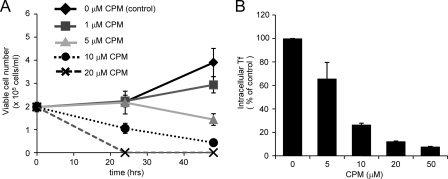
Tf and TfR complexes internalized via clathrin-coated pits pass through early endosomes and recycling endosomes, and this process is important for cell proliferation. To examine the involvement of clathrin-dependent internalization of Tf in cell proliferation, we employed a pharmacological approach using chlorpromazine (36, 37), which is known to suppress clathrin-dependent internalization. Chlorpromazine treatment inhibited Tf-induced cell proliferation of SMS1-expressing and SM-rich WR19L/Fas-SM(+) cells in a dose-dependent manner (Fig. 3A), and the treatment blocked Tf internalization (Fig. 3B). The TfR family consists of two isoforms, TfR1 and TfR2 (1, 38). As we failed to detect significant expression of TfR2 in WR19L/Fas-SM(+) cells (supplemental Fig. S1), TfR1 is likely the major isoform in these cells, accounting for Tf-induced cell proliferation. Thus, the data suggest that clathrin-dependent Tf-TfR1 internalization is important for cell proliferation.
FIGURE 3.
Inhibition of Tf-induced cell growth by chlorpromazine, an inhibitor of clathrin-dependent Tf internalization. A, WR19L/Fas-SM(+) cells (2 × 105 cells/ml) were treated with the indicated concentrations of chlorpromazine (CPM). After 24 and 48 h, viable cell numbers were counted by the trypan blue exclusion method. B, cells were treated with the indicated concentrations of CPM and then incubated with [125I]Tf at 37 °C for 10 min. Cells were washed with acidic buffer (pH 5.0). Radioactivity of intracellular [125I]Tf was counted by a gamma counter and expressed as the percentage of total [125I]Tf radioactivity (mean ± S.D.).
Effects of Individual SMS Isoforms on Tf Internalization and Recycling
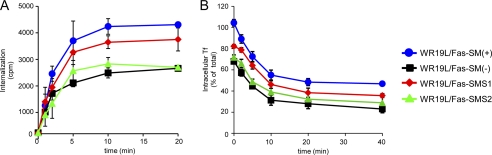
A recent study of trafficking organelles using synaptic vesicles as a model revealed that clathrin-containing vesicles are composed of various lipids, including SM (39). Thus, SM-deficient cells lacking SMSs were presumed to have impairments in clathrin-coated vesicle. Indeed, SMS-deficient cells were impaired in Tf-mediated cell proliferation (Fig. 2). We measured Tf uptake and recycling in all cell types. Cells were incubated with [125I]Tf for the indicated time periods. Tf uptake increased with time and then plateaued after 10 min, but amounts of Tf uptake after 10 and 20 min were significantly reduced in SM-deficient cells compared with SM-rich cells expressing SMS1 (Fig. 4A). Although cellular and/or cell surface expression of TfR1 possibly affected Tf uptake, there were no significant differences in expression levels between the WR19L/Fas variants (supplemental Fig. S1). These results suggest that SMS1-mediated SM synthesis is crucial for Tf uptake.
FIGURE 4.
Kinetics of Tf internalization and recycling. A, cells were kept in serum-free medium for 60 min at 37 °C and then incubated with 1 μg/ml Tf containing [125I]Tf (40,000 cpm) at 37 °C. After Tf treatment, cells were harvested at the indicated time points (0, 1, 2, 5, 10, and 20 min). Internalized [125I]Tf was expressed as cpm (n = 4; mean ± S.D.). B, cells were incubated with [125I]Tf at 37 °C for 10 min and washed twice with PBS and with acidic buffer (pH 5.0) to remove plasma membrane-associated [125I]Tf. Cells were then incubated for the indicated time periods, and the radioactivity of intracellular [125I]Tf was measured with a gamma counter and expressed as a percentage of total internalized [125I]Tf (mean ± S.D.).
We also determined the rate of Tf recycling in these cells. Cells were incubated for 10 min at 37 °C, and cell surface [125I]Tf was dissociated from plasma membrane TfR1 using an acidic buffer at 4 °C. Recycling of internalized [125I]Tf into the medium was measured during re-incubation of the cells at 37 °C. Although [125I]Tf was inefficiently internalized by SMS-SM-deficient WR19L/Fas-SM(−) cells (Fig. 4A), the amount of intracellular label sufficed for recycling measurements. The rate of recycling was not significantly affected by deficits in SMS1-mediated SM synthesis (Fig. 4B).
Requirement of SMS1-mediated SM Synthesis for Clathrin-dependent Internalization of Tf
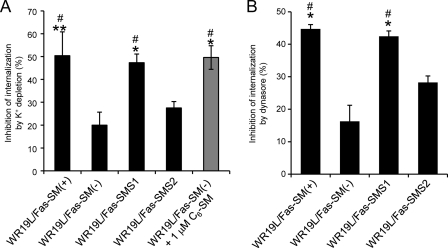
In light of the results above, SMS1-mediated SM synthesis likely controlled clathrin-dependent Tf internalization. To examine this possibility, we tested the effects of SM on clathrin-dependent endocytosis of Tf (Fig. 5A). K+ depletion (40) is known to abolish clathrin-dependent endocytosis, similar to chlorpromazine, thereby inhibiting Tf uptake. More than 50% of Tf internalization was sensitive to K+ depletion in both SM-rich WR19L/Fas-SM(+) and WR19L/Fas-SMS1 cells, whereas SM-deficient cells such as WR19L/Fas-SM(−) and WR19L/Fas-SMS2 showed less sensitivity to K+ depletion. Most importantly, the addition of 1 μm C6-SM significantly restored the susceptibility of SM-deficient WR19L/Fas-SM(−) cells to inhibition of Tf internalization by K+ depletion by more than 2-fold, suggesting that SM is a key lipid that allows Tf to be internalized through clathrin-coated vesicles.
FIGURE 5.
SMS1-mediated SM synthesis promotes clathrin-dependent Tf internalization. A, WR19L cells were kept in serum-free medium at 37 °C for 60 min, and then the medium was replaced with K+ depletion buffer or 10 mm KCl-containing buffer. After 30 min, cells were treated with or without 10 μm C6-SM for 10 min and then incubated with [125I]Tf at 37 °C for 10 min. After washing with PBS and acidic buffer (pH 5.0), the radioactivity of intracellular [125I]Tf was counted with a gamma counter. The values are expressed as the inhibition rate of [125I]Tf internalization by 10 mm KCl (means ± S.D.). B, cells were kept in serum-free medium at 37 °C for 60 min and then pretreated with 80 μm dynasore at 37 °C for 10 min. Cells were incubated with 2 μg/ml Alexa Fluor 488-conjugated Tf at 37 °C for 10 min. Internalization was measured with FACS. The values are expressed as the inhibition rate of intracellular Alexa Fluor 488-conjugated Tf in vehicle treated cells (mean ± S.D.). *, p < 0.005; **, p < 0.05 versus WR19L/Fas-SM(−) cells. #. p < 0.005 versus WR19L/Fas-SMS2 cells.
Dynamin is a GTPase that is essential for clathrin-dependent vesicle formation (41), and the selective inhibitor dynasore has been shown to inhibit the clathrin-dependent internalization of Tf (42). Similar to K+ depletion, Tf internalization in SM-rich WR/Fas-SM(+) and SMS1 cells was sensitive to dynasore treatment compared with SM-deficient cells (Fig. 5B). Thus, SM appears to be required for dynamin-dependent internalization of Tf, and dynamin is unlikely to significantly contribute to the clathrin-independent pathway.
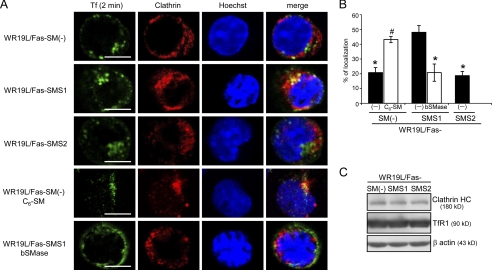
When internalized, Tf-TfR complexes are packed into clathrin-coated vesicles. To examine the effects of SMS1 introduction on the compartmentalization of Tf-TfR complexes into clathrin-coated vesicles, we performed confocal microscopy using Alexa Fluor 488-conjugated Tf and clathrin heavy chain antibodies. After 2 min of Tf chase, the colocalization of Tf with clathrin heavy chain in SM-deficient cells was 20.8%, and the restoration of SM by SMS1 introduction significantly increased colocalization (WR19L/Fas-SMS1 cells, 48.1%; Fig. 6, A and B). No colocalization was observed after 5 min (data not shown), which matches the mode of Tf internalization and traffic in other cell types (41). Furthermore, we tested whether manipulating SM content at the plasma membrane affected the internalization of Tf with clathrin. The loading of exogenous C6-SM onto WR19L/Fas-SM(−) cells significantly restored early Tf/clathrin colocalization to 43.2% from 20.8%, and conversely, SM hydrolysis by bSMase led to the loss of early colocalization in WR19L/Fas-SMS1 cells, down to 20.8% from 48.1% (Fig. 6, A and B). No significant difference in protein levels of TfR1 and clathrin heavy chain between SM-rich and -deficient cells was found, suggesting that changes in protein expression were not involved in regulating the Tf internalization pathway from being primarily clathrin-dependent to being clathrin-independent or vice versa. Tf was colocalized with TfR1 in all WR19L/Fas variants (supplemental Fig. S1C), indicating that Tf compartmentalization is similar to that of TfR1. These data suggest that SMS1-mediated SM synthesis plays a key role in promoting the packaging of Tf-TfR1 complexes into clathrin-coated vesicles at the plasma membrane. Consistent with the results in Fig. 5, these results also suggest that SM deficiency directs Tf to the endocytic pathway instead of the clathrin pathway.
FIGURE 6.
Involvement of SMS1-mediated SM synthesis on the sorting of Tf into clathrin-positive compartments. A, cells were treated with or without 10 μm C6-SM or 50 milliunits/ml bSMase for 10 min at room temperature and then incubated with Alexa Fluor 488-conjugated Tf (green) for 2 min. Cells were fixed, stained with clathrin heavy chain antibody (red), and analyzed by confocal microscopy. Nuclei were stained with Hoechst33342 (blue). Scale bars, 5 μm. B, Tf colocalization with clathrin was determined using the Manders' colocalization coefficient as described under “Experimental Procedures.” The values are expressed as percentages of colocalized Tf and clathrin heavy chain. Error bars, S.D. *, p < 0.001 versus WR19L/Fas-SMS1 cells; #, p < 0.001 versus WR19L/Fas-SM(−) cells. C, TfR and clathrin heavy chain levels in cells were determined by Western blot analysis.
SM Synthesis by SMS1 Promotes Tf Sorting into Recycling Endosomes
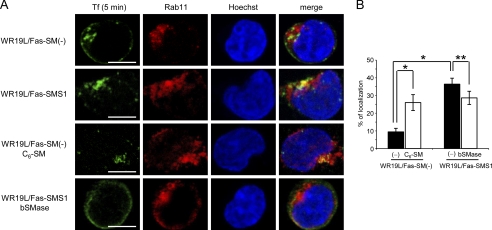
Tf internalized along with other clathrin-coated cargo is sequentially sorted to Rab11-positive recycling endosomes (43). We examined the involvement of SMS1-mediated SM synthesis in the sequestration of Tf into these recycling endosomes. In SM-deficient WR19L/Fas-SM(−) cells, 9.4% of internalized Tf was colocalized with Rab11 at 5 min (Fig. 7), and no significant colocalization was observed at later time points (7 and 10 min, data not shown). Surprisingly, SM-rich WR19L/Fas-SMS1 cells showed significantly increased colocalization compared with SM-deficient WR19L/Fas-SM(−) cells. Those results show that introduction of the SMS1 gene promoted the sorting of Tf into Rab11-positive compartments.
FIGURE 7.
Involvement of SMS1-mediated SM synthesis on the sorting of Tf into Rab11-positive recycling endosomes. A, WR19L/Fas-SM(−) or WR19L/Fas-SMS1 cells were treated with or without 10 μm C6-SM or 50 milliunits/ml bSMase for 10 min. Cells were then incubated with Alexa Fluor 488-conjugated Tf (green) for 5 min, fixed, and stained with Rab11 antibody (red). Nuclei were counterstained with Hoechst33342 (blue). Images were obtained by confocal microscopy. Scale bars, 5 μm. B, shown are percentages of colocalized Tf and Rab11. Error bars, S.D. *, p < 0.005; **, p < 0.005.
Treatment of SM-deficient WR19L/Fas-SM(−) cells with exogenous C6-SM restored the colocalization of Tf with Rab11 by 28.7%, whereas bSMase hydrolysis of cell surface SM led to a decrease in Tf-Rab11 colocalization down to 26.0% from 36.4% in SM-rich WR19L/Fas-SMS1 cells, suggesting that SM promotes the sequestration of Tf to Rab11-positive recycling endosomes (Fig. 7). These results demonstrate that not only is SMS1-mediated SM synthesis required for promoting Tf passage thorough recycling endosomes but also that Tf exits from the recycling endosome pathway in SM-deficient cells.
Tf Trafficking to the Lysosomal Degradation Pathway in SM Deficiency
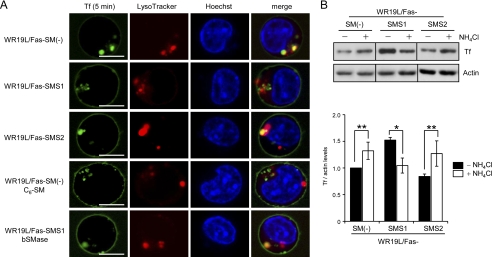
Cargo-specific sorting leads to distinct cargo trafficking. Cargo can be routed from early endosomes to late endosomes and lysosomes for degradation or to recycling endosomes that bring the cargo back to the plasma membrane. In SM-deficient cells, internalized Tf was less colocalized with Rab11-positive compartments compared with SM-rich cells, thereby passing into the degradation pathway. We examined possible routes of Tf movement to lysosomes in SM-deficient cells. Tf was colocalized with lysosomes (LysoTracker Red) in SM-deficient cells, and treatment of those cells with C6-SM decreased colocalization (Fig. 8A), whereas bSMase treatment of SM-rich cells increased colocalization. Moreover, approximately half of the Tf was colocalized with Lamp2 in SM-deficient cells at 5 min (SM(−), 46.8%; SMS2, 52.1% colocalization), although SM-rich WR19L/Fas-SMS1 cells displayed only 26.3% colocalization (supplemental Fig. S2), suggesting that SM deficiency promotes the re-directing of Tf to lysosomes.
FIGURE 8.
The sorting of Tf into LysoTracker-positive endolysosomal compartments in SM-deficient cells. A, cells were preincubated with 200 ng/ml LysoTracker Red DND99 (red) at 37 °C for 60 min and then treated with or without 10 μm C6-SM or 50 milliunits/ml bSMase for 10 min. The cells were treated with Alexa Fluor 488-conjugated Tf (green) and Hoechst3342 (blue) for 5 min. Living cells were analyzed by confocal microscopy. Scale bars, 5 μm. B, effects of NH4Cl on Tf trafficking are shown. Cells were treated with 20 mm NH4Cl for 10 min and then incubated with 50 μg/ml Tf for 10 min. After washing with ice-cold PBS and acidic buffer (pH 5.0) to remove membrane-bond Tf, intracellular Tf was detected by Western blot analysis and quantified with Image J 1.43. Value are expressed as the -fold increase over vehicle-treated WR19L/Fas-SM(−) cells (n = 3, mean ± S.D.). *, p < 0.005; **, p < 0.005.
Lysosomes are acidic organelles that play an important role in the degradation of proteins and lipids. Inhibition of lysosomal function by NH4Cl (44) increased cellular Tf in SM-deficient cells but decreased cellular Tf in SM-rich cells (Fig. 8B), indicating that Tf might be degraded in the lysosomal compartment of SM-deficient cells. These results suggest that SM deficiency induces the sequestration of internalized Tf into the lysosomal compartment for degradation. Why the reduction by NH4Cl treatment occurs in SM-rich cells remains unclear, but pH neutralization of the early endosomes by NH4Cl is thought to inhibit the internalization of Tf.
DISCUSSION
We previously reported on the biological role of SMS, which is one of most important enzymes regulating hematopoietic cell proliferation, differentiation, and death through “SM cycle” (21, 45–47). Up-regulation of SMS promotes cell proliferation in lymphocytes, hepatocytes, astrocytes, and fibroblasts (48–50). In contrast, inhibition of SMS activity is involved in inducing apoptosis of TNF-α-treated Kym-1 cells (51) and Fas-cross-linked Jurkat cells (47). Our group (25) and that of Holthuis and co-workers (24) discovered the SMS genes, but the biological role of SMS1 and SMS2 in Tf-mediated cell proliferation remained poorly understood. Here we demonstrate a role for SMS1-mediated SM synthesis in clathrin-dependent endocytosis of Tf-TfR1 complexes and cell proliferation. It also appears that the complexes on SM-deficient membranes could be re-directed to lysosomal compartments for degradation.
Ding et al. (52) demonstrated that both SMS1 and SMS2 in THP1-derived macrophages are involved in generating plasma membrane SM. However, SMS2 introduction into WR19L/Fas-SM(−) cells showed little increase in in vivo SMS activity and SM levels, whereas enrichment of cellular and plasma membrane SM in SMS1-overexpressing WR19L/Fas-SMS1 cells resulted in increases in both in vivo and in vitro SMS activity (Fig. 1). Consistent with those results, mouse embryonic fibroblasts obtained from SMS2 knock-out mice, which still expressed SMS1, showed higher amounts of SM in the plasma membrane compared with embryonic fibroblasts from SMS1 knock-out mice (data not shown), suggesting that SMS1 is the primary enzyme maintaining SM levels in the plasma membrane.
Why SMS2 overexpression in WR19L/Fas-SM(−) cells failed to fully generate SM in the plasma membrane remains unclear. It is possible that SMS2 activity is suppressed by an inhibitory molecule(s) in lymphoma cells, although the biological regulation of SMS2 activity remains poorly understood.
Tf and TfR are involved in the regulation of cell proliferation (53). Cells treated with anti-TfR antibodies could internalize Tf via the non-TfR/clathrin pathway, inhibiting cell proliferation (53). In addition, the inhibition of clathrin-dependent internalization of Tf suppressed cell proliferation (Fig. 3). Tf-TfR1 complexes are delivered to early endosomes where the low pH facilitates the dissociation of iron from receptor/ligand complexes (54), making iron biologically available. Iron-containing proteins (e.g. cytochromes, mitochondrial aconitase, and Fe-S proteins of the electron transport chain) catalyze key reactions involved in energy metabolism and DNA synthesis (e.g. ribonucleotide reductase) (54). In light of the requirement for iron in cell proliferation, the biological availability of this metal plays an important role in Tf-induced cell proliferation. Therefore, the present study suggests that SMS1-mediated SM synthesis plays a key role in generating the biological activity of Tf.
SM deficiency is likely to decrease Tf colocalization with Rab11 (Fig. 7) and redirects Tf to the lysosomal compartment (Fig. 8) without affecting the rate of Tf recycling (Fig. 4). Although these results seem to be inconsistent, we can speculate why SM deficiency had no effect on the Tf recycling rate. Internalized Tf may simply not pass through early endosomes. It has been shown that (a) internalized Tf is recycled directly back from early endosomes to the plasma membrane (55), and (b) Tf is recycled in the absence of perinuclear recycling endosomes (44). In WR lymphoid cells, the majority of internalized Tf might be transported from early endosomes to the plasma membrane. Thus, SM deficiency is suggested to have no effects on the rate of Tf recycling.
We propose that SMS1 activity accounts for the majority of SM synthesis. Although SM is a possible constituent of transport vesicles and plays a role in the clathrin-dependent trafficking of Tf, its mechanism of function still remains largely unknown. SMase hydrolyzes SM, generating ceramide. The levels of ceramide in SM-deficient cells were similar to those in SM-rich cells (Fig. 1). Furthermore, in contrast to SM in the present study, ceramide has been shown to prevent classical PKC-dependent sequestration of Tf into the recycling endosome-like compartment, the pericentrion, by activating ceramide-activated protein phosphatases (56). Those rule out the involvement of ceramide. Thus, SM, but not ceramide, is likely to be the key lipid regulating the clathrin-dependent internalization of Tf.
SM deficiency disrupts the clathrin-dependent uptake of Tf. LDL and cholera toxin subunit B are internalized through clathrin-dependent and independent pathways, respectively (57). SM deficiency had no significant effects on their uptake (supplemental Fig. S3). Thus, SM is specifically required for the clathrin-dependent internalization of Tf in mouse lymphoma cells.
Plasma membrane receptors are not always degraded by lysosomes; some are recycled back to the plasma membrane after endocytosis (58). For example, Tf and TfR are internalized and recycled back to the cell surface through recycling endosomes. However, this does not imply that Tf-TfR complexes are always recycled and used endlessly. TfR itself undergoes degradation and is degraded by a Rab12-dependent pathway (59). Tf is directed to lysosomal compartments close to perinuclear regions in SM-deficient cells (Fig. 8A). Although we failed to detect the degradation products of Tf and/or TfR1 (data not shown), Tf-TfR1 complexes are presumed to undergo degradation. The molecular mechanisms underlying the redirection of Tf to lysosomal compartments in SM-deficient cells remain unknown.
In summary, SM synthesized by SMS1 plays a pivotal role in the clathrin-dependent internalization of Tf-TfR and promotes cell proliferation. SM deficiency (e.g. due to a lack of SMS1) facilitates the sequestration of Tf into endolysosomal compartments, thereby reducing the biological availability of Tf-derived iron and its ability to enhance cell division.
Supplementary Material
This study was supported in part by the Sapporo Biocluster “Bio-S,” the Knowledge Cluster Initiative of the Ministry of Education, Culture, Sports, Science, and Technology (Japan), Japan Society for the Promotion of Science Grants-in-aid for Young Scientists 21890144 and 23790366 (to K. K.), the Sumitomo Foundation (to K. K.), Japanese Ministry of Education, Culture, Sports, Science, and Technology Grants 22249041 (to H. U.) and 21113530 and 22390018 (to T. K.), Lipid Dynamics Program of RIKEN (to T. K.), the Uehara Memorial Foundation (to H. U.), the Vehicle Racing Commemorative Foundation (to H. U.), and the Kanazawa Medical University Research Foundation (to H. U.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Tf
- transferrin
- TfR
- transferrin receptor
- SM
- sphingomyelin
- SMS1
- sphingomyelin synthase 1
- SMS2
- sphingomyelin synthase 2
- SMase
- sphingomyelinase
- bSMase
- bacterial SMase
- NBD
- 12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl))
- MBP
- myelin binding protein.
REFERENCES
- 1. Gomme P. T., McCann K. B., Bertolini J. (2005) Drug Discov. Today 10, 267–273 [DOI] [PubMed] [Google Scholar]
- 2. Gatter K. C., Brown G., Trowbridge I. S., Woolston R. E., Mason D. Y. (1983) J. Clin. Pathol. 36, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jefferies W. A., Brandon M. R., Williams A. F., Hunt S. V. (1985) Immunology 54, 333–341 [PMC free article] [PubMed] [Google Scholar]
- 4. Panaccio M., Zalcberg J. R., Thompson C. H., Leyden M. J., Sullivan J. R., Lichtenstein M., McKenzie I. F. (1987) Immunol. Cell Biol. 65, 461–472 [DOI] [PubMed] [Google Scholar]
- 5. Cano E., Pizarro A., Redondo J. M., Sánchez-Madrid F., Bernabeu C., Fresno M. (1990) Eur. J. Immunol. 20, 765–770 [DOI] [PubMed] [Google Scholar]
- 6. Manger B., Weiss A., Hardy K. J., Stobo J. D. (1986) J. Immunol. 136, 532–538 [PubMed] [Google Scholar]
- 7. Moura I. C., Centelles M. N., Arcos-Fajardo M., Malheiros D. M., Collawn J. F., Cooper M. D., Monteiro R. C. (2001) J. Exp. Med. 194, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Roy C., Wrana J. L. (2005) Nat. Rev. Mol. Cell Biol. 6, 112–126 [DOI] [PubMed] [Google Scholar]
- 9. Kaksonen M. (2008) J. Cell Biol. 180, 1059–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higgins M. K., McMahon H. T. (2002) Trends Biochem. Sci. 27, 257–263 [DOI] [PubMed] [Google Scholar]
- 11. Grant B. D., Donaldson J. G. (2009) Nat. Rev. Mol. Cell Biol. 10, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Traub L. M. (2009) Nat. Rev. Mol. Cell Biol. 10, 583–596 [DOI] [PubMed] [Google Scholar]
- 13. De Camilli P., Emr S. D., McPherson P. S., Novick P. (1996) Science 271, 1533–1539 [DOI] [PubMed] [Google Scholar]
- 14. Padrón D., Tall R. D., Roth M. G. (2006) Mol. Biol. Cell 17, 598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roth M. G. (1999) Trends Cell Biol. 9, 174–179 [DOI] [PubMed] [Google Scholar]
- 16. Du G., Altshuller Y. M., Vitale N., Huang P., Chasserot-Golaz S., Morris A. J., Bader M. F., Frohman M. A. (2003) J. Cell Biol. 162, 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Du G., Huang P., Liang B. T., Frohman M. A. (2004) Mol. Biol. Cell 15, 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hughes W. E., Parker P. J. (2001) Biochem. J. 356, 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demmel L., Gravert M., Ercan E., Habermann B., Müller-Reichert T., Kukhtina V., Haucke V., Baust T., Sohrmann M., Kalaidzidis Y., Klose C., Beck M., Peter M., Walch-Solimena C. (2008) Mol. Biol. Cell 19, 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 21. Okazaki T., Bell R. M., Hannun Y. A. (1989) J. Biol. Chem. 264, 19076–19080 [PubMed] [Google Scholar]
- 22. Hannun Y. A. (1994) J. Biol. Chem. 269, 3125–3128 [PubMed] [Google Scholar]
- 23. Voelker D. R., Kennedy E. P. (1982) Biochemistry 21, 2753–2759 [DOI] [PubMed] [Google Scholar]
- 24. Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. (2004) EMBO J. 23, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. (2004) J. Biol. Chem. 279, 18688–18693 [DOI] [PubMed] [Google Scholar]
- 26. Rietveld A., Neutz S., Simons K., Eaton S. (1999) J. Biol. Chem. 274, 12049–12054 [DOI] [PubMed] [Google Scholar]
- 27. Vacaru A. M., Tafesse F. G., Ternes P., Kondylis V., Hermansson M., Brouwers J. F., Somerharju P., Rabouille C., Holthuis J. C. (2009) J. Cell Biol. 185, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hanada K., Nishijima M., Kiso M., Hasegawa A., Fujita S., Ogawa T., Akamatsu Y. (1992) J. Biol. Chem. 267, 23527–23533 [PubMed] [Google Scholar]
- 29. Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J. C. (2007) J. Biol. Chem. 282, 17537–17547 [DOI] [PubMed] [Google Scholar]
- 30. Puri V., Watanabe R., Singh R. D., Dominguez M., Brown J. C., Wheatley C. L., Marks D. L., Pagano R. E. (2001) J. Cell Biol. 154, 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koval M., Pagano R. E. (1990) J. Cell Biol. 111, 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zha X., Pierini L. M., Leopold P. L., Skiba P. J., Tabas I., Maxfield F. R. (1998) J. Cell Biol. 140, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 34. Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. (2006) Methods 39, 82–91 [DOI] [PubMed] [Google Scholar]
- 35. Miyaji M., Jin Z. X., Yamaoka S., Amakawa R., Fukuhara S., Sato S. B., Kobayashi T., Domae N., Mimori T., Bloom E. T., Okazaki T., Umehara H. (2005) J. Exp. Med. 202, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hernaez B., Alonso C. (2010) J. Virol. 84, 2100–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Subtil A., Hémar A., Dautry-Varsat A. (1994) J. Cell Sci. 107, 3461–3468 [DOI] [PubMed] [Google Scholar]
- 38. Kawabata H., Yang R., Hirama T., Vuong P. T., Kawano S., Gombart A. F., Koeffler H. P. (1999) J. Biol. Chem. 274, 20826–20832 [DOI] [PubMed] [Google Scholar]
- 39. Takamori S., Holt M., Stenius K., Lemke E. A., Grønborg M., Riedel D., Urlaub H., Schenck S., Brügger B., Ringler P., Müller S. A., Rammner B., Gräter F., Hub J. S., De Groot B. L., Mieskes G., Moriyama Y., Klingauf J., Grubmüller H., Heuser J., Wieland F., Jahn R. (2006) Cell 127, 831–846 [DOI] [PubMed] [Google Scholar]
- 40. Larkin J. M., Brown M. S., Goldstein J. L., Anderson R. G. (1983) Cell 33, 273–285 [DOI] [PubMed] [Google Scholar]
- 41. van Dam E. M., Stoorvogel W. (2002) Mol. Biol. Cell 13, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. (2006) Dev. Cell 10, 839–850 [DOI] [PubMed] [Google Scholar]
- 43. Trischler M., Stoorvogel W., Ullrich O. (1999) J. Cell Sci. 112, 4773–4783 [DOI] [PubMed] [Google Scholar]
- 44. Liebl D., Difato F., Horníková L., Mannová P., Stokrová J., Forstová J. (2006) J. Virol. 80, 4610–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Itoh M., Kitano T., Watanabe M., Kondo T., Yabu T., Taguchi Y., Iwai K., Tashima M., Uchiyama T., Okazaki T. (2003) Clin. Cancer Res. 9, 415–423 [PubMed] [Google Scholar]
- 46. Taguchi Y., Kondo T., Watanabe M., Miyaji M., Umehara H., Kozutsumi Y., Okazaki T. (2004) Blood 104, 3285–3293 [DOI] [PubMed] [Google Scholar]
- 47. Watanabe M., Kitano T., Kondo T., Yabu T., Taguchi Y., Tashima M., Umehara H., Domae N., Uchiyama T., Okazaki T. (2004) Cancer Res. 64, 1000–1007 [DOI] [PubMed] [Google Scholar]
- 48. Luberto C., Hannun Y. A. (1998) J. Biol. Chem. 273, 14550–14559 [DOI] [PubMed] [Google Scholar]
- 49. Miró-Obradors M. J., Osada J., Aylagas H., Sánchez-Vegazo I., Palacios-Alaiz E. (1993) Carcinogenesis 14, 941–946 [DOI] [PubMed] [Google Scholar]
- 50. Riboni L., Viani P., Bassi R., Giussani P., Tettamanti G. (2001) J. Biol. Chem. 276, 12797–12804 [DOI] [PubMed] [Google Scholar]
- 51. Bourteele S., Hausser A., Döppler H., Horn-Müller J., Röpke C., Schwarzmann G., Pfizenmaier K., Müller G. (1998) J. Biol. Chem. 273, 31245–31251 [DOI] [PubMed] [Google Scholar]
- 52. Ding T., Li Z., Hailemariam T., Mukherjee S., Maxfield F. R., Wu M. P., Jiang X. C. (2008) J. Lipid Res. 49, 376–385 [DOI] [PubMed] [Google Scholar]
- 53. Daniels T. R., Delgado T., Rodriguez J. A., Helguera G., Penichet M. L. (2006) Clin. Immunol. 121, 144–158 [DOI] [PubMed] [Google Scholar]
- 54. Richardson D. R., Ponka P. (1997) Biochim. Biophys. Acta 1331, 1–40 [DOI] [PubMed] [Google Scholar]
- 55. Sheff D. R., Daro E. A., Hull M., Mellman I. (1999) J. Cell Biol. 145, 123–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kitatani K., Idkowiak-Baldys J., Hannun Y. A. (2007) J. Biol. Chem. 282, 20647–20656 [DOI] [PubMed] [Google Scholar]
- 57. Cheng Z. J., Singh R. D., Sharma D. K., Holicky E. L., Hanada K., Marks D. L., Pagano R. E. (2006) Mol. Biol. Cell 17, 3197–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maxfield F. R., McGraw T. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 59. Matsui T., Itoh T., Fukuda M. (2011) Traffic, in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.