Background: Glycoglycerolipids, synthesized by glycosyltransferases (glycolipid synthases), are key structural membrane components in mycoplasma.
Results: M. genitalium GT MG517 has been recombinantly expressed and kinetically characterized. It is a processive GT transferring glucosyl or galactosyl residues to membrane-bound diacylglycerol, with higher monoglycosyl- than diglycosyltransferase activity. Inhibition of the enzyme results in mycoplasma growth inhibition.
Conclusion: The membrane glycoglycerolipids of M. genitalium are synthesized by a single processive glycosyltransferase, GT MG517. It is proposed as a potential therapeutic target against mycoplasma infections.
Significance: The essential function of GT MG517 for mycoplasma viability and the absence of glycoglycerolipids in animal host cells of mycoplasma infections make the enzyme a target for drug discovery by designing specific inhibitors.
Keywords: Enzyme Kinetics, Glycolipids, Glycosylation, Membrane Enzymes, Membrane Lipids, Glycosyltransferase, Infectious Diseases, Mycoplasma, Therapeutic Target
Abstract
Mycoplasmas contain glycoglycerolipids in their plasma membrane as key structural components involved in bilayer properties and stability. A membrane-associated glycosyltransferase (GT), GT MG517, has been identified in Mycoplasma genitalium, which sequentially produces monoglycosyl- and diglycosyldiacylglycerols. When recombinantly expressed in Escherichia coli, the enzyme was functional in vivo and yielded membrane glycolipids from which Glcβ1,6GlcβDAG was identified as the main product. A chaperone co-expression system and extraction with CHAPS detergent afforded soluble protein that was purified by affinity chromatography. GT MG517 transfers glucosyl and galactosyl residues from UDP-Glc and UDP-Gal to dioleoylglycerol (DOG) acceptor to form the corresponding β-glycosyl-DOG, which then acts as acceptor to give β-diglycosyl-DOG products. The enzyme (GT2 family) follows Michaelis-Menten kinetics. kcat is about 5-fold higher for UDP-Gal with either DOG or monoglucosyldioleoylglycerol acceptors, but it shows better binding for UDP-Glc than UDP-Gal, as reflected by the lower Km, which results in similar kcat/Km values for both donors. Although sequentially adding glycosyl residues with β-1,6 connectivity, the first glycosyltransferase activity (to DOG) is about 1 order of magnitude higher than the second (to monoglucosyldioleoylglycerol). Because the ratio between the non-bilayer-forming monoglycosyldiacylglycerols and the bilayer-prone diglycosyldiacylglycerols contributes to regulate the properties of the plasma membrane, both synthase activities are probably regulated. Dioleoylphosphatidylglycerol (anionic phospholipid) activates the enzyme, kcat linearly increasing with dioleoylphosphatidylglycerol concentration. GT MG517 is shown to be encoded by an essential gene, and the addition of GT inhibitors results in cell growth inhibition. It is proposed that glycolipid synthases are potential targets for drug discovery against infections by mycoplasmas.
Introduction
Glycoglycerolipids (galactolipids) are found in plant chloroplasts and cyanobacteria as membrane components associated with photosynthetic tissues (1, 2). Similar glycolipids with a wider structural diversity are widespread in Gram-positive bacteria, in which the headgroups linked to diacylglycerol contain glucosyl, galactosyl, or mannosyl units with diverse glycosidic linkages (1). Their functions expand from membrane anchors of other biomolecules (i.e. lipoteichoic acids) (3, 4) to free components involved in membrane bilayer stability (1). Within the latter role, incorporation of glycosyl residues switches a non-bilayer-forming monoglycosyldiacylglycerol into a bilayer-forming diglycosyldiacylglycerol (5–7).
Glycoglycerolipids are synthesized by the action of glycosyltransferases (GTs)4 on a diacylglycerol acceptor. Glycolipid synthases are classified in different sequence-based GT families (carbohydrate-active enzyme classification (8)). Although not many have been enzymologically studied, most of them have either mono- or disynthase activity, one enzyme for the first glycosylation of diacylglycerol, and a different enzyme for further glycosylation leading to the diglycosyldiacylglycerol product. Table 1 summarizes currently identified glycolipid synthases classified by GT families. In plants, monogalactosyldiacylglycerol (MGalDAG) synthases belong to family 28 glycosyltransferases (GT28) (9–13, 15, 16), whereas the digalactosyldiacylglycerol (DGalDAG) synthases are classified in family GT4 (17). In cyanobacteria, MGlcDAG synthases are family GT2 enzymes (18), whereas DGalDAG synthases are tentatively assigned to family GT4 (19). In non-photosynthetic Gram-positive bacteria, sequentially acting GTs are also found, in which a single enzyme transfers the first and second (and eventually a third or more) glycosyl unit. The enzymes from Bacillus and Staphylococcus belong to family GT28 and have been shown to produce mono-, di-, and oligoglucoglycerolipids starting with DAG as primary acceptor (3, 20, 21). The Staphylococcus aureus GT activity (YpfP) is localized on the cytoplasmic side of the plasma membrane, and the diglucosyldiacylglycerol product is translocated to the extracellular side, serving as a priming structure for lipotheicoic acid synthesis (4). Also, processive bacterial enzymes are found in family GT21, which mainly contains mammalian glucosylceramide synthases (22).
TABLE 1.
Characterized glycosyldiacylglycerol synthases grouped by CAZyme GT families
Phylogenetic trees for glycosyldiacylglycerol synthases (UNIPROT sequences, both characterized and uncharacterized) are provided in supplemental Fig. S4).
| Family | Activity/EC | Organism | Enzyme | UNIPROT | Product | Reference |
|---|---|---|---|---|---|---|
| GT28a | MGalDAG synthase/EC 2.41.1.46 | Arabidopsis thaliana | Mgd1 | O81770 | Galβ3DAG | 10, 15 |
| A. thaliana | Mgd2 | O82730 | Galβ3DAG | 10, 12 | ||
| A. thaliana | Mgd3 | Q9SI93 | Galβ3DAG | 10, 12 | ||
| Cucumis sativus | MGD1 | P93115 | Galβ3DAG | 9 | ||
| Spinacia aleracea | MGD1 | Q9SM44 | Galβ3DAG | 13 | ||
| MGalD synthase/EC 2.41.1.46 | Chloroflexus aurantiacus | MGD | A9WFF3 | Galβ3DAG | 14 | |
| DGlcDAG/EC 2.4.1.208 | C. aurantiacus | DGD | A9WFF4 | Glcβ6Galβ3DAG | 14 | |
| Processive GlcDAG synthase | Bacillus subtilis | YpfP | P54166 | Glcβ3DAG, Glcβ6Glcβ3DAG | 20 | |
| S. aureus str. RN4220 | YpfP | Q5HH69 | Glcβ3DAG, Glcβ6Glcβ3DAG | 21 | ||
| GT21 | Procesive Glc/Gal synthase | Agrobacterium tumefaciens | AtGcs | Q7CYH2 | Glcβ3DAG, Galβ3DAG, Glcβ6Glcβ3DAG, Glcβ6Galβ3DAG, Galβ6Galβ3DAG | 22 |
| Mesorhizobium loti | MlGcs | Q98BB3 | Same as above | 22 | ||
| GT4b | DGalDAG synthase/EC 2.4.1.241 | A. thaliana | DGDG1 | Q9S7D1 | Galα6Galβ3DAG | 17 |
| MGlcDAG synthase/EC 2.4.1.157 | A. laidlawii | AlMgs | Q93P60 | Glcα3DAG | 33 | |
| Streptococcus pneumoniae | SpMgs | O06453 | Glcα3DAG | 33 | ||
| Deinococcus radiodurans | DrMgs | Q9RV05 | Glcα3DAG | 14 | ||
| Thermotoga maritima | TrMgs | Q9WZK3 | Glcα3DAG | 14 | ||
| DGlcDAG synthase/EC 2.4.1.208 | A. laidlawii | AlDgs | Q8KQL6 | Glcα2Glcα3DAG | 36 | |
| S. pneumoniae | CpoA | O06452 | Galα2Glcα3DAG | 52 | ||
| GT2 | MGlcDAG synthase/EC 2.4.1.157 | Synechocystis sp. | MGlcDG | P74165 | Glcβ3DAG | 18 |
| Nostoc sp. PCC 7120 | MGlcDG | Q8YMK0 | Glcβ3DAG | 18 | ||
| Procesive Glc/Gal synthase | M. pneumoniae | MPN483 | P75302 | Galβ3DAG, Glcβ6Galβ3DAG | 38 | |
| M. genitalium | MG517 | Q9ZB73 | Glcβ3DAG, Galβ3DAG, Glcβ6Glcβ3DAG | This work |
a Other annotated (UNIPROT), not characterized GT28 (plant): Glycine max (Q9FZL4), Nicotina tabacum (Q9FZL3), Oryza sativa subsp. Japonica (type 1, Q69QJ7 ; type 2, Q6UTZ2 ; type 3, Q0DWQ1), O. sativa subsp. Indica (type 2, A2YTP9), Vigna unguiculata (type A, Q07A03; type B, A4UNX3, fragment).
b Other annotated (UNIPROT), not characterized GT4 (plant): DGalDAG synthase: A. thaliana (type 2, Q8W1S1), Lotus japonicus (type 2, Q6DW73), L. japonicus (type 1, Q6DW74), G. max (type 1, Q6DW76), G. max (type 2, Q6DW75), N. tabacum (Q6DQ98), V. unguiculata (type 1, Q07A01), V. unguiculata (type 2, A4UNX4, fragment); Xerophyta humilis (Q84XL8). Other annotated (UNIPROT), not characterized GT4 (cyanobacteria): DGalDAG synthase Synechocystis sp. PCC 6803 (P73948).
Mollicutes are a class of bacteria devoid of cell wall, which include several genera, such as Acholeplasma, Anaeroplasma, and Mycoplasma. They are related to Gram-positive bacteria and contain free glycoglycerolipids as key structural membrane components (23, 24). Acholeplasma laidlawii is one of the best investigated non-photosynthetic bacteria with regard to the function of glycolipids in biological membranes (7, 25–29, 31). Two different GTs belonging to family GT4 have been identified as mono- and diglucosyldiacylglycerol synthases, respectively, responsible for the sequential glycosylation of the precursor diacylglycerol and regulation of the ratio between non-bilayer and bilayer-forming glycolipids (32–37). More recently, Mycoplasma pneumoniae has been shown to contain a processive GT2 producing mainly galactoglycerolipids (38).
Due to the fundamental role of glycoglycerolipids in membrane stability in mycoplasmas and the fact that these lipid structures are absent in the animal host cells of mycoplasma infections, we hypothesize that GTs involved in their biosynthesis are potential targets for drug discovery. GT4 enzymes from A. laidlawii have been well characterized (32–37), producing α-linked glycolipids (Table 1). However, genome-sequenced mycoplasmas (more than 18 different species) contain mainly GT2 enzymes. Experimentally, only the MPN483 gene product from M. pneumoniae has been identified as a glycolipid synthase (38). It is responsible for the synthesis of most of the glycoglycerolipids found in the plasma membrane (processive enzyme with broad specificity), but this study characterized the enzyme in crude membrane extracts, where other membrane components may interfere or regulate the observed activity.
Because GT2 enzymes are the most common glycosyltransferases in pathogenic mycoplasmas and are encoded by essential genes (as shown by gene inactivation in Mycoplasma genitalium (39)), they are potential targets for enzyme inhibition in the design of new antibiotics. With the aim of better understanding the kinetic properties of processive (or sequentially acting) GT2 enzymes, we here address the recombinant expression and purification of a novel GT from M. genitalium for detailed kinetic characterization of the isolated enzyme. M. genitalium is the smallest free-living organism considered a minimal cell model (39, 40) and the subject of intense research in mycoplasma systems biology (41–43). Due to its reduced genome of only 580 kb and about 480 genes encoding for proteins, it lacks many metabolic pathways like amino acid, nucleotide, fatty acid, and cholesterol biosynthesis and maintains a reduced energy metabolism devoid of Krebs cycle (23, 44). M. genitalium is a human pathogen involved in urogenital diseases such as acute and chronic non-gonococcal urethritis, cervicitis, and pelvic inflammation (45, 46). Here we report a chaperone co-expression system that has allowed the overproduction and subsequent purification of the membrane-associated GT MG517. We provide for the first time the kinetic parameters for the first (monoglycosyldiacylglycerol synthase) and second (diglycosyldiacylglycerol synthase) glycosyl transfer activities catalyzed by a GT2 glycolipid synthase in isolated form. Inhibition of this GT activity is shown to inhibit mycoplasma growth, supporting our hypothesis of GT2 glycolipid synthases as potential therapeutic targets against mycoplasma infections.
EXPERIMENTAL PROCEDURES
Mycoplasma Growth Conditions and Glycolipid Labeling
20 ml of SP4 medium (47) supplemented with 5 g/liter glucose (or [14C6]glucose, 15 μCi, in labeling experiments) in 150-cm2 Falcon flasks were inoculated with 50 μl of M. genitalium glycerinate stock and incubated at 37 °C in 5% CO2 atmosphere until exponential (3.5 days) or stationary (6 days) phase, depending on its use for GT activity assays or lipid extraction, respectively. Cells were washed with 5 ml of PBS and resuspended in 5 ml of PBS by direct scraping of the surface. After centrifugation, the cellular pellet was resuspended in reaction buffer when used for activity assays or extracted with chloroform/methanol (2:1) when used for lipid analysis.
For cell growth inhibition assays, PPMP and N-(n-nonyl)-deoxygalactonojirimycin (C9J) (inhibitors of GT MG517, as shown below) dissolved in 100 μl of ethanol were added to 5 ml of culture medium (in 25-cm2 Falcon flasks) before inoculation (with 7 μl of M. genitalium glycerinate) to give final inhibitor concentrations of 0–500 μm. Cell growth was followed by monitoring the pH of the medium, which drops from 7 to 5 within 6 days for a control M. genitalium culture (without inhibitor) (43).
Cloning of M. genitalium ORFs mg025, mg060, and mg517
M. genitalium cells grown at 37 °C, 5% CO2 to stationary phase (6 days) were scraped, washed, and resuspended in Lysis buffer A (67 mm Tris-Cl, pH 8.8, 16.7 mm ammonium sulfate, 5 mm β-mercaptoethanol, 6.7 mm MgCl2, 6.7 mm EDTA, pH 8.0, 1.67 μm SDS, and 50 μg/ml proteinase K). The cell lysate was directly used as template in hot start polymerase chain reactions (PCRs) to amplify the three ORFs, mg025, mg060, and mg517. After restriction digestion and ligation to a pUC18 vector, electrocompetent Escherichia coli DH5α cells were transformed with the ligation mixtures, and positive transformants were verified by DNA sequencing. All Trp-coding UGA codons present in mg025 (two codons; Trp-218 and Trp-273), mg060 (five codons; Trp-88, Trp-126, Trp-131, Trp-227, and Trp-287) and mg517 (four codons; Trp-170, Trp-197, Trp-268, and Trp-275) were mutated to UGG for recombinant protein expression in E. coli. Next, the codon-adapted genes in pUC18 vectors were subcloned into pET38b(+) between NdeI and AvrII sites for protein expression of the full-length proteins under the control of a T7 promoter. The final constructs were verified by DNA sequencing. E. coli BL21(DE3) cells were transformed with each vector for protein expression. Detailed protocols and primer sequences are given in the supplemental material (Table S1).
Expression of mg025, mg060, and mg517 Gene Products in Recombinant E. coli
E. coli BL21(DE3) cells harboring each plasmid (pET38b(+)-mg025, pET38b(+)-mg060, or pET38b(+)-mg517) were grown in LB medium supplemented with 50 μg/ml ampicillin at 37 °C. When the culture reached an OD of 0.3, a heat shock at 42 °C for 30 min was applied. Then IPTG (1 mm) was added, and cells were grown for 16 h at 20 °C. After centrifugation at 4 °C, the cell pellet was directly used for lipid analysis (in vivo glycolipid biosynthesis) or for the preparation of spheroblasts (for in vitro GT activity assays).
Lipid Extraction and TLC Analysis
The cellular pellet of M. genitalium or recombinant E. coli BL21(DE3) expressing the corresponding genes was subjected to lipid extraction with chloroform/methanol (2:1) (v/v). The organic fraction, dried with MgSO4 and concentrated under vacuum, was analyzed by TLC developed with chloroform/methanol/water 65:35:4 (v/v) and stained with different reagents for lipid identification (48): (a) α-naphtol in sulfuric acid/methanol for general staining, where different lipid families develop a characteristic color (red-brown for cholesterol and apolar lipids; blue for glycolipids; brown for phospholipids); (b) ninhydrin for phosphatidylethanolamine; (c) N,N-diphenylamine for glycolipid staining; (d) Draggendorf staining for phosphatidylcholine; and (d) sulfuric/methanol/water (45:45:10, v/v/v) for general visualization. In the case of 14C-labeled glycolipids, developed TLC plates were visualized by autoradiography.
Glycosyltransferase Activity Assay on Cells
M. genitalium cells or E. coli spheroblasts were resuspended in reaction buffer with acceptor lipid premix to a final concentration of 120 mm Tris-maleate buffer, pH 8, 20 mm MgCl2, 1 mm dioleoylglycerol (DOG), and 0.2 mm SDS. After preincubation at 4 °C for 30 min, reactions were started by the addition of UDP-Glc donor substrate (0.2 μCi of UDP-[14C6]Glc) to a final 5 mm concentration (100-μl total volume). Reactions were incubated for 1 h at 28 °C and stopped by the addition of 1 ml of chloroform/methanol (2:1, v/v) or 1 ml of scintillation liquid (Econofluor®, PerkinElmer Life Sciences). In the first case, the organic fraction was separated, and lipids were analyzed by TLC-autoradiography. In the second case, the reaction mixture was added to the scintillation liquid, which acted as an organic extractant. The aqueous phase containing an excess of UDP-[14C6]Glc was decanted in the scintillation vial, and radioactivity in the organic phase was quantified by scintillation counting (Tri-carb 2100 TR, Packard).
Isolation, Methanolysis, and Acetylation of GL2 from Recombinant E. coli
E. coli BL21(DE3)-pET38b(+)-mg517 cells expressing GT MG517 were harvested and then subjected to continuous extraction (Soxhlet) with acetone. The lipid extract was dried, redissolved in chloroform, and fractionated by silica gel chromatography sequentially eluted with chloroform, acetone, and methanol. The acetone fraction containing GL2 was collected, and the solvent was evaporated under vacuum. The product was subjected to methanolysis (NaOH, 0.2 n in methanol) for 1 h, chloroform and water (1:1) were added, and the aqueous phase was neutralized by adding Amberlist IR-120 resin. After filtration, the aqueous solution was freeze-dried. The resulting solid was acetylated with acetic anhydride in pyridine (1:1) and catalytic amounts of dimethylaminopyridine for 16 h at room temperature. After work-up, the acetylated product was purified by silica gel chromatography using ethyl acetate/hexane (1:1) as eluent. The structure of the purified product dissolved in CDCl3 was elucidated by NMR experiments as described under “Results.”
In Vitro Synthesis of GL1 and GL2 with Purified GT MG517
25 mg of DOG and 2.3 mg of SDS were mixed with 4 ml of Reaction buffer (1.2 m Tris-malate, pH 8.0, 0.2 m MgCl2) and 2 ml of H2O. Micelles were formed by extensive vortexing, and then 1 mg of purified GT MG517 protein (see below) in dialysis buffer was added. After a 1-h incubation at 4 °C, UDP-Glc was added to get the final reaction conditions: 5 mm UDP-Glc, 1 mm DOG, 0.2 mm SDS, 120 mm Tris-maleate, pH 8, 20 mm MgCl2 in a 40-ml total volume. After 48 h of incubation at 28 °C in an orbital shaker, glycolipid products were extracted with 150 ml of chloroform/methanol (2:1), and the organic phase was dried over MgSO4 and concentrated in a rotatory evaporator under vacuum. The lipid mixture was then fractionated by silica gel chromatography column (sequential elution with ethyl acetate, chloroform, acetone, and methanol). MGlcDOG and DGlcDOG were isolated after solvent evaporation pure enough for NMR analysis.
Recombinant Expression of GT MG517
Gene mg517 in pET38b(+) vector was subcloned into pET44b(+) to introduce a His tag at the C terminus of the expressed protein. First, a new restriction site was introduced by replacing the stop codon in plasmid pET38b(+)-mg517 for the XhoI restriction sequence (therefore adding two codons for Leu-Glu at the C terminus of the encoded protein) in order to ligate the gene in frame to the XhoI site upstream of the His tag coding sequence of the vector. The final pET44b(+)-mg517 construct was verified by DNA sequencing (for details, see the supplemental material).
E. coli BL21(DE3star) cells were co-transformed with pET44b(+)-mg517 and pGro7 (from Takara Bio, Ltd.), a plasmid encoding for the E. coli chaperones GroEL and GroES under the control of an araB promoter and containing a chloramphenicol resistance gene (49). Cells were grown in LB medium containing 50 μg/ml ampicillin and 170 μg/ml chloramphenicol at 37 °C. When the optical density of the culture reached 0.4, expression of chaperones was induced by adding l-arabinose (final concentration 0.2%). The culture was further incubated for 30 min at 37 °C before IPTG (1 mm) induction of GT-MG517 expression. Cells continued to be grown for 16 h at 30 °C before harvesting.
GT MG517 Protein Purification
Cell Lysis Buffer Selection
Cells from 50-ml cultures were harvested by centrifugation (9000 × g for 10 min at 4 °C), washed with 5 ml of 0.9% NaCl, and resuspended in 2.5 ml of different lysis buffers (Table 2). Bacteria were lysed by sonication, and the resulting suspension was incubated for 2 h at 4 °C with continuous stirring. Separation of the solubilized protein fraction was achieved by centrifugation (16,000 × g for 30 min at 4 °C). Total protein concentration and glycolipid synthase activity of the resulting supernatant was determined by the BCA assay and radiometric activity assay described below, respectively. A buffer containing CHAPS and glycerol was selected for protein purification.
TABLE 2.
Buffers for solubilization of recombinant E. coli cell extracts for GT MG517 purification
| Buffer | Total proteina | Specific activityb |
|---|---|---|
| mg/ml | units/mg | |
| Buffer A: glycerol (20%) in 0.5 m NaCl, 50 mm phosphate, pH 7 | 4.1 | 0.083 |
| Buffer B: Triton X-100 (0.5%) + glycerol (5%) in 0.5 m NaCl, 50 mm phosphate, pH 7 | 6.3 | 0.17 |
| Buffer C: CHAPS (20 mm) in 0.5 m NaCl, 20 mm HEPES, pH 8 | 4.8 | 0.076 |
| Buffer D: CHAPS (20 mm) + glycerol (10%) in 0.5 m NaCl, 20 mm HEPES, pH 8 | 7.0 | 0.24 |
a Total protein in the soluble fraction after extraction, expressed as mg of protein/ml of cell extract.
b Specific activity of the solubilized protein extract determined by the radiometric assay. Conditions were as follows: 1 mm UDPGal, 1.25 mm DOG, 20 mm HEPES, 20 mm MgCl2, 20 mm CHAPS, 12.5 mm DOPG, pH 8.0, 35 °C. Units/mg, μmol of [14C]Gal incorporated/min/mg of total protein.
Protein Purification by Metal Affinity Chromatography
Cells coming from 1 liter of culture were harvested by centrifugation (9000 × g for 10 min at 4 °C), washed in 100 ml of 0.9% NaCl, and resuspended in 50 ml of lysis buffer (20 mm HEPES, pH 8, 500 mm NaCl, 10 mm CHAPS, 20% glycerol). Bacteria were lysed by a single passage through a cell disrupter (Constant Cell Disruption Systems Ltd.) at a cell pressure of 20.5 kilopascals/inch2. After washing the cells with an additional 50 ml of lysis buffer, the final 100-ml mixture was centrifuged at 16,000 × g for 30 min at 4 °C. After pellet removal, viscosity was decreased by sonication, and the resulting suspension was centrifuged (16,000 × g for 30 min at 4 °C) and filtered through a 0.22-μm filter (Millipore) to obtain the final soluble protein fraction. It was loaded to a prepacked 1-ml HisTrapTM HP column (GE Healthcare), previously equilibrated with lysis buffer in an ÄKTApurifierTM FPLC system (GE Healthcare). Non-specifically bound proteins were eluted first by a step gradient up to 8–9% of CHAPS-containing elution buffer (20 mm HEPES, pH 8, 500 mm NaCl, 10 mm CHAPS, 20% glycerol, 500 mm imidazole). Then the column was extensively washed with washing buffer (20 mm HEPES, pH 8, 500 mm NaCl, 20% glycerol) to ensure CHAPS detergent removal. The protein was finally eluted by a second step-gradient up to 80% of glycerol-containing elution buffer (20 mm HEPES, pH 8, 500 mm NaCl, 20% glycerol, 500 mm imidazole). Collected fractions were analyzed by SDS-PAGE, and fractions containing GT-MG517 were combined and dialyzed against 20 mm HEPES, pH 8, 500 mm NaCl, 20% glycerol to remove the imidazole. The final protein was free of lipidic contaminants as shown by chloroform extraction and TLC analysis. The protein was quantified by the BCA assay and was used for activity assays within 48 h of storage at 4 °C.
Radiometric Glycosyltransferase Activity Assay
The radiometric assay developed for GT-MG517 activity monitoring was based on the protocol by Shimojima et al. (9).
Solubilization of Substrate and Matrix Lipids
DOG (acceptor substrate), dioleoylphosphatidylglycerol (DOPG) (enzyme activator), and (when indicated) dioleoylphosphatidylcholine (DOPC) (matrix lipid) were dissolved in chloroform using the required amounts to give the final concentrations for each assay as indicated. The solvent was evaporated under a steam of N2, and the lipid mixture was dried under vacuum for 1 h. The mixture was then solubilized to homogeneity (mixed micelles) in 20 mm CHAPS, 20 mm HEPES, pH 8.0, 20 mm MgCl2, by extensive vortexing and bath sonication for 5 min.
Activity Assay
The glycolipid synthase activity assay was performed with both protein extract obtained as described under “Cell Lysis Buffer Selection” and purified GT-MG517. To prepare the reaction mixture, 10 μl of enzyme source (protein extract or pure protein, 1–10 μm assay concentration) were added to 60 μl of freshly prepared micellar solution containing the acceptor substrate as indicated above. The preparation was kept for 30 min on ice and preincubated for 10 min at 35 °C. Reactions were started by adding the labeled donor (UDP-[14C6]Glc or UDP-[14C6]Gal, 1–5 μCi). Final buffer composition was 20 mm HEPES, pH 8, 20 mm MgCl2, 20 mm CHAPS in a 110-μl reaction volume. Five 20-μl samples were taken at different times and immediately quenched by the addition of 500 μl of ethyl acetate and 250 μl of 0.45% NaCl. After vortexing and centrifugation (2500 rpm for 5 min), 350 μl of the organic phase were withdrawn and mixed with 4 ml of ACS-II scintillation liquid (GE Healthcare) and 500 μl of methanol. The amount of the produced radiolabeled glycolipids was determined with a TRI-CARB 2100 TR scintillation counter (Packard). Initial rates were calculated from the slopes of the linear time course (μmol of Glc or Gal incorporated to products) versus time.
For specific activity determination (protein extracts or purified protein) reaction conditions were as follows: 1 mm UDP-Glc, 1.25 mm (3.7 mol %) DOG, 12.5 mm (37 mol %) DOPG, 20 mm CHAPS, 20 mm MgCl2, 20 mm HEPES, pH 8.0, at 35 °C. For determination of kinetic parameters with purified enzyme, donor (0.5 μm to 2 mm), acceptor (50 μm to 1.25 mm), and DOPG (0–12.5 mm) were individually varied, maintaining constant the other components. Initial rates versus donor or acceptor concentrations were fitted to a Michaelis-Menten equation by non-linear regression, from which the kinetic parameters were derived.
Enzyme Inhibition
For inhibition measurements, PPMP was included in the mixed micelles with DOG and DOPG in CHAPS. Reaction conditions were 300 μm UDP-Glc, 300 μm DOG, 12.5 mm DOPG, 0–200 μm PPMP, 20 mm CHAPS, 20 mm MgCl2, 20 mm HEPES, pH 8.0, at 35 °C. Enzyme activity at each inhibitor concentration was determined as above.
HPLC-MS Time Course Monitoring of GT Activity
Reactions prepared as above (with cold UDP-Glc and UDP-Gal) were monitored by taking 20-μl aliquots at different time intervals, which were added to 200 μl of methanol and analyzed by HPLC-MS. The equipment was an Acquity UPLC system (Waters) coupled to a ZMD mass spectrometer (Waters) as detector (ESI+, SIM mode, 20-V cone voltage). The ions analyzed were m/z 644 [M + Na+] for DOG, 806 [M + Na+] for MGlcDOG, and 968 [M + Na+] for DGlcDOG. Chromatographic separation was done on a Nova-Pack C18 column (Waters), eluted with MeOH/H2O (98:2) at a 1-ml/min flow rate, 10-μl injection. Calibration curves (peak area versus concentration) were obtained using GL1 and GL2 glycolipid products from in vitro synthesis (see above).
RESULTS
Functional GT Activity in M. genitalium
In addition to phospholipids and cholesterol, two major glycolipids (namely GL1 and GL2) were observed in the plasma membrane of M. genitalium cells grown to stationary phase in [14C]glucose-enriched medium (supplemental Fig. S1). They were sequentially synthesized from diacylglycerol by the action of GT activities. When incubating M. genitalium cells with a mixture of UDP-[14C6]Glc and DOG as donor and acceptor substrates in the presence of SDS as solubilizing detergent, the lipid extract showed two labeled spots corresponding to GL1 and GL2 (supplemental Fig. S1). Their relative intensities remarkably differ from those obtained in the in vivo labeling experiment, indicating that the relative glycosyltransferase activities are regulated in vivo. The isolated 14C-labeled GL1 product was then used as acceptor and incubated with the M. genitalium cell pellet in the presence of SDS and cold UDP-Glc, showing that GL1 is converted to GL2, thus demonstrating the sequential action of the two GT activities. They are localized in the plasma membrane because activity was only observed in the membrane fraction.
Only three putative GT genes have been annotated in the M. genitalium genome. Based on their translated amino acid sequences, all three (mg025, mg060, and mg517) are classified in family GT2 in the CAZY data base (8). BLAST screening identified homologues in other Mycoplasma species, being those from M. pneumoniae the closest relatives. The M. genitalium mg517 gene product shares 77% similarity with the M. pneumoniae MPN483 protein, recently identified as a processive GT-synthesizing galactosyl and glucosyl-β1,6-galactosyldiacylglycerols (38). The other two genes, mg025 and mg060 (orthologs of mpn028 and mpn075), have unknown function. None of them showed homology with the best characterized GTs involved in glycolipid biosynthesis in A. laidlawii (33, 50), where both synthases belong to CAZY family GT4. The three ORFs were cloned in E. coli for recombinant expression and activity characterization (see “Experimental Procedures”).
Glycolipid Synthesis in Recombinant E. coli Cells
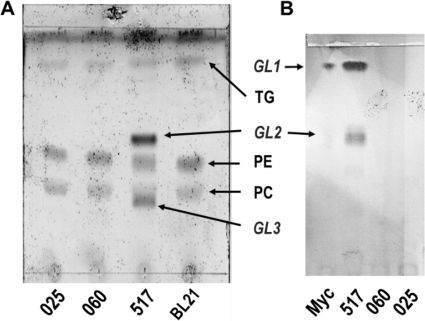
Gram-negative bacteria, such as E. coli, do not have glycoglycerolipids, so the functionality of the M. genitalium genes can be detected by analyzing formation of new glycolipids in the plasma membrane of recombinant E. coli cells expressing the cloned genes. Total lipid extracts of recombinant E. coli BL21(DE3) cells expressing genes mg025, mg060, and mg517 were prepared and analyzed by TLC (Fig. 1a). Only E. coli cells expressing mg517 showed two new spots not present in control cells and in cells expressing mg025 and mg060, which were identified as glycolipids by specific staining with N,N-diphenylamine. When compared with the glycolipids observed in the membrane of M. genitalium (GL1 and GL2), the less polar glycolipid corresponded to GL2, whereas GL1 was not observed, and the more polar glycolipid (GL3) had no counterpart in Mycoplasma membrane lipids.
FIGURE 1.
Glycolipid synthesis in recombinant E. coli. A, TLC of lipid extracts from E. coli cells expressing mg517, mg060, and mg025 genes, respectively. BL21 are control cells transformed with the pET38b(+) with no insert. B, TLC-autoradiography of in vivo glycolipids from M. genitalium grown in [14C]glucose medium (lane 1 labeled Myc) and in vitro products from GT assays with spheroblasts of E. coli cell expressing mg517, mg060, and mg025 genes incubated with UDP-[14C6]Glc, DOG, and SDS. GL1, GL2, and GL3, glycolipids; TG, triglycerides.
To confirm further that only MG517 and not MG025 and MG060 produce glycolipids, GT activity assays were performed on isolated spheroblasts from the three recombinant E. coli cells. After a 1-h incubation with UDP-[14C6]Glc and DOG in the presence of SDS, the lipid extracts were analyzed by TLC-autoradiography (Fig. 1b). Lanes for mg025 and mg060 did not show any radioactive spots, confirming that UDP-Glc was not incorporated into lipophilic metabolites. Spheroblasts of transformed E. coli cells with the mg517-containing plasmid yielded two major glycolipids in the GT activity assay, which, compared with M. genitalium glycolipids, correspond to GL1 and GL2. Other minor spots for more polar glycolipids were also observed with much lower intensity.
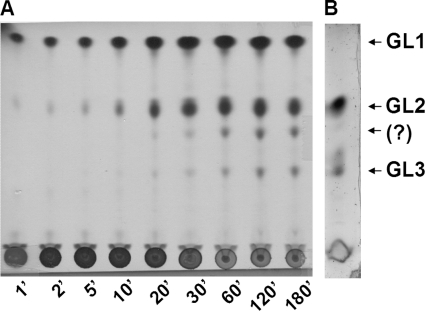
Time course monitoring of the GT activity of E. coli (mg517) spheroblasts is shown in Fig. 2. TLC-autoradiography analysis of the lipid extracts at increasing reaction times shows the sequential behavior of the GT activity. Initially, GL1 was formed, followed by formation of GL2. After longer reaction times (>20 min under these experimental conditions), two more polar glycolipids were produced, the lower spot corresponding to the previously seen GL3, whereas the other spot was not identified.
FIGURE 2.
Time course monitoring of GT activity in recombinant E. coli spheroblasts expressing mg517. A, in vitro GT assay: TLC-autoradiography of the reaction mixture containing E. coli-pET38b(+)-mg517, UDP-[14C6]Glc, and DOG in SDS at increasing reaction times. B, in vivo glycolipid biosynthesis; TLC of a lipid extract of E. coli cell expressing mg517 stained for glycolipids with N,N-diphenylamine.
Structure Determination of Glycolipids GL1 and GL2
For structural elucidation of the glycolipids by spectroscopic analysis, two different experiments were performed. First, the glycosidic moiety of GL2 produced in vivo by recombinant E. coli cells expressing mg517 was determined. For that, the ester linkages were hydrolyzed to simplify the analysis due to the heterogeneity of the fatty acids that come from E. coli. Second, GL1 and GL2 were produced in vitro using the purified GT MG517 enzyme with dioleoylglycerol as acceptor and UDP-Glc as donor, allowing structure elucidation of the glycolipids with homogeneous oleoyl fatty acid chains.
To identify the glycosidic moiety of GL2 produced in vivo, recombinant E. coli (mg517) cells were subjected to acetone extraction, yielding a glycolipid-enriched extract that was then fractionated by chromatography. Purified GL2 was treated with sodium hydroxide in methanol to release the fatty acid side chains, and the resulting glycosylglycerol was acetylated with acetic anhydride in pyridine. After chromatographic purification, the structure of the peracetylated glycosylglycerol product was elucidated by NMR experiments (see supplemental Table S1, Fig. S2, and Fig. S3). The coupling constants J1,2 = 8.1 Hz in both sugar rings indicates a glucose-glucose-glycerol structure with β linkages, consistent with the inverting mechanism proposed for this family 2 glycosyltransferase. The up-field shift of both 1H and 13C signals for H6 and C6 compared with H6′ and C6′ signals indicates a 1→6 linkage that was confirmed by NOESY experiments, where H1′ showed spatial correlation with H6a and H6b signals. Therefore, the polar head structure of glycolipid GL2 was identified as 3-O-β-(d-glucopyranosyl-β(1→6)-d-glucopyranosyl)-syn-glycerol.
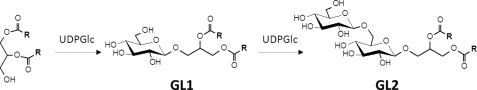
Next, glycolipids GL1 and GL2 were synthesized in vitro by incubating purified enzyme (see below) with UDP-Glc and preformed micelles of DOG and SDS at pH 8.0 and 28 °C. When the reaction was completed, lipophilic products were extracted with chloroform/methanol and fractionated by column chromatography. The structures of isolated GL1 and GL2 were determined by 1H and 13C NMR experiments (see supplemental Tables S3 and S4). GL1 was identified as 3-O-β-d-glucopyranosyl-1,2-dioleoyl-syn-glycerol, and GL2 was identified as 3-O-β-(d-glucopyranosyl-β(1→6)-d-glucopyranosyl)-1,2-dioleoyl-syn-glycerol (Fig. 3).
FIGURE 3.
Structure of glycolipids GL1 and GL2 produced by in vitro synthesis with purified GT MG517 with UDP-Glc and DOG in SDS.
Expression and Purification of GT MG517
Once it was demonstrated that glycolipid biosynthesis in M. genitalium was mediated by the mg517 gene product, the GT enzyme was isolated and purified from recombinant E. coli for characterization. To simplify the purification protocol, the gene was subcloned into the pET44b(+) expression vector in order to introduce a His tag at the C terminus of the protein for metal affinity chromatography separation. Bioinformatics analysis of the translated amino acid sequence (341 amino acids) identified the GT family 2 fold in the N terminus (from amino acid 6 to 192) and a C terminus region with no homology to other GT2 proteins except with its Mycoplasma orthologs. Although the enzyme activity is associated with the membrane fraction, no transmembrane domain was predicted, but the hydrophobicity profile suggested that it may be membrane-associated through the C terminus region.
Protein expression in E. coli by standard IPTG induction of transformed cells yielded mainly insoluble protein. After observing that a heat shock at 42 °C for 30 min just before IPTG induction and subsequent growth at 20 °C afforded higher yields of soluble protein, a chaperone co-expression system was attempted. E. coli cells were co-transformed with the pET vector containing the mg517 gene (T7 promoter, Amp resistance) and the pGro7 vector (49) encoding for GroEL and GroES chaperones (araB promoter, Chl resistance). In this way, expression yields were notably improved.
Solubilization of the cell extract to isolate solubilized protein for purification was further explored. Cells were lysed by sonication in different buffers (Table 2). Based on the amount of total protein extracted and specific activity, a buffer containing CHAPS and glycerol in 20 mm HEPES, pH 8.0, 0.5 m NaCl was selected. Although glycerol alone (buffer A) was able to solubilize the protein, it was observed that the protein co-eluted with membrane lipids when purified by metal affinity chromatography. With the chaperone co-expression system and protein extraction with a CHAPS/glycerol buffer, ∼60% of the GT MG517 protein was obtained in the soluble fraction.
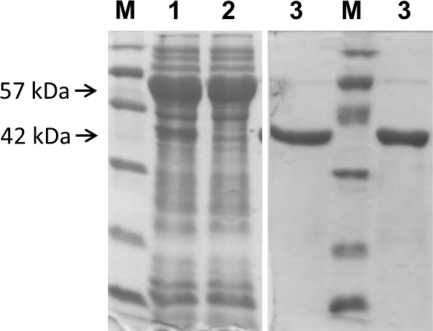
For preparative purification, cells suspended in CHAPS/glycerol extraction buffer were lysed in a cell disrupter. The solubilized protein fraction was applied to an immobilized metal affinity chromatograph. Detergent was removed, and the protein eluted in a 20% glycerol buffer with imidazole. After dialysis, the protein was about 90–95% homogeneous as judged by SDS-PAGE (Fig. 4) and was free of lipidic contaminants. The protein remained soluble in 20% glycerol buffer up to 5 mg/ml concentration, and the specific activity (radiometric assay) with UDP-Glc as donor and DOG as acceptor was 0.34 unit/mg, with an overall yield of 9 mg of purified protein/liter of initial culture.
FIGURE 4.
SDS-PAGE (Coomassie staining) of expressed GT MG517. M, molecular weight marker; 1, solubilized cell extract with CHAPS/glycerol; 2, flow-through in IMAC; 3, isolated MG517 after IMAC. The 52 kDa band corresponds to GroEL, and the 42 kDa band is GT MG517 (identified by MS fingerprinting after trypsin digestion).
Kinetic Characterization
Enzyme activity was analyzed with UDP-Glc and UDP-Gal as donors and DOG as acceptor in mixed micelles with CHAPS and DOPG as activating lipid. Although SDS was used as solubilizing detergent in the initial in vitro assays with cells (see above), CHAPS proved to give higher specific activity in conjunction with the phospholipid DOPG in mixed micelles with the acceptor substrate.
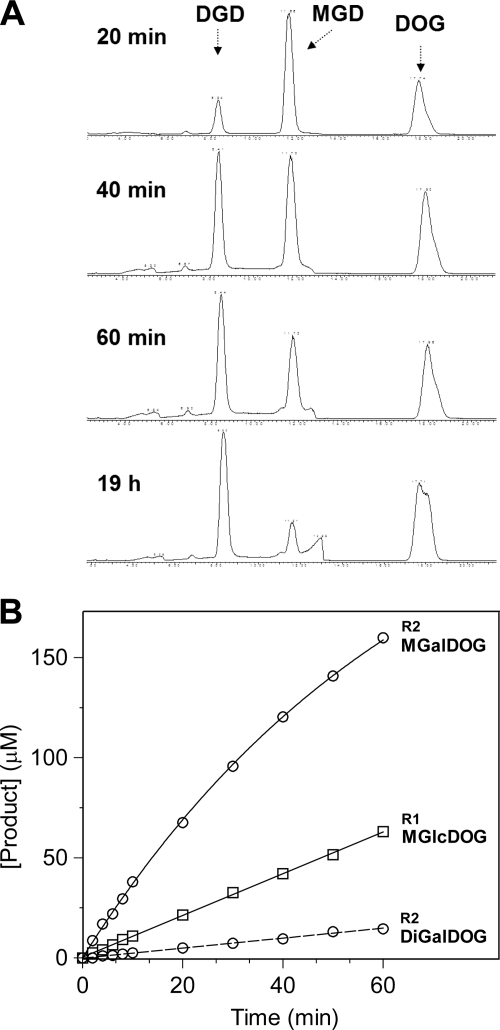
First, monoglycosyl- and diglycosyltransferase activities of GT MG517 were monitored by HPLC-MS. Fig. 5A shows the time course at 1 mm UDP-Glc donor and 1.25 mm (3.7 mol %) DOG acceptor substrates with a solubilized E. coli cell extract in CHAPS/glycerol/NaCl buffer. MGlcDOG was formed fast and then converted to DGlcDOG, the reaction being completed after 200 min. To set up initial rate conditions for kinetics, dilutions of purified GT MG517 were assayed. As shown in Fig. 5B, at 0.1 μm enzyme, the reaction was slow enough to monitor MGlcDOG (from UDP-Glc) or MGalDOG (from UDP-Gal) formation up to ∼10–15% conversion before any diglycosylated products (or only traces) were formed.
FIGURE 5.
HPLC-MS monitoring of the GT MG517 activity. A, reaction monitoring with recombinant E. coli cell extracts (1.3 mg/ml total (solubilized) protein, 0.01 unit based on radiometric assay) with 1 mm UDP-Glc, 1.25 mm DOG. MGD, MGlcDOG m/z 806 [M + Na]+; DGD, DGlcDOG m/z 968 [M + Na]+; DOG, dioleoylglycerol m/z 644 [M + Na]+. B, reaction monitoring with purified GT MG517 (0.1 μm) under initial rate conditions. R1, reaction with 1 mm UDP-Glc, 1.25 mm DOG, where only MGlcDOG product is detected. R2, reaction with 1 mm UDP-Gal, 1.25 mm DOG, where MGalDOG is formed and a trace amount of DGalDOG. Conditions were as follows: 1 mm UDP-Glc or UDP-Gal, 1.25 mm (3.7 mol %) DOG, 12.5 mm (37 mol %) DOPG in 20 mm HEPES, 20 mm CHAPS, 20 mm MgCl2, pH 8.0, at 35 °C.
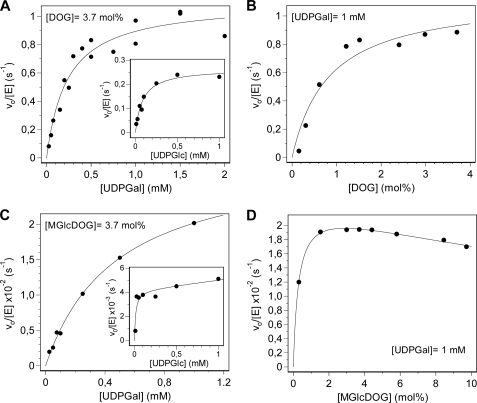
Kinetic Parameters for Donor and Acceptor Substrates
Using the above conditions (initial rates for less than 15% conversion), kinetic parameters were determined by a radiometric assay that monitors 14C label incorporation from UDP-[14C6]Glc or UDP-[14C6]Gal to the acceptor in mixed micelles. Kinetics varying one substrate at saturating concentration of the other obeyed Michaelis-Menten kinetics (Fig. 6), and kinetic parameters are summarized in Table 3. Parameters for UDP-Glc and UDP-Gal donors at saturating DOG correspond to the first glycosyl transfer to give monoglycosyldioleoylglycerol products because no diglycosylated products were observed under these conditions by HPLC-MS monitoring. Km is lower for UDP-Glc, but kcat is higher for UDP-Gal, rendering similar kcat/Km values for both donor substrates. Kinetic parameters for the acceptor DOG may not only reflect the first monoglycosyltransferase activity because at low DOG concentration (high donor/acceptor ratio) some diglycosylated products might be formed even under initial rate conditions. MGlcDOG is also an acceptor to yield the diglycosylated products (Fig. 6, c and d), and kinetic parameters for donor substrates are assignable to the diglycosyltransferase activity of GT MG517. Again, UDP-Gal is a better donor than UDP-Glc in terms of kcat, but Km for UDP-Glc is much lower, rendering similar kcat/Km values for both donor substrates (Table 3). DOG is a better acceptor than MGlcDOG in terms of kcat and kcat/Km, indicating that the diglycosyltransferase activity is significantly slower than the monoglycosyltransferase under these conditions (12.5 mm DOPG as activating anionic lipid), in agreement with the time course monitoring by HPLC-MS (Fig. 5).
FIGURE 6.
GT MG517 kinetics with UDP-Glc and UDP-Gal donors and DOG and MGlcDOG acceptors at pH 8.0, 35 °C. A and B, monoglycosyltransferase activity with DOG acceptor. A, varying UDP-Gal (or UDP-Glc; inset) at constant 1.25 mm (3.7 mol%) DOG. B, dependence on DOG concentration at constant 1 mm UDP-Gal. C and D, diglycosyltransferase activity with MGlcDOG acceptor. C, varying UDP-Gal (or UDP-Glc; inset) at constant 1.25 mm (3.7 mol %) MGlcDOG. D, dependence on MGlcDOG concentration at constant 1 mm UDP-Gal. Fitted parameters are given in Table 3.
TABLE 3.
Kinetic parameters of GT MG517 for donor and acceptor substrates at pH 8.0, 35 °C
Conditions were as follows: 20 mm HEPES, 20 mm MgCl2, 20 mm CHAPS, 12.5 mm DOPG, [GT MG517] = 50–350 nm, at pH 8.0, 35 °C.
| Donor | Acceptor | kcat | Km | kcat/Km | |
|---|---|---|---|---|---|
| s−1 | μm | m−1·s−1 | |||
| Donora parameters | UDP-Glc | DOG | 0.27 ± 0.02 | 87 ± 16 | (3.1 ± 0.8) × 103 |
| UDP-Gal | DOG | 1.10 ± 0.06 | 234 ± 45 | (5 ± 1) × 103 | |
| UDP-Glc | MGlcDOG | 0.004 ± 0.001 | <50 | >80 | |
| UDP-Gal | MGlcDOG | 0.030 ± 0.001 | 479 ± 50 | 63 ± 9 | |
| Acceptorb parameters | UDP-Gal | DOG | 1.13 ± 0.1 | 270 ± 90 (0.83 mol%) | (4 ± 1) × 103 |
| UDP-Gal | MGlcDOG | 0.023 ± 0.001 | ∼90 (0.3 mol %) | >30 |
a Donor parameters at 1.25 mm (3.7 mol%) acceptor (DOG or MGlcDOG, saturating).
b Acceptor parameters at 1 mm donor (UDP-Gal, saturating).
Specific Activity for Donor/Acceptor Pairs
Specific activities of purified GT M517 at 1 mm/1.25 mm donor/acceptor pairs (saturating conditions) are summarized in Table 4. The enzyme has higher activity with UDP-Gal than UDP-Glc using any acceptor. With both donors, DOG is the preferred acceptor, having a 45-fold higher specific activity than MGlcDOG under these conditions, which reflect kcat values (saturation for both donor and acceptor). A monogalactosylated analog, galactosyldistearylglycerol (MGalDStG) from plant chloroplasts was also assayed. It was shown to be a better acceptor than MGlcDOG but still with a 10-fold lower specific activity than DOG. With both donors, the diglycosyl synthase activity (with MGlcDOG acceptor) is about 2.2–2.3% relative to the monosynthase activity (with DOG acceptor), but it increases to 9–14% with MGalDStG acceptor.
TABLE 4.
Specific activities of GT MG517 for donor/acceptor pairs at pH 8.0, 35 °C
Conditions were as follows: 1 mm donor, 1.25 mm (3.7 mol%) acceptor, 20 mm HEPES, 20 mm MgCl2, 20 mm CHAPS, 12.5 mm (37 mol %) DOPG, at pH 8.0, 35 °C.
| Donor | Acceptor | Unitsa/mg | Percentage specific activityb | Percentage specific activityc |
|---|---|---|---|---|
| % | % | |||
| UDP-Gal | DOG | 1.27 | 100 | |
| MGlcDOG | 0.029 | 2.3 | ||
| MGalDStG | 0.18 | 14 | ||
| UDP-Glc | DOG | 0.34 | 27 | 100 |
| MGlcDOG | 0.0073 | 0.6 | 2.2 | |
| MGalDStG | 0.030 | 2.4 | 9 |
a Units, μmol/min product formed, as determined by the radiometric assay.
b Percentage specific activity relative to UDGGal.
c Percentage specific activity relative to UDPGlc.
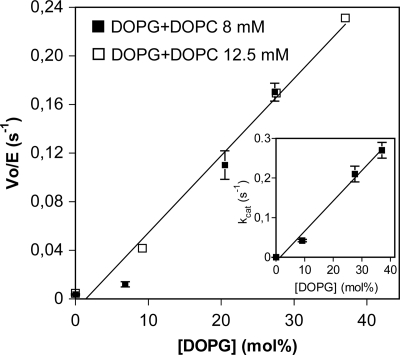
Activation by Anionic Lipids
As shown for bacterial and plant glycolipid synthases (i.e. see Refs. 13 and 28), GT activity requires anionic phospholipids for activation. All of the above kinetics with GT MG517 were performed in the presence of 12.5 mm (37 mol %) DOPG as part of mixed micelles with the acceptor (DOG or MGlcDOG) and the detergent (CHAPS). The effect of anionic DOPG was evaluated by varying DOPG concentration while maintaining constant the total lipid concentration in the mixed micelles with added DOPC as matrix (neutral) lipid (Fig. 7). In the absence of DOPG, essentially no activity was detected, and kcat for UDP-Glc increased with DOPG concentration. The activation was only due to DOPG because experiments at two different total lipid concentrations (DOPG + DOPC) yielded equivalent dependence on DOPG concentration.
FIGURE 7.
GT MG517 activation by DOPG. Dependence of initial rates on DOPG concentration at two different lipid concentrations (DOPG + DOPC as neutral lipid). Conditions were as follows: 1 mm UDP-Glc, 1.25 mm (3.7 mol %) DOG in 20 mm HEPES, pH 8.0, 20 mm MgCl2, 20 mm CHAPS, 35 °C. Inset, dependence of kcat on [DOPG] at 12.5 mm (37 mol %) total DOPG + DOPC concentration.
Essential Function of GT MG517 for Cell Viability
To evaluate the hypothesis of GT MG517 as a potential target against mycoplasma infections, a knock-out was prepared to verify its essential function, and enzyme inhibition was tested for cell growth inhibition.
mg517 Knock-out
Gene mg517 was proposed to be essential by global transposon mutagenesis in the M. genitalium genome (39). To confirm its essential function, a single gene knock-out was prepared by homologous recombination. After verifying that mg517 is not part of an operon and that it does not overlap with neighboring genes, the full coding sequence was replaced by the tetM438 selection marker (tetracycline resistance). This was achieved by using a suicidal plasmid that contains the tetM438 marker (51) enclosed by the flanking regions of the mg517 gene for transformation of M. genitalium cells as previously used for the preparation of other M. genitalium null mutants (52). Upon recombination, only colonies from single recombination events were obtained, where the tetM marker was inserted but mg517 was not deleted (supplemental material). The lack of transformants from double recombination events reinforces the essential role of the mg517 gene. The same experiment was performed on mg025 and mg060 genes (the other two annotated putative glycosyltransferases with unknown function), showing that mg060 is also an essential gene, but mg025 is not. This is in contrast with the earlier proposal by global transposon mutagenesis (39) in which all three putative GTs were assigned as essential genes.
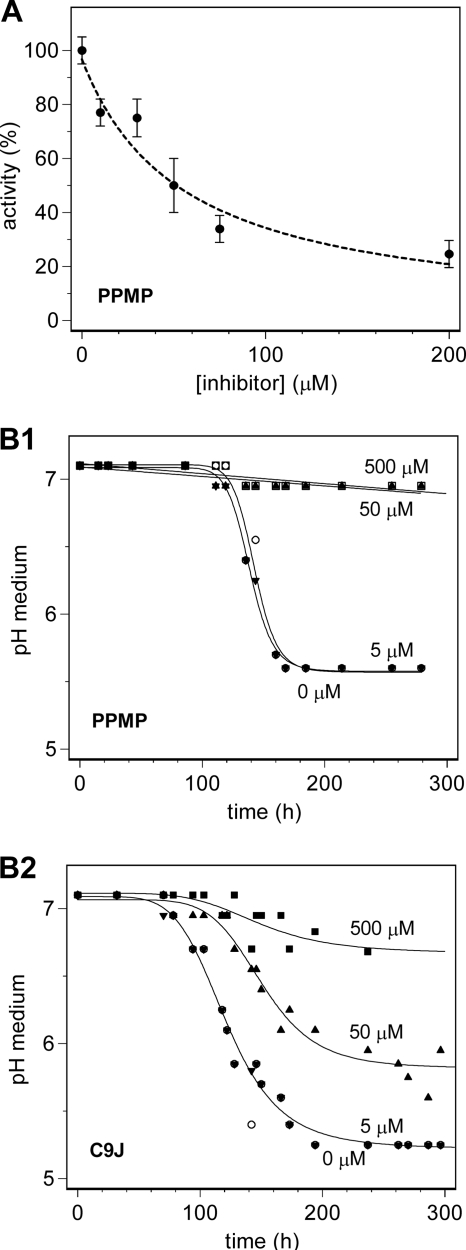
Inhibition of GT MG517 Activity
PPMP (threo-1-phenyl-2-palmitoylamino-3-morpholino-1-propanol), a known inhibitor of glucosylceramide synthases (53), was tested as an inhibitor of GT MG517. The activity assay was performed at 300 μm UDP-Gal, 300 μm DOG acceptor, and increasing concentrations of PPMP inhibitor under the same experimental conditions used for kinetic characterization. Fig. 8A shows the activity dependence on inhibitor concentration. An IC50 value of about 50 μm was obtained, but no full inhibition was observed up to 200 μm inhibitor. Likewise, C9J, another inhibitor of glucosylceramide synthases (30), was shown to inhibit GT MG517 (data not shown).
FIGURE 8.
Inhibition by PPMP and C9J. A, GT M517 enzyme inhibition by PPMP at 300 μm UDP-Gal, 300 μm DOG. B, M. genitalium cell growth inhibition by PPMP (B1) and C9J (B2) at increasing inhibitor concentration (0, 5, 50, and 500 μm) by monitoring the pH of the medium.
Mycoplasma Cell Growth Inhibition
Both PPMP and C9J were assayed as inhibitors of M. genitalium cell growth. In control cultures, the pH of the medium drops from 7 to 5 in 6 days due to the release of low molecular weight acids (44). Growth is inhibited by the low pH, and turbidity is observed arising from the slow release of cells from the attached monolayer on the flask surface. When PPMP or C9J was added to the medium before inoculation, a concentration-dependent inhibition was observed (Fig. 8B). Both compounds had essentially no effect at 5 μm concentration. For C9J, the growth was slower at 50 μm with a t50 (midpoint of the pH change) of 150 h as compared with 120 h in the absence of inhibitor. No growth was observed at 500 μm. For PPMP, full inhibition was already obtained at 50 μm compound.
DISCUSSION
Membrane Glycoglycerolipids in M. genitalium Are Synthesized by a Single Glycosyltransferase
M. genitalium contains two major glycolipids in the plasma membrane synthesized by a single GT enzyme encoded by mg517, which sequentially transfers glycosyl units to the initial diacylglycerol acceptor. Recombinant expression of GT MG517 in E. coli showed that the major in vivo product accumulated in the plasma membrane was a diglucosylated DAG, with a polar headgroup Glcβ1,6Glcβ-. The in vitro activity of the isolated enzyme with UDP-Glc and DOG as donor and acceptor substrates produced the monoglucosylated (GlcβDOG) and diglucosylated (Glcβ1,6GlcβDOG) dioleoylglycerols.
Processive (or sequentially acting) glycoglycerolipid synthases have only been reported in bacteria (Table 1): in family GT28 for Gram-positive bacteria (i.e. Bacillus and Staphylococcus), in family 21 (i.e. Agrobacterium), and in family GT2 for mycoplasmas (M. pneumoniae and M. genitalium here reported). A phylogenetic analysis based on amino acid sequence similarity of characterized and annotated glycoglycerolipid synthases (supplemental Fig. S4) shows that, in GT2, processive mycoplasma enzymes are closely related to cyanobacterial MGlcDAG synthases, whereas GT28 processive bacterial enzymes are close to plant MGalDAG synthases and also to bacterial GT4 enzymes.
GT MG517 is homologous to the M. pneumoniae enzyme encoded by mpn483 gene, reported to produce similar glycolipids (38). However, both enzymes show subtle differences in substrate specificity. Only galactolipids are synthesized in M. pneumoniae cells, and in vitro assays with solubilized M. pneumoniae cells and solubilized recombinant E. coli cells expressing mpn483 gave a variety of glycolipids. With UDP-Gal as glycosyl donor, DAG and GalβDAG were good acceptors, the latter being preferred to give Galβ1,6GalβDAG. With UDP-Glc as donor, DAG and GalβDAG were acceptors, but not GlcβDAG (38). Moreover, the main glycolipid synthesized in vivo by recombinant E. coli cells expressing mpn483 was identified as Glcβ1,6GalβDAG. It was concluded that the M. pneumoniae enzyme was mainly a galactosyltransferase, with higher diglycosyltransferase activity on GalβDAG as acceptor, whereas it did not accept GlcβDAG as substrate. This is remarkably in contrast with the main glycolipid product identified for the M. genitalium enzyme (Glcβ1,6GlcβDAG). Such in vivo specificity resembles more closely that of the B. subtilis and Staphylococcus GT28 glycosyltransferases, which produce diglucosylated diacylglycerols with the same β1,6 connectivity (20, 21). To get a better insight on the specificity of the new GT MG517, the M. genitalium enzyme was purified to homogeneity for detailed kinetic characterization.
The Monoglycosyl- and Diglycosyltransferase Activities of GT MG517 Are Regulated by Other Membrane Components
The monosynthase (UDP-Glc + DAG → MGlcDAG (GL1)) and disynthase (UDP-Glc + MGlcDAG → DGlcDAG (GL2)) activities (as well as further transfer activity leading to GL3, tentatively assigned to triglucosyldiacylglycerol) should be regulated because the relative amounts of glycolipid products strongly depend on the experimental system. The in vivo activity in M. genitalium produced GL1 as a major compound and a lower amount of GL2, whereas the GT activity assay with isolated M. genitalium cells rendered similar amounts of GL1 and GL2 (supplemental Fig. S1). In recombinant E. coli cells expressing mg517, GL2 was the major component in vivo, GL1 not being detected or being detected in trace amounts, and GL3 was also formed in lower amounts (Fig. 2). However, the GT activity assay on isolated spheroblasts produced all three glycolipids (and the unknown compound (?)) after a long reaction time (Fig. 2), and the time course showed the sequential appearance of these glycolipids.
The mechanisms by which other membrane components regulate the activities and therefore the ratio of glycolipid products remain to be studied. The different composition of the E. coli membrane seems to displace the glycolipid ratio toward DGlcDAG as compared with M. genitalium. Because monoglycosylated diacylglycerols are non-bilayer-forming and diglycosylated diacylglycerols are bilayer-forming lipids, their relative concentration (monoglycosyldiacylglycerol/diglycosyldiacylglycerol ratio) contributes to regulate the properties and stability of the plasma membrane, as it has been best studied in A. laidlawii (25–31). As discussed below, anionic lipids strongly activate the enzyme, and the amount and ratio of glycolipids are probably regulated by the lipidic environment of the enzyme.
Chaperone Co-expression and Extraction with Detergents Are Required to Isolate Soluble Recombinant GT MG517
To further characterize GT MG517, the enzyme was overexpressed in E. coli and purified. Although standard recombinant expression in a pET vector produced functional protein, as confirmed by the formation of glycolipid products in the E. coli membrane in vivo, attempts to isolate the enzyme mainly yielded insoluble protein. Higher yields of soluble protein were obtained with a chaperone coexpression system. A lysis buffer containing CHAPS detergent and glycerol gave the best results, with about 60% protein recovery in the soluble fraction. A buffer with glycerol alone was also able to extract and solubilize the protein, but the protein retained bound membrane lipids that could not be released by subsequent column chromatography. A buffer containing detergent had to be included not only to increase the extraction yield but also to displace bound membrane lipids. These results strongly indicate that GT MG517 is a membrane-associated protein, probably attached to the membrane through hydrophobic regions or amphipathic helixes at the C terminus of the protein because no transmembrane domains are suggested by bioinformatic tools based on amino acid sequence. The protein was further purified by metal affinity chromatography, kept soluble in a buffer containing glycerol, and proved to be stable for at least 1 week with less than 10% reduction in specific activity.
GT MG517 Has Broad Donor and Acceptor Specificity
GT MG517 sequentially transfers Glc or Gal from the sugar nucleotide donor to DAG acceptor first, forming monoglycosyl-DAG, which then acts as acceptor for a second Glc or Gal transfer to produce diglycosyl-DAG (Figs. 2 and 5). It is an intriguing feature of processive glycosyltransferases that they attach a glycosyl residue to hydroxyl groups of a hydrophobic acceptor (DAG) embedded in the surface layer of the membrane, and then they glycosylate a hydrophilic glycosyl residue. However, in both transferase reactions, the acceptor hydroxyl group is a primary alcohol (3-OH in DAG and 6-OH in monoglycosyldiacylglycerol). This is the case seen for the M. genitalium enzyme here reported, the M. pneumoniae enzyme (38), and other “processive” glycolipid synthases, such as those from Bacillus subtilis and S. aureus, all of which form β1,6 linkages (20, 21). Interestingly, in other organisms that produce glycolipids with connectivities different from β1,6 between glycosyl units, the monoglycosyl- and diglycosyltransferase activities are catalyzed by different enzymes (i.e. A. laidlawii, where the diglycosylated product is Glcα1,2Glc-DAG or plant GTs with α1,6 linkage (1)). This is an intriguing feature that will deserve structural interpretation when a three-dimensional structure of any glycolipid synthase becomes available.
For both monoglycosyltransferase and diglycosyltransferase activities of GT MG517, the enzyme follows hyperbolic (Michaelis-Menten) kinetics when varying one substrate at saturating concentration of the other (Fig. 6). An exception seems to be the reaction with MGlcDOG as acceptor and UDP-Gal as donor, where some substrate inhibition at high acceptor concentration becomes apparent (Fig. 6d). Initial rates for UDP-Gal donor are faster than for UDP-Glc. kcat is about 5-fold higher for UDP-Gal with either DOG or MGlcDOG acceptors, but the enzyme shows better binding for the glucoside donor, as reflected by the lower Km values for UDP-Glc than UDP-Gal. It results in similar kcat/Km values for both donor substrates. With regard to acceptor, DOG reacts faster than MGlcDOG with both UDP-Glc and UDP-Gal donors. kcat is about 50-fold higher, whereas Km is just 5-fold higher for DOG than MGlcDOG, respectively. The same trend in reactivity and specificity is observed when comparing specific activities (Table 4). In conclusion, under these experimental conditions, GT MG517 has a higher monoglycosyl- than diglycosyltransferase activity and has higher kcat values for UDP-Gal than UDP-Glc, but kcat/Km values are similar with both donors and 1 order of magnitude higher for the monoglycosyl- than diglycosyltransferase activity.
Activity and specificity of GT MG517 differ from its ortholog MPN483 from M. pneumoniae, although direct comparison is tentative because only relative activities for solubilized cell extracts have been reported for the M. pneumoniae enzyme (38). When comparing specific activities of GT MG517 (Table 4) and MPN483, the later has higher diglycosyltransferase (with GalβDAG as acceptor) than monoglycosyltransferase activity, as opposed to the M. genitalium enzyme for which the second transferase activity is about 2% (MGlcDOG) or 10% (MGalDStG) of the monoglycosyltransferase activity (with DOG as acceptor). Moreover, GlcDAG is not even an acceptor for the M. pneumoniae enzyme, and the main glycolipid obtained in vivo from recombinant E. coli cells was Glcβ1,6GalβDAG. In contrast, Glcβ1,6GlcβDAG was the main product isolated from recombinant E. coli expressing GT MG517. Therefore, the M. genitalium GT has a broader specificity, with similar kcat/Km values for both UDP-Glc and UDP-Gal. The higher intracellular availability of UDP-Glc than UDP-Gal explains why the main product detected in vivo was the diglucosylated diacylglycerol due to the significantly lower Km for UDP-Glc in both transferase activities.
Anionic Lipids Are Required for Enzyme Activation
GT MG517 activity required the presence of DOPG (anionic phospholipid) in mixed micelles of acceptor substrate (DOG or MGlcDOG) with detergent (CHAPS). As shown in Fig. 7, activity linearly increases with DOPG concentration, and it was independent of total lipid concentration because approximately the same dependence was observed at two different total lipid concentrations (with added DOPC as neutral lipid). With only DOPC in CHAPS or with CHAPS alone to solubilize DOG, essentially no activity was detected. Also, SDS as anionic detergent to solubilize the DOG acceptor was activating the enzyme (conditions used for the preparative synthesis of glycolipid products with purified enzyme or with cell extracts). Enzyme regulation by anionic lipids is known for membrane-associated glycolipid synthases, such as those from A. laidlawii (28) and plant galactosyltransferases (13). Whether they are required not only for activation but also to modulate the ratio between monoglycosyl- and diglycosyltransferase activity remains to be analyzed for the M. genitalium enzyme. However, indirect evidence is provided by the observation that the lipidic environment modifies the ratio of MGlcDAG and DGlcDAG products obtained in vivo and in vivo as discussed above.
GT MG517 Is a Potential Therapeutic Target
Three lines of evidence allow us to propose mycoplasma glycolipid synthases as potential therapeutic targets against mycoplasma infection. First, mg517 is an essential gene for M. genitalium viability; second, glycoglycerolipids are absent in animal host cells of mycoplasma infections (because they only contain glycosphingolipids); and third, inhibition of GT MG517 activity results in cell growth inhibition. Glucosylceramide synthase inhibitors have here been preliminarily tested as a proof of concept, but specific inhibitors based on acceptor specificity may render selective compounds toward the design of novel antibiotics.
Conclusion
A new membrane-associated, sequentially acting glycosyltransferase (glycolipid synthase, family GT2) from M. genitalium has been cloned, recombinantly expressed, and characterized. It has broad substrate specificity, producing glucosyl and galactosyl diacylglycerols with similar kcat/Km values for UDP-Glc and UDP-Gal but higher kcat values for the latter. Although sequentially adding glycosyl residues with β-1,6 connectivity, the first glycosyltransferase activity (to DAG acceptor) is about 1 order of magnitude higher than the second (to MGlcDOG as acceptor) under the tested experimental conditions. Because the ratio between the non-bilayer-forming monoglycosyl DAG and the bilayer-prone diglycosyl DAG contribute to regulate the properties and stability of the plasma membrane, both synthase activities should be regulated. In the best characterized microorganism, A. laidlawii, two different enzymes (both from family GT4) are responsible for each activity, and they are independently regulated by anionic phospholipids and other metabolites. In the case here reported for M. genitalium, a single enzyme is synthesizing both types of glycolipids by sequential glycosyl transfer. DOPG (anionic phospholipid) has been shown to be required for enzyme activation, but its function in regulating the ratio between both transferase activities is unknown. Whether other membrane components or metabolites are involved in the enzyme regulation remains to be studied, as well as the interaction with the membrane and its role in activity.
Because animal cells are devoid of glycoglycerolipids, mycoplasma glycolipid synthases are potential targets for drug discovery against infections by mycoplasma and Gram-positive bacteria. Structural and mechanistic knowledge about activity, specificity, and regulation will provide the background for the design of specific inhibitors.
Supplementary Material
Acknowledgments
We thank Oriol Julve for the HPLC-MS experiments and Dr. Xevi Biarnés for bioinformatics analysis.
This work was supported by Ministerio de Ciencia e Innovación (MICINN), Spain, Grants BIO2007-67904-C02-02 and BFU2010-22209-C02-02.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1–S4.
- GT
- glycosyltransferase
- C9J
- N-(n-nonyl)deoxygalactonojirimycin
- DAG
- diacylglycerol
- DOG
- dioleoylglycerol
- DOPC
- dioleoylphosphatidylcholine
- DOPG
- dioleoylphosphatidylglycerol
- DGlcDAG
- diglucosyldiacylglycerol
- MGlcDOG
- monoglucosyldioleoylglycerol
- DGlcDOG
- diglucosyldioleoylglycerol
- MGalDAG
- monogalactosyldiacylglycerol
- MGalDOG
- monogalactosyldioleoylglycerol
- DGalDAG
- digalactosyldiacylglycerol
- DGalDOG
- digalactosyldioleoylglycerol
- MGlcDAG
- monoglucosyldiacylglycerol
- PPMP
- threo-1-phenyl-2-palmitoylamino-3-morpholino-1-propanol
- MGalDStG
- galactosyldistearylglycerol
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
REFERENCES
- 1. Hölzl G., Dörmann P. (2007) Prog. Lipid Res. 46, 225–243 [DOI] [PubMed] [Google Scholar]
- 2. Benning C. (2008) Prog. Lipid Res. 47, 381–389 [DOI] [PubMed] [Google Scholar]
- 3. Kiriukhin M. Y., Debabov D. V., Shinabarger D. L., Neuhaus F. C. (2001) J. Bacteriol. 183, 3506–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gründling A., Schneewind O. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8478–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wieslander A., Christiansson A., Rilfors L., Lindblom G. (1980) Biochemistry 19, 3650–3655 [DOI] [PubMed] [Google Scholar]
- 6. Lindblom G., Brentel I., Sjölund M., Wikander G., Wieslander A. (1986) Biochemistry 25, 7502–7510 [DOI] [PubMed] [Google Scholar]
- 7. Dahlqvist A., Nordström S., Karlsson O. P., Mannock D. A., McElhaney R. N., Wieslander A. (1995) Biochemistry 34, 13381–13389 [DOI] [PubMed] [Google Scholar]
- 8. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimojima M., Ohta H., Iwamatsu A., Masuda T., Shioi Y., Takamiya K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maréchal E., Awai K., Block M. A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2000) Biochem. Soc. Trans. 28, 732–738 [PubMed] [Google Scholar]
- 11. Awai K., Maréchal E., Block MA., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobayashi K., Nakamura Y., Ohta H. (2009) Plant Physiol. Biochem. 47, 518–525 [DOI] [PubMed] [Google Scholar]
- 13. Botté C., Jeanneau C., Snajdrova L., Bastien O., Imberty A., Breton C., Maréchal E. (2005) J. Biol. Chem. 280, 34691–34701 [DOI] [PubMed] [Google Scholar]
- 14. Hölzl G., Zähringer U., Warnecke D., Heinz E. (2005) Plant Cell Physiol. 46, 1766–1778 [DOI] [PubMed] [Google Scholar]
- 15. Dubots E., Audry M., Yamaryo Y., Bastien O., Ohta H., Breton C., Maréchal E., Block M. A. (2010) J. Biol. Chem. 285, 6003–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamaryo Y., Motohashi K., Takamiya K., Hisabori T., Ohta H. (2006) FEBS Lett. 580, 4086–4090 [DOI] [PubMed] [Google Scholar]
- 17. Dörmann P., Balbo I., Benning C. (1999) Science 284, 2181–2184 [DOI] [PubMed] [Google Scholar]
- 18. Awai K., Kakimoto T., Awai C., Kaneko T., Nakamura Y., Takamiya K., Wada H., Ohta H. (2006) Plant Physiol. 141, 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakurai I., Mizusawa N., Wada H., Sato N. (2007) Plant Physiol. 145, 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jorasch P., Wolter F. P., Zähringer U., Heinz E. (1998) Mol. Microbiol. 29, 419–430 [DOI] [PubMed] [Google Scholar]
- 21. Jorasch P., Warnecke D. C., Lindner B., Zähringer U., Heinz E. (2000) Eur. J. Biochem. 267, 3770–3783 [DOI] [PubMed] [Google Scholar]
- 22. Hölzl G., Leipelt M., Ott C., Zähringer U., Lindner B., Warnecke D., Heinz E. (2005) Glycobiology 15, 874–886 [DOI] [PubMed] [Google Scholar]
- 23. Razin S., Yogev D., Naot Y. (1998) Microbiol. Mol. Biol. Rev. 62, 1094–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Razin S., Hayflick L. (2010) Biologicals 38, 183–190 [DOI] [PubMed] [Google Scholar]
- 25. Wieslander A., Rilfors L. (1977) Biochim. Biophys. Acta 466, 336–346 [DOI] [PubMed] [Google Scholar]
- 26. Dahlqvist A., Andersson S., Wieslander A. (1992) Biochim. Biophys. Acta 1105, 131–140 [DOI] [PubMed] [Google Scholar]
- 27. Lindblom G., Hauksson J. B., Rilfors L., Bergenståhl B., Wieslander A., Eriksson P. O. (1993) J. Biol. Chem. 268, 16198–16207 [PubMed] [Google Scholar]
- 28. Karlsson O. P., Dahlqvist A., Wieslander A. (1994) J. Biol. Chem. 269, 23484–23490 [PubMed] [Google Scholar]
- 29. Osterberg F., Rilfors L., Wieslander A., Lindblom G., Gruner S. M. (1995) Biochim. Biophys. Acta 1257, 18–24 [DOI] [PubMed] [Google Scholar]
- 30. Platt F. M., Neises G. R., Dwek R. A., Butters T. D. (1994) J. Biol. Chem. 269, 8362–8365 [PubMed] [Google Scholar]
- 31. Andersson A. S., Rilfors L., Bergqvist M., Persson S., Lindblom G. (1996) Biochemistry 35, 11119–11130 [DOI] [PubMed] [Google Scholar]
- 32. Karlsson O. P., Dahlqvist A., Vikström S., Wieslander A. (1997) J. Biol. Chem. 272, 929–936 [DOI] [PubMed] [Google Scholar]
- 33. Berg S., Edman M., Li L., Wikström M., Wieslander A. (2001) J. Biol. Chem. 276, 22056–22063 [DOI] [PubMed] [Google Scholar]
- 34. Li L., Storm P., Karlsson O. P., Berg S., Wieslander A. (2003) Biochemistry 42, 9677–9686 [DOI] [PubMed] [Google Scholar]
- 35. Vikström S., Li L., Karlsson O. P., Wieslander A. (1999) Biochemistry 38, 5511–5520 [DOI] [PubMed] [Google Scholar]
- 36. Vikström S., Li L., Wieslander A. (2000) J. Biol. Chem. 275, 9296–9302 [DOI] [PubMed] [Google Scholar]
- 37. Lind J., Rämö T., Klement M. L., Bárány-Wallje E., Epand R. M., Epand R. F., Mäler L., Wieslander A. (2007) Biochemistry 46, 5664–5677 [DOI] [PubMed] [Google Scholar]
- 38. Klement M. L., Ojemyr L., Tagscherer K. E., Widmalm G., Wieslander A. (2007) Mol. Microbiol. 65, 1444–1457 [DOI] [PubMed] [Google Scholar]
- 39. Glass J. I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M. R., Maruf M., Hutchison C. A., 3rd, Smith H. O., Venter J. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hutchison C. A., Peterson S. N., Gill S. R., Cline R. T., White O., Fraser C. M., Smith H. O., Venter J. C. (1999) Science 286, 2165–2169 [DOI] [PubMed] [Google Scholar]
- 41. Gibson D. G., Benders G. A., Andrews-Pfannkoch C., Denisova E. A., Baden-Tillson H., Zaveri J., Stockwell T. B., Brownley A., Thomas D. W., Algire M. A., Merryman C., Young L., Noskov V. N., Glass J. I., Venter J. C., Hutchison C. A., 3rd, Smith H. O. (2008) Science 319, 1215–1220 [DOI] [PubMed] [Google Scholar]
- 42. Yus E., Maier T., Michalodimitrakis K., van Noort V., Yamada T., Chen W. H., Wodke J. A., Güell M., Martínez S., Bourgeois R., Kühner S., Raineri E., Letunic I., Kalinina O. V., Rode M., Herrmann R., Gutiérrez-Gallego R., Russell R. B., Gavin A. C., Bork P., Serrano L. (2009) Science 326, 1263–1268 [DOI] [PubMed] [Google Scholar]
- 43. Güell M., van Noort V., Yus E., Chen W. H., Leigh-Bell J., Michalodimitrakis K., Yamada T., Arumugam M., Doerks T., Kühner S., Rode M., Suyama M., Schmidt S., Gavin A. C., Bork P., Serrano L. (2009) Science 326, 1268–1271 [DOI] [PubMed] [Google Scholar]
- 44. Pollack J. D., Williams M. V., McElhaney R. N. (1997) Crit. Rev. Microbiol. 23, 269–354 [DOI] [PubMed] [Google Scholar]
- 45. Horner P. J., Gilroy C. B., Thomas B. J., Naidoo R. O., Taylor-Robinson D. (1993) Lancet 342, 582–585 [DOI] [PubMed] [Google Scholar]
- 46. Falk L., Fredlund H., Jensen J. S. (2005) Sex. Transm. Infect. 81, 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trully J. G. (1975) in Molecular and Diagnostic Procedures in Mycoplasmology (Razin S., Trully J. G. eds) pp. 33–39, Vol. 1, Academic Press, Inc., San Diego [Google Scholar]
- 48. Zweig C., Sherma J. (1972) in Handbook of Chromatography, Vol. 2, pp. 107–173, CRC Press, Inc., Cleveland, OH [Google Scholar]
- 49. Nishihara K., Kanemori M., Kitagawa M., Yanagi H., Yura T. (1998) Appl. Environ. Microbiol. 64, 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edman M., Berg S., Storm P., Wikström M., Vikström S., Ohman A., Wieslander A. (2003) J. Biol. Chem. 278, 8420–8428 [DOI] [PubMed] [Google Scholar]
- 51. Pich O. Q., Burgos R., Planell R., Querol E., Piñol J. (2006) Microbiology 152, 519–527 [DOI] [PubMed] [Google Scholar]
- 52. Burgos R., Pich O. Q., Querol E., Piñol J. (2007) J. Bacteriol. 189, 7014–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abe A., Inokuchi J., Jimbo M., Shimeno H., Nagamatsu A., Shayman J. A., Shukla G. S., Radin N. S. (1992) J. Biochem. 111, 191–196 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.