Abstract
The human bitter taste receptors (T2Rs) are non-Class A members of the G-protein-coupled receptor (GPCR) superfamily, with very limited structural information. Amino acid sequence analysis reveals that most of the important motifs present in the transmembrane helices (TM1–TM7) of the well studied Class A GPCRs are absent in T2Rs, raising fundamental questions regarding the mechanisms of activation and how T2Rs recognize bitter ligands with diverse chemical structures. In this study, the bitter receptor T2R1 was used to systematically investigate the role of 15 transmembrane amino acids in T2Rs, including 13 highly conserved residues, by amino acid replacements guided by molecular modeling. Functional analysis of the mutants by calcium imaging analysis revealed that replacement of Asn-662.65 and the highly conserved Asn-241.50 resulted in greater than 90% loss of agonist-induced signaling. Our results show that Asn-241.50 plays a crucial role in receptor activation by mediating an hydrogen bond network connecting TM1-TM2-TM7, whereas Asn-662.65 is essential for binding to the agonist dextromethorphan. The interhelical hydrogen bond between Asn-241.50 and Arg-552.54 restrains T2R receptor activity because loss of this bond in I27A and R55A mutants results in hyperactive receptor. The conserved amino acids Leu-1975.50, Ser-2005.53, and Leu-2015.54 form a putative LXXSL motif which performs predominantly a structural role by stabilizing the helical conformation of TM5 at the cytoplasmic end. This study provides for the first time mechanistic insights into the roles of the conserved transmembrane residues in T2Rs and allows comparison of the activation mechanisms of T2Rs with the Class A GPCRs.
Keywords: Calcium Imaging, G-protein-coupled Receptors (GPCRs), Membrane Proteins, Receptor Structure-Function, Rhodopsin, Bitter Taste Receptors (T2Rs), Molecular Modeling
Introduction
The mammalian taste sensation provides valuable information about the nature and quality of food. Taste transduction predominantly involves the interaction of molecules (i.e. tastants) with taste receptor-expressing cells that reside in the taste buds located on the papillae of the tongue. Taste buds relay information to the brain on the nutrient content of food. At present, there are five basic tastes, sweet, salt, sour, umami, and bitter, and among these, sweet, umami, and bitter taste sensations are sensed by G-protein-coupled receptors (GPCRs).2 Sweet and umami tastes are encoded by three GPCRs, with sweet taste sensed by a heterodimer of T1R2 and T1R3 receptors, whereas umami is sensed by a heterodimer of T1R1 and T1R3 receptors. Bitter taste, which is sensed by bitter taste receptors (referred to as T2Rs), is one of the five basic taste modalities. Bitter sensing serves as a central warning signal against the ingestion of potentially harmful substances. The human genome encodes 25 T2Rs localized as clusters on chromosomes 5p15, 7q31, and 12p13 (1–3). T2Rs are between 290 and 333 amino acids long and have seven transmembrane helices (TM1–TM7), a short extracellular N terminus, and an intracellular C terminus. The ligands that activate these T2Rs have diverse chemical structures and include natural alkaloids, such as quinine, caffeine, nicotine, and morphine.
Previously, it was shown using the well characterized Class A GPCRs, rhodopsin and β2-adrenergic receptor (β2-AR), that there are at least three levels of amino acid conservation that can be considered in understanding the structure and function of a given receptor or subfamily of receptors within the GPCR superfamily (4, 5), the most important being the highly conserved signature residues present in each helix, such as Asn1.50, Leu2.50, Arg3.50, Trp4.50, Tyr5.50, Pro6.50, and Pro7.50 (4, 5). In addition, results from mutational studies and crystal structures of rhodopsin and β2-AR show that TM1–TM4 form a helical bundle core, with other helices moving around this core upon activation (6–8).
Outside of Class A, the other well studied GPCRs belong to Class C, which include the mammalian T1Rs, metabotropic glutamate receptors, and the GABAB receptors (9). However, the classification of T2Rs is not clear, with some classification systems describing them as a putative separate family (10), whereas others have grouped them with the frizzled receptors (11). In addition, structure-function studies on T2Rs are very limited, and only recently have a few studies focused on elucidating the ligand binding mechanisms of T2Rs been published (12–14).
Fig. 1 presents a two-dimensional representation of the T2R1 amino acid sequence. Amino acid sequence analysis of 188 T2Rs shows high conservation of amino acid residues in the transmembrane domains, which are distinct from the Class A GPCRs (Table 1). The amino acid motifs in TM1–TM7 of Class A GPCRs, such as the LXXXD in TM2, (D/E)RY in TM3, CWXP in TM6, and NPXXY in TM7, are absent in T2Rs. In addition, in T2Rs, the intracellular loops (ICLs) have a higher degree of sequence similarity, and the most divergent parts in T2R sequences are the extracellular loop (ECL) regions. In general, T2Rs have a shorter ECL-2 and ICL-3 than Class A GPCRs.
FIGURE 1.
Two-dimensional representation of the T2R1 amino acid sequence. The receptor has seven transmembrane helices (TM1–TM7). Amino acids are shown in single-letter codes. Red circles denote residues subjected to site-directed mutagenesis in this study. Ballesteros and Weinstein (24) numbering for the highly conserved residue in each helix in T2R1 is shown in italic type. In T2Rs, the ICLs have a higher degree of sequence similarity, and the most divergent parts in T2R sequences are the ECL regions. The rhodopsin octapeptide tag (ETSQVAPA) inserted into the C terminus to facilitate detection of the protein using the monoclonal antibody rho-1D4 is shown.
TABLE 1.
Conserved transmembrane amino acid residue in Class A GPCRs and bitter taste receptors (T2Rs)
| Transmembrane helix | Highly conserved residue in Class A GPCRsa | T2Rsb |
||||
|---|---|---|---|---|---|---|
| Amino acid residue in T2R1 | Humans [25] | Rat [31] | Mouse [29] | Total [188] | ||
| % | % | % | % | |||
| I | Asn1.50 | Asn-241.50 | 92 | 90.3 | 89.7 | 89.9 (19) |
| Ile-271.53 | 92 | 83.9 | 82.8 | 85.6 (27) | ||
| II | Leu2.50 | Leu-512.50 | 100 | 96.8 | 93.1 | 96.8 (6) |
| Arg-552.54 | 96 | 96.8 | 96.6 | 93.6 (12) | ||
| Asn-662.65 | 16 | 12.9 | 13.7 | 12.7 (164) | ||
| Glu-742.73 | 4 | 2.1 (184) | ||||
| III | Arg3.50 | Asn-893.45 | 84 | 80.6 | 72.4 | 82.0 (34) |
| Trp-943.50 | 100 | 96.8 | 96.6 | 97.3 (5) | ||
| Leu-993.55 | 96 | 96.8 | 96.6 | 97.8 (4) | ||
| IV | Trp4.50 | Leu-1254.50 | 88 | 83.9 | 82.8 | 88.8 (21) |
| V | Tyr5.50 | Leu-1975.50 | 100 | 100 | 100 | 98.2 (3) |
| Ser-2005.53 | 100 | 96.8 | 100 | 97.9 (4) | ||
| Leu-2015.54 | 96 | 100 | 100 | 98.9 (2) | ||
| VI | Pro6.50 | Phe-2336.50 | 100 | 87.1 | 93.1 | 93.1 (13) |
| VII | Pro7.50 | His-2737.46 | 96 | 90.3 | 89.7 | 89.4 (20) |
| Leu-2777.50 | 96 | 93.5 | 93.1 | 93.1 (13) | ||
| Ile-2787.51 | 92 | 87.1 | 82.8 | 85.6 (27) | ||
a Numbers in superscript correspond to the Ballesteros and Weinstein (24) nomenclature for GPCRs.
b The conserved amino acids from the sequence analysis were expressed as a percentage. A total of 188 T2R amino acid sequences were analyzed (shown in the last column). Only Asn-66 and Glu-74 are T2R1-specific residues. Numbers in brackets correspond to the total number of receptors analyzed, and numbers in parentheses in the last column correspond to number of receptors in which the amino acid residues are not conserved (e.g. in the case of Leu-512.50 of the 188 T2R amino acid sequences analyzed (brackets), this residue was absent in six sequences (parentheses)). Trp-943.50 and Leu-993.55 and similarly Leu-1975.50 and Leu-2015.54 show very close sequence conservation among the 188 T2Rs analyzed; however, Trp-943.50 and Leu-1975.50 are 100% conserved in all the 25 human T2Rs and hence were designated as residues 0.50 in the respective TM helices.
To elucidate the signal transduction mechanism of T2Rs at the molecular level, we carried out amino acid replacements guided by molecular modeling of 15 transmembrane residues, including 13 highly conserved amino acids present in TM1, TM2, TM3, TM5, and TM7 of T2R1. All of the mutants were functionally characterized based on their ability to be activated by the bitter agonist, dextromethorphan (DXM), a commonly used antitussive drug. Our results show that replacement of Asn-662.65 and the highly conserved Asn-241.50 and Asn-893.45 resulted in greater than 90% loss of agonist-induced signaling. We identify the interactions between Asn-241.50 and Arg-552.54 as playing a crucial role in receptor activation. Furthermore, we identify a conserved LXXSL motif, which performs a structural role in T2Rs by stabilizing the helical conformation of TM5 at the cytoplasmic end. Replacement of the conserved residues in this motif with the bulky β-branched amino acids results in protein misfolding and/or a non-functional receptor. We discuss the varied roles of these conserved TM residues in T2Rs and, where possible, compare and contrast with those of the Class A GPCRs.
EXPERIMENTAL PROCEDURES
Materials
Fetal bovine serum and DMEM high glucose were purchased from Sigma and Invitrogen. Common chemicals and bitter compounds were purchased either from Fisher or Sigma. Fluo-4NW was purchased from Invitrogen. Acetaminophen, caffeine monohydrate, quinine hydrochloride, leporamide hydrochloride, and femotidine were purchased from MP Biomedicals (Solon, OH). HCl (0.1 and 1 m), sodium chloride (NaCl, 1 m), and monosodium glutamate (0.1 m) solutions were provided by Alpha M.O.S (Hanover, MD). All chemicals were of analytical grade and used without further purification.
Molecular Biology and Cell Culture
Amino acid substitutions were introduced into the synthetic T2R1-rho1D4 gene carried by the expression vector pcDNA3.1 (Invitrogen) as described previously (15, 16) or synthesized commercially (GenScript USA Inc.). DNA sequences of all of the mutated genes were verified by automated DNA sequencing (MICB DNA Sequencing Facility, Winnipeg). The wild type and mutant T2R1 genes in pcDNA3.1 were transiently expressed in C6-glioma cells using Lipofectamine 2000 (Invitrogen) according to previously published methods (15).
Taste Sensory Analysis by Electronic Tongue (E-Tongue)
Taste sensory analysis of compounds used in this study was performed using the analytical instrument E-Tongue from Alpha M.O.S. (Toulouse, France). Each experiment consisted of three main steps: E-Tongue conditioning and calibration, sample preparation and analysis, and data processing. The pass criterion for E-Tongue conditioning was to achieve stable signals for all seven sensors with minimal or no noise or drift. Due to the chemical nature of the samples and the sensitivity of the sensor array used in this study, the conditioning step was repeated 12 times at the beginning of every working week following more than 2 days of sensor storage in the dry state. To ensure consistency and reproducibility of data, each individual sensor was calibrated to a known numerical value and a previously defined error limit. This calibration step ensured that the output response of each sensor did not exceed the maximum error allowed. The calibration was performed after every successful conditioning step. The pass criterion for the calibration step was to have all sensors adjusted to their target values within the specified error limit. Next, to ensure that the E-Tongue was able to identify distinctive tastes, a diagnostic test using HCl, NaCl, and monosodium glutamate each at a concentration 0.1 m representing sourness, saltiness, and umami tastes, respectively, was performed. The pass criterion required a discrimination index of at least 0.94 with compound clusters being visibly separated from each other on a principal component analysis map (data not shown). After this diagnostic step, sensor calibration and validation was done using known reference compounds suggested by Alpha M.O.S (supplemental Table 1). To predict the bitterness scores of test compounds, 3 mm solutions of the compounds were prepared in water using a sonic bath incubator for 10 min at 37 °C and were analyzed by the E-Tongue against known reference solutions of caffeine and quinine.
Calcium Assays
Glial cells have comparatively lower basal calcium noise compared with HEK293 for DXM, and thus we were able to do a dose-response analysis for T2R1 activation by DXM in glial cells, according to previously published protocols (15). In addition, we have shown that glial cells do not expresses endogenous T2R1 (15). The measurement of intracellular calcium produced was carried out as follows. Approximately 1.0 × 105 C6-glioma cells were plated into each well of 96-well tissue culture-treated BD-Falcon Biolux plates, and plasmid DNA (0.2 μg/well) was transfected using Lipofectamine 2000. Following 24 h after transfection, the medium was removed, and cells were washed once with PBS and incubated with the calcium-sensitive dye Fluo-4NW (Invitrogen) for 1 h. Receptor activation was determined by measuring changes in intracellular calcium after application of different concentrations of DXM or buffer alone (for measuring basal activity) using a Flexstation-3 fluorescence plate reader (Molecular Devices) at 525 nm following excitation at 494 nm. The data presented were from 2–5 independent transfections in duplicate. Dose-response curves were generated, and EC50 values were calculated by nonlinear regression analysis using PRISM software version 4.03 (GraphPad Software Inc., San Diego, CA) after subtracting the responses of mock-transfected cells.
Immunofluorescence
HEK293T cells were seeded into 6-well tissue culture plates containing sterilized poly-l-lysine (Sigma)-coated glass coverslips and transiently transfected with wild-type T2R1 or the mutant constructs according to published procedures (5). All steps were carried out at room temperature. 22–24 h post-transfection, the cells were washed with 1× PBS and fixed in 3.7% formaldehyde, 1× PBS buffer for 15 min and then permeabilized with 0.05% Triton X-100, 1× PBS buffer for 20 min. The cells were washed with PBS and blocked with 2% bovine serum albumin (IgG- and protease-free) in 1× PBS for 90 min. T2R1 and the mutants were labeled for 90 min using a 1:500 dilution of the mouse-anti-rho-1D4 monoclonal antibody (C-terminal tagged T2R1) and a 1:100 dilution of rabbit anti-calnexin polyclonal antibody (Abcam; endoplasmic reticulum marker). The transfected cells were washed and incubated with fluorophore-conjugated secondary antibodies using a 1:2000 dilution of goat anti-mouse Alexafluor 488 (Invitrogen) and a 1:300 dilution of goat anti-rabbit Alexafluor 594 (Invitrogen) for 60 min. Following washing, the nuclei were stained with a 1:1000 dilution of Hoechst-33342 stain for 10 s, and the coverslips were dried for 30–45 min. Prolong-antifade gold (Invitrogen) was used to mount the coverslips on slides, and the edges were sealed with nail polish. Representative cells were selected and visualized using an Olympus BX81 microscope for cytoplasmic or plasma membrane localization.
Molecular Modeling of the T2R1 Receptor
The model of T2R1 receptor was built by multiple-threading alignments with different GPCR crystal structures using the I-TASSER server (17). The transmembrane regions of T2R1 were predicted by TMpred and HMMTOP servers(18). Loop regions of the receptor were modeled using the ModLoop server (19). Side chains of the amino acids were refined with SCWRL4 (20). Then the whole molecule was energy-minimized by 1000 steps of steepest descent and 1000 steps of conjugate gradients using SPDBV 4.0.1. Molecular dynamics simulations for 10 ps were performed for the active model using OpenMM Zephyr (21). The quality of the model was verified by using the program PROCHEK (22), and 98.2% of the residues were in allowed regions of the Ramachandran plot.
The ligands were docked to the receptor using the program AutoDock Vina (23). The binding site for the ligand on T2R1 was defined by forming a cube with the dimensions 60 × 80 × 70 around the protein with a grid point spacing of 0.375 Å and center grid boxes −51.807, −12.467, and 38.921 in x, y, and z dimensions, respectively. We performed 50 genetic algorithm runs for each ligand. In each run, the best pose was saved. Finally, all poses were superimposed, and the most frequent orientation of the ligand was taken as the final pose. Then the receptor-ligand complex was energy-minimized by 2000 steps of conjugate gradients. The T2R1 mutants were built by using the “mutate residue” application in PyMOL using a basal T2R1 three-dimensional model. Then the model was energy-minimized, followed by molecular dynamics simulations using OpenMM Zephyr. The mutants were then docked with dextromethorphan. Molecular dynamics simulations were performed on all of the protein ligand complexes using Desmond 2.4.2.1.
RESULTS
Taste Sensory Analysis
The bitterness of the compounds yohimbine, DXM, salicin, and thiamine was analyzed by E-Tongue against known reference solutions of caffeine and quinine, as described under “Experimental Procedures.” Sensory analysis of the compounds tested shows that DXM has the most intense bitterness, with a predicted bitterness score of 12.87 (supplemental Table 1). In addition, our previous study on T2R1 using different ligands showed that DXM activates T2R1 with the highest potency (15). Based on the sensory analysis and EC50 value for T2R1, DXM was selected as the ligand in the present study.
Generic Amino Acid Numbering System for T2Rs and Identification of Amino Acids for Mutational Analysis
Sequence analysis of the Class A GPCRs reveals that most of the highly conserved sites are located in the transmembrane helices. Ballesteros and Weinstein (24) have utilized this feature to propose a numbering scheme for amino acids in GPCRs, where the helix number is followed by the sequence position relative to the most conserved residue in the helix, designated as 50. This highly accepted generic numbering system for GPCRs was recently updated for the Class A GPCRs, where it was shown that leucine (94.0% conserved) rather than aspartate (89.4% conserved) is the most highly conserved amino acid in TM2, and they are now designated as Leu-2.50 and Asp-2.54 (4).
We carried out a multiple sequence analysis of 188 T2R amino acid sequences present in the NCBI data base and aligned them with the Class A GPCR sequences using Clustal version 2.1 (25). Based on the amino acid sequence analysis (supplemental Fig. 1), we found that only Asn-1.50 and Leu-2.50 are the highly conserved transmembrane residues that are common between Class A GPCRs and T2Rs (Table 1). This interesting result shows that most of the highly conserved and important motifs in Class A GPCRs, such as the LXXXD in TM2, (D/E)RY in TM3, CWXP in TM6, and NPXXY in TM7 are absent in T2Rs and brings up the question of how similar or dissimilar are the activation mechanisms of T2Rs and the Class A GPCRs. In general, T2Rs showed very low similarity with the Class A GPCRs within the transmembrane regions analyzed.
Amino acid replacements guided by molecular modeling were carried out at key transmembrane positions, Asn-241.50, Iso-271.53, Leu-512.50, Arg-552.54, Asn-662.65, Glu-742.73, Asn-893.45, Trp-943.50, Leu-993.55, Leu-1975.50, Ser-2005.53, Leu-2015.54, His-2737.46, Leu-2777.50, and Iso-2787.51 (Table 1). Two types of mutations were made at each position. First, mutations were made to a smaller amino acid, such as alanine; the hypothesis is that this substitution will have a minimal effect on receptor folding and ligand binding. Second, guided by a molecular model of T2R1, mutations were made with either a non-conserved substitution, such as asparagine to aspartate, or with conserved substitutions, such as arginine to lysine.
Analysis of TM1 Mutants
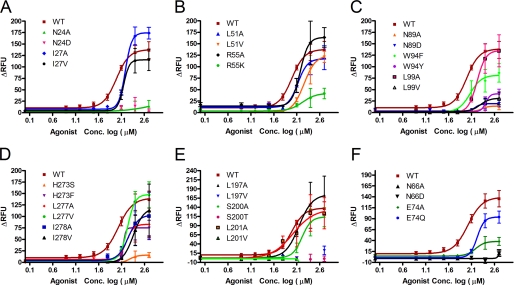
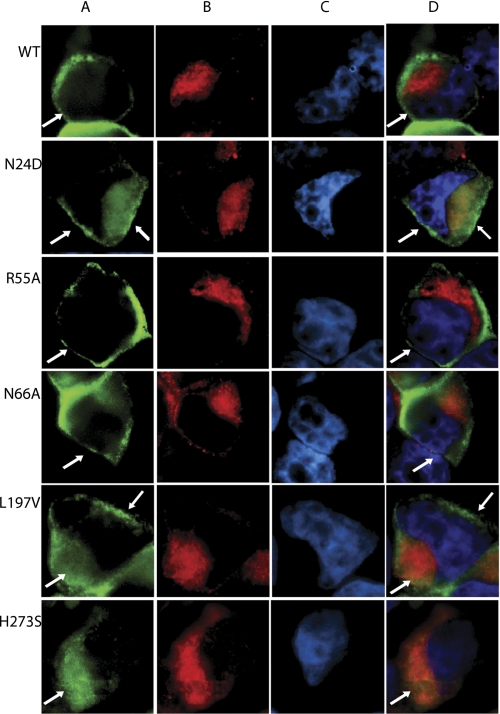
Functional analysis of wild type and mutant T2R1 receptors was determined by measuring changes in intracellular calcium of glial cells transiently expressing these receptors, after application of different concentrations of the T2R1 agonist DXM (15). The most highly conserved amino acid residue in GPCRs is Asn at position 1.50, and this residue is conserved as Asn-241.50 in T2R1. Replacement of Asn-24 with either alanine or aspartate drastically reduced by >90% agonist-induced signaling by the respective mutants (Fig. 2A). To elucidate if this loss in signaling is due to poor cell surface expression of the receptors, subcellular localization of the N24A and N24D mutants using immunofluorescence microscopy was carried out in the HEK293T cells. Results from immunofluorescence microscopy show that N24A is predominantly internalized, whereas N24D mutant is partially localized on the cell surface (Fig. 3 and supplemental Fig. 2).
FIGURE 2.
Functional characterization of T2R1 mutants. Concentration-dependent changes in intracellular calcium [Ca2+] induced by bitter ligand DXM in C6 glial cells transfected with T2R1 or mutants. Data were collected from at least three independent experiments carried out in duplicate. Dose-response curves were generated using Graph Pad Prism software, after subtracting the responses of mock-transfected cells. A, TM1 mutants; B, TM2 mutants; C, TM3 mutants; D, TM5 mutants; E, TM7 mutants; F, mutants validating the molecular model(s). RFU, relative fluorescence units.
FIGURE 3.
Immunofluorescence microscopy showing localization of wild-type T2R1 and mutants expressed in HEK293T cells. Double label immunofluorescence was performed using mouse monoclonal anti-rho-1D4 antibody, which recognizes the C-terminal octapeptide tag on the expressed receptors, and rabbit polyclonal anti-calnexin antibody, which localizes to the endoplasmic reticulum. The WT and mutant T2R1 receptor was visualized using goat anti-mouse Alexafluor 488 secondary antibody (A, green), and the endoplasmic reticulum was visualized with goat anti-rabbit Alexafluor 594 secondary antibody (B, red). The nucleus stained with Hoechst-33342 dye is shown in blue (C). The overlay of the receptor, endoplasmic reticulum, and nucleus is shown in D. The merged images show that in the wild type, R55A, and N66A, the receptors were predominantly localized at the plasma membrane (white arrows), in N24D and L197V partly localized on the cell surface, whereas in H273S, the receptors appear to be located in intracellular compartments.
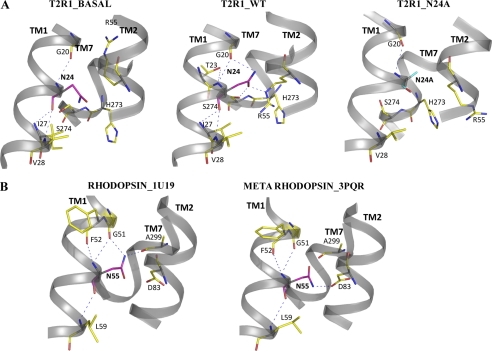
Molecular modeling analysis of T2R1 bound to the agonist DXM shows that Asn-241.50 is situated outside the ligand binding pocket and mediates a network of inter- and intrahelical H-bonds (Fig. 4). Close analysis of residues within 4 Å of Asn-241.50 shows that Asn-241.50 establishes backbone contacts with Gly-201.46, Iso-271.53, and Val-281.54 on TM1, its side chain carbonyl is H-bonded to the side chain functional groups of Arg-552.54 and Ser-2747.47 on TM2 and TM7, and its side chain amide is H-bonded to the backbone carbonyl of Gly-201.46, respectively (Fig. 4). This intricate H-bond network is absent in the unbound T2R1 and N24A mutant, whereas it is significantly modified in the N24D mutant (Fig. 4). The N24D mutant loses backbone contacts with Iso-27 and Val-28, whereas its side chain carboxyl oxygen forms a new bond with Gln-592.58 on TM2. These interactions cause relaxation in the helical backbone around the residue 1.50 in the N24D mutant, causing it to move closer to TM2 than TM7 (supplemental Fig. 3). These results show that Asn-241.50 is involved in mediating an H-bonding network connecting TM1-TM2-TM7 in T2Rs that is important for receptor activation.
FIGURE 4.
The molecular contacts of Asn-241.50 in T2R1 and the Class A GPCR, rhodopsin. A, T2R1_BASAL represents the molecular contacts of Asn-24 in the T2R1 unbound model; T2R1_WT represents the DXM-bound model of T2R1; and T2R1_N24A represents the DXM-bound N24A mutant. Asn-241.50 is represented in cyan, and the blue dotted lines represent hydrogen bond contacts. In all three models, Gly-201.46 and Asn-241.50 residues have backbone-backbone contacts. In T2R1_basal and T2R1_WT, the backbone contacts of Asn-241.50 are very similar, but in the basal form, the side chain of Asn-241.50 is not involved in H-bonding. In T2R1_WT, the side chain amine of Asn-241.50 is interacting with the backbone carbonyl of Gly-201.46 and carbonyl with side chain amine of Arg-552.54 and the oxyl group of Ser-2747.47. B, RHODOPSIN_1U19 represents the inactive structure of rhodopsin (Protein Data Bank code 1U19) showing the interactions of Asn-551.50, and METARHODOPSIN_3PQR shows the interactions of Asn-551.50 in the active structure of metarhodopsin (Protein Data Bank code 3PQR). In rhodopsin upon activation, Asn-551.50 loses contact with Gly-511.46 and Ala-2997.46 and forms a new contact, Asn-832.54. In T2R1, upon activation Asn-241.50 moves into space between TM2 and TM7 and establishes an H-bond network connecting Arg-552.54 and Ser-2747.47. The interhelical H-bond between Asn-241.50 and Arg-552.54 restrains the activity of the receptor because loss of this in I27A and R55A mutants results in hyperactivity.
Another highly conserved residue in TM1 is Iso-271.53; the I27A and I27V mutants showed robust intracellular calcium responses to DXM with EC50 values of 140 ± 11 μm and 153 ± 17 μm, respectively (Table 2). The I27A mutant displayed hyperactivity as shown by the upward shift of its dose-response curve (Fig. 2A). In wild type, Iso-271.53 makes multiple backbone-backbone interactions with both residues that are one turn above (i.e. Asn-241.50 and Thr-231.49) and one turn below (with Asn-31) on the cytoplasmic side (supplemental Fig. 3). However, in the hyperactive I27A mutant, Ala-27 loses the backbone contact with Asn-241.50; in addition, another key interaction involving Arg-552.54 with Asn-241.50 was lost. Interestingly, this H-bonding involving Arg-552.54 with Asn-241.50 was present in the I27V mutant, which displays wild type activity (Fig. 2A and supplemental Fig. 3). Thus, from our models of I27A and I27V mutants, it was apparent that the loss of the restraining interaction of Arg-552.54 with Asn-241.50 rather than loss of the backbone contact of Ala-27 with Asn-241.50 was responsible for the observed hyperactivity of I27A mutant. To validate our models and elucidate the role of the highly conserved Arg-552.54, which forms part of the L2.50XXXR2.54 motif in T2Rs, it was subjected to mutational analysis (see below).
TABLE 2.
Summary of signaling properties (EC50 values) of highly conserved and T2R1-specific transmembrane mutant receptors obtained from cell-based calcium assay
The values are expressed as the mean ± S.E. (n = 2–5 experiments) in duplicate, using the agonist dextromethorphan.
| Receptor | Transmembrane helix | EC50 | EC50 mutant/EC50 WT |
|---|---|---|---|
| μm | |||
| Wild type | 97 ± 21 | 1.0 | |
| N24A | I | No responsea | |
| N24D | I | No response | |
| I27A | I | 153 ± 17 | 1.59 |
| I27V | I | 140 ± 11 | 1.45 |
| L51A | II | 133 ± 20 | 1.38 |
| L51V | II | 180 ± 6 | 1.85 |
| R55A | II | 155 ± 13 | 1.59 |
| R55K | II | 168 ± 50 | 1.73 |
| N66A | II | No response | |
| N66D | II | No response | |
| E74A | II | 154 ± 20 | 1.58 |
| E74Q | II | 166 ± 7 | 1.71 |
| N89A | III | No response | |
| N89D | III | No response | |
| W94F | III | 118 ± 40 | 1.21 |
| W94Y | III | 249 ± 33 | 2.56 |
| L99A | III | 168 ± 9 | 1.73 |
| L99V | III | 211 ± 45 | 2.17 |
| L197A | V | 135.3 ± 9 | 1.39 |
| L197V | V | No response | |
| S200A | V | 147 ± 10 | 1.51 |
| S200T | V | No response | |
| L201A | V | 104.2 ± 40 | 1.07 |
| L201V | V | No response | |
| H273S | VII | No response | |
| H273F | VII | 164 ± 18 | 1.69 |
| L277A | VII | 158.9 ± 33 | 1.62 |
| L277V | VII | 168 ± 21 | 1.73 |
| I278A | VII | 174.1 ± 30 | 1.79 |
| I278V | VII | 226 ± 8 | 2.32 |
a No response, those mutants where there is no agonist concentration-dependent response, or the observed specific response is below the base-line activity of wild type T2R1.
Analysis of TM2 Mutants
The next highly conserved amino acid that is common between the Class A GPCRs and T2Rs is Leu2.50, present on TM2. Whereas Class A GPCRs have an L2.50XXXD2.54 motif on TM2, this is conserved as a L2.50XXXR2.54 motif in T2Rs (Table 1). To elucidate the role of the conserved TM2 residues, both Leu-51 and Arg-55 of the LXXXR motif in T2R1 were replaced with alanine, valine, or lysine, and the mutants were analyzed. From the dose-response assays using the agonist DXM, L51A was similar to wild type with an EC50 of 133 ± 20 μm, whereas L51V showed a reduced response to DXM with an EC50 of 180 ± 6 μm (Table 2). Both the R55A and R55K had higher EC50 mutant/wild type ratios of 1.6 and 1.7. Immunofluorescence microscopy shows that the R55K mutant was poorly expressed on the cell surface, which would explain the low activity observed (Fig. 2B). Similar to the I27A replacement, the R55A mutant also displayed hyperactivity (i.e. an upward shift in the dose-response curve).
Analysis of the molecular models of R55A and R55K mutants shows that both mutants lose the H-bonding with Asn-241.50 and Ser-2747.47, whereas the side chain of the Lys-55 mutant is restrained with new H-bonding contacts with the side chain functional group of Glu-92 and backbone carbonyl of Leu-51 (supplemental Fig. 4). The H-bonds connecting Arg-55 seem to restrain the receptor because loss of these contacts as in the R55A and I27A mutants causes the receptor to exhibit hyperactivity. No major changes were noticed in the molecular models of L51A and L51V mutants, and immunofluorescence microscopy shows that these were properly expressed on the cell surface (supplemental Fig. 2).
Analysis of TM3 Mutants
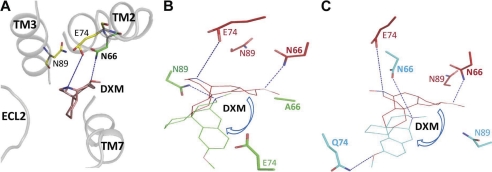
Sequence analysis of TM3 of T2Rs reveals three conserved amino acids, Asn-893.45, Trp-943.50, and Leu-993.55, with sequence conservation of 82, 97, and 97%, respectively. No significant DXM-dependent calcium signaling was observed for the N89A and N89D mutants (Fig. 2C). The Trp-943.50 and Leu-993.55 mutants displayed a dose-dependent response to DXM, although the W94Y and L99V had a 2-fold increase in EC50 of 249 ± 33 and 211 ± 45 μm compared with EC50 of 97 ± 21 μm for wild type. The W94Y mutant shows defective ligand binding with the highest EC50 mutant/wild type ratios of 2.56 (Table 2). Immunofluorescence microscopy of the N89A, N89D, W94Y, and L99V mutants shows that they are poorly expressed on the cell surface (supplemental Fig. 2). In our T2R1 models, Trp-943.50 is buried and facing away from the ligand binding pocket, whereas Leu-993.55 is present toward the cytoplasmic end of TM3. Only Asn-893.45 is in the ligand binding pocket (Fig. 5).
FIGURE 5.
Molecular model of T2R1 bound to agonist DXM. The T2R1 receptor was built by homology modeling using the crystal structure of active opsin as template (Protein Data Bank code 3DQB). Shown is an extracellular view of the ligand binding pocket of T2R1 bound to DXM (red). The residues within the 4 Å region, Asn-662.65, Glu-742.73, and Asn-893.45, are shown. In our molecular models, the orientation of DXM in WT_T2R1 is different from the Asn-662.65 and Glu-742.73 mutants; the direction of change in orientation is indicated by blue arrows. A, WT_T2R1 bound to DXM. The amine nitrogen of Asn-662.65 makes a hydrogen bond with the 3-methoxyl group, and Glu-742.73 makes a hydrogen bond with the tertiary amine functional groups of DXM. B, models of WT_T2R1 (red) and N66A mutant (green) docked with DXM are superimposed; replacement of asparagine at position 2.65 with alanine causes loss of H-bond contacts of residues 2.65 and 2.73 with DXM, and a new H-bond with Asn-89 was formed. C, models of WT_T2R1 (red) and E74Q mutant (cyan) docked with DXM are superimposed. Replacement of glutamate at position 2.73 with glutamine restores H-bond contacts of residues 2.65 and 2.73 with DXM; in contrast to wild type, Gln-74 makes H-bond contact with the 3-methoxyl group, and Asn-66 makes H-bond contact with the tertiary amine of DXM. The orientation of DXM in both N66A and E74Q is similar. Both Asn-662.65 and Glu-742.73 play an important role in T2R1 binding to the bitter agonist DXM.
Analysis of TM5 Mutants
From the sequence analysis of T2Rs, the most conserved amino acids are present in TM5. These residues form a putative LXXSL motif at the cytoplasmic end of TM5 (Fig. 6). The amino acids Leu-1975.50, Ser-2005.53, and Leu-2015.54 are more than 97% conserved in T2Rs (Table 1). Alanine substitutions of all of the three amino acids were tolerated, with L197A, S200A, and L201A showing EC50 values comparable with that of wild type (Table 2). Unexpectedly, dramatic changes in agonist-induced activity were observed with conservative substitutions (leucine to valine or serine to threonine) at all of the three positions. The L197V, S200T, and L201V mutants displayed complete loss of agonist induced dose-dependent signaling. (Fig. 2D). Immunofluorescence microscopy showed that the L197A, S200A, and L201A mutants were expressed on the cell surface; the L197V was partly localized on the cell surface (Fig. 3); and the S200T and L201V mutants appeared misfolded and were predominantly localized in the intracellular compartments (supplemental Fig. 2).
FIGURE 6.
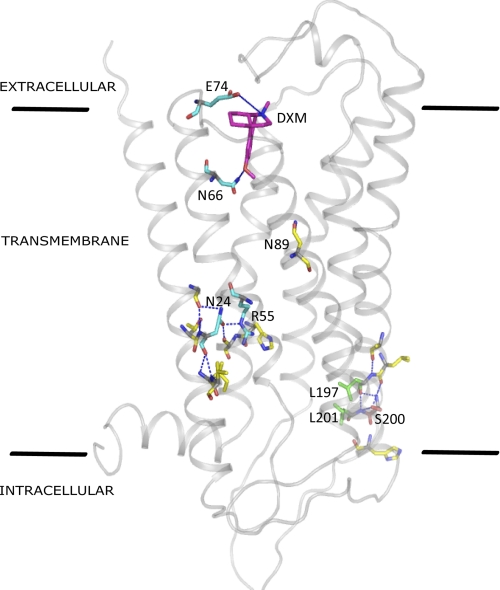
Transmembrane view of DXM-bound T2R1. The transmembrane amino acids identified in this study to be important for structure and function in T2Rs are highlighted. Blue lines indicate hydrogen bond interactions.
Molecular model of wild type T2R1 shows a network of intrahelical H-bond interactions between backbones of the three residues, Leu-1975.50, Ser-2005.53, and Leu-2015.54, and mutations that cause a significant loss of these interactions, such as L197V, S200T, and L201V, disturb the helical structure of TM5 at the cytoplasmic end, which may have caused protein misfolding and/or non-functional receptor (supplemental Fig. 6). Although L197V, S200T, and L201V can be considered conservative mutations with respect to functionality (serine and threonine are polar; leucine and valine are hydrophobic), structurally they are non-conserved because both threonine and valine are β-branched amino acids. β-Branched amino acids, such as valine or threonine, can cause bulkiness in close proximity to the protein backbone, restricting the conformation the main chain can adopt, as observed in the case of the L197V, S200T, and L201V mutants. The presence of valine at position 5.50 in the L197V mutant causes a change in conformation of the protein backbone, resulting in loss of local helical structure and intrahelical backbone contacts with Ala-193 and Ser-200. In the case of the S200T mutant, an additional interaction with the backbone of His-204 was lost. The His-204 of ICL3 is highly conserved (96%) across T2Rs, although its role in T2R structure and function remains to be determined.
Analysis of TM7 Mutants
The three conserved amino acids present on TM7, His-2737.46, Leu-2777.50, and Iso-2787.51, which have >85% sequence conservation among the T2R subfamily, were initially replaced with smaller amino acids, such as serine or alanine. The H273S mutant displayed a complete loss of agonist-induced signaling, whereas the L277A, L277V, I278A, and I278V were functional (Fig. 2E). Immunofluorescence microscopy of these mutants demonstrates that they are expressed predominantly on the cell surface (supplemental Fig. 2).
In wild type T2R1, the imidazole group of His-273 is less than 2 Å facing the side chain of Arg-55, and this distance slightly increases in the H273F mutant; however, in the H273S mutant, the side chain of serine moves away and is at a distance of 6 Å from Arg-55, resulting in the loss of TM2-TM7 interactions (supplemental Fig. 4). Although the relationship between His-273 and Arg-55 is difficult to interpret from our model, we hypothesize that His-273 might play an indirect role in maintaining TM1-TM2-TM7 contacts (e.g. in helping Ser-274 to make contacts with TM2 and TM1 because these were lost in the H273S mutant); in addition, the restraining interaction of Arg-552.54 with Asn-241.50 was lost in the H273F mutant. Interestingly, whereas the H273F mutant is partially localized on the cell surface, which might explain its reduced activity, the H273S mutant exhibited defective folding (Fig. 3); this can be partly explained based on conformational and folding tendencies of amino acid side chains in proteins. Histidine exhibits a high degree of similarity with phenylalanine but very low similarity with serine (26).
Mutations Based on Molecular Models
To further validate the T2R1 molecular models, we targeted amino acids predicted to be important for ligand binding and activation. Our molecular model of agonist DXM-bound T2R1 showed Asn-662.65, Glu-742.73, and Asn-893.45 to be present within the binding site (Fig. 5). However, except for Asn-893.45, the other residues are not conserved in T2Rs (Table 1). This is not surprising because the bitter compounds recognized by T2Rs are very diverse with no common functional group associated with the ligand(s), removing the need for a single highly conserved residue for ligand recognition in T2Rs.
Replacement of Asn-662.65 with either alanine or aspartate caused greater than 90% loss in agonist-induced signaling (Fig. 2F). No significant DXM-dependent calcium signaling was observed for the N66A and N66D mutants (Table 2). Both the E74A and E74Q mutants had higher EC50 mutant/wild type ratios of 1.6 and 1.7; however, the E74Q mutant showed up to 80% of wild type response (supplemental Table 2). Results from immunofluorescence experiments showed that the mutants N66A and E74Q are properly folded and expressed on the cell surface, whereas the N66D and E74A mutants are poorly expressed (Fig. 3 and supplemental Fig. 2).
Based on molecular studies, in wild type T2R1 bound to DXM, the amine nitrogen of Asn-662.65 makes a hydrogen bond with the 3-methoxyl and Glu-742.73 with the tertiary amine functional groups of DXM (Fig. 5A). This contact was lost in the case of both of the mutants, N66A and N66D. In N66A, the Ala residue was not in the 4-Å region of DXM (Fig. 5B). In our molecular models, within the same binding site, the orientation of DXM in wild type T2R1 is different from the Asn-662.65 and Glu-742.73 mutants. In E74Q mutant, DXM binds in a different orientation, but the H-bond interactions of Asn-662.65 and Gln-742.73 with the two functional groups of DXM are still conserved (Fig. 5C). Gln-74 makes H-bond contact with the 3-methoxyl group, and Asn-66 makes H-bond contact with the tertiary amine of DXM. These interactions are absent in the E74A mutant (supplemental Fig. 5). This shows that the capability to hydrogen-bond rather than charge the residue at position 2.73 is important in DXM binding.
DISCUSSION
Functional analysis of T2Rs have shown that some T2Rs recognize a wide range of ligands, whereas others like T2R1 are very specific and recognize only selective ligands (27). However, the mechanisms underlying these differences in specificities have so far not been elucidated. There are very limited numbers of structure-function studies on T2Rs, and thus far none have addressed the roles of the highly conserved transmembrane T2R amino acids in receptor activation or ligand binding. Even in the Class A GPCRs, although some of the highly conserved residues present in TM helices, such as Asn1.50, Arg3.50, and Pro7.50, are well studied, the hydrophobic residues Leu2.50 and Trp4.50 are poorly studied because it was not clear how they influence receptor structure and function (28–30).
From the results presented in this paper, we can compare and contrast the role played by the conserved Asn1.50 in Class A GCPRs and T2Rs. In Class A GPCRs, Asn1.50 is involved in intrahelical hydrogen bonding with Gly1.46 and interhelical hydrogen bonding with the side chain functional group of Asp2.54 and backbone carbonyl of amino acid at position 7.46 (5). In inactive rhodopsin (Protein Data Bank entry 1U19), Asn1.50 is hydrogen-bonded with Gly1.46 and Ala7.46; upon activation of rhodopsin (Protein Data Bank entry 3PQR), these bonds are broken, and Asn1.50 forms a new H-bond with the carbonyl of Asp2.54 (Fig. 4B). The interhelical H-bond interactions connecting Asn1.50 with Gly1.46 and Ala7.46 are thought to restrain rhodopsin in the inactive state (4). In contrast, in T2Rs, the residues on TM2 and TM7 interacting with Asn-241.50 are Arg-552.54 and Ser-2747.47 (Fig. 4A). In the agonist-unbound (inactive) model of T2R1 and the inactive N24A mutant, the side chain of Asn-241.50 is not making any H-bonding contacts with Gly-201.46, Arg-552.54 and Ser-2747.47, whereas in the agonist bound model (active), it forms an H-bonding network connecting TM1-TM2-TM7, which appears necessary for receptor activity (Fig. 4). In the gonadotropin-releasing hormone receptor, mutation of Asn1.50 resulted in the mutants being poorly expressed and unable to bind to the ligand (28). Although our study is the first to target Asn1.50 in T2Rs, we observed the same non-functional phenotype as was observed in Class A GPCRs. In T2Rs, Asn-241.50 is also involved in mediating an H-bonding network connecting TM1-TM2-TM7, which is important for receptor function. However, the residues on TM2 and TM7 interacting with Asn-241.50 are Arg-552.54 and Ser-2747.47 in T2Rs.
The recent crystal structures of the active states of Class A GPCRs show that amino acid residues at positions 2.54 and 7.46 along with structurally conserved water molecules form an extended H-bond network, which plays an important role in receptor activation (6, 31). We speculate the existence of a similar network involving Arg2.54, Ser7.47, and structural waters in T2Rs. However, in the absence of a T2R crystal structure and because of the very low amino acid sequence similarity between T2Rs and Class A GPCRs, we did not introduce conserved structural waters into our T2R1 molecular models.
In our previous docking studies on T2R1, we showed that few amino acid residues present in TM2, TM3, and TM7 predominantly contribute to ligand binding; all of them except for Asn-893.45 are non-conserved residues. A very recent study proposed that Asn-893.45 is involved in salicin recognition in T2R16 and that Asn-893.45 mutants failed to bind to salicin (12); however, the cell surface localization of the mutants was not elucidated. In this study, we found that although T2R1 Asn-893.45 mutants failed to bind to DXM, they were predominantly localized in the intracellular compartments, precluding any assessment of the role of Asn-893.45 in ligand binding. In Class A GPCRs, the key ligand recognition amino acids are highly conserved across a specific subfamily, for example, Asp-1133.32 in β2-AR is 92% conserved in the amine subfamily. However, except for Asn-893.45, which is only 82% conserved in T2Rs, the rest are mostly non-conserved residues. A major factor responsible for the low sequence conservation of ligand binding residues can be the vast diversity of the exogenous bitter ligands recognized by T2Rs.
The putative LXXSL motif in T2Rs has a predominantly structural role in stabilizing the helical conformation of TM5 at the cytoplasmic end and a functional role by influencing the conformation of ICL3 (Fig. 6). It was previously shown in a number of GPCRs that the proper conformation of ICL3 is important for receptor folding (32, 33). The residues in the LXXSL motif have extensive intrahelical interactions as well as interhelical interactions with TM3 and with the amino acids in ICL3, which appears to be crucial for proper receptor folding and function. Helix-destabilizing mutations in the LXXSL motif cause extensive loss of these interactions. However, alanine substitutions of the residues in the LXXSL motif resulted in mutants that displayed cell surface expression and signaling properties similar to the wild-type receptor, which suggests that the mutations did not significantly perturb the structure of the receptor.
Although the role of His-2737.46 is not clear from our studies, recent mutational analysis of the residues present on the extracellular side of TM7 showed that they are involved in agonist selectivity in T2R31, T2R43, and T2R46 (14). In Class A GPCRs, the residue at position 7.46 is a small amino acid, such as alanine or serine. Recently, it was shown in β2-AR that Ser-3197.46 is involved in maintaining a hydrogen bond network connecting structural waters and amino acids on TM1-TM2-TM7 that restrain the receptor in the inactive state (5, 34). Furthermore, mutation of the β2-AR Ser-3197.46 results in constitutive activity (5, 34). Under our assay conditions, we did not observe constitutive activity upon mutation of the His-2737.46; further structure-function studies are needed to elucidate whether structural waters can act as allosteric regulators of T2R function.
In conclusion, our mutational analysis of the transmembrane residues in T2R1 aided by molecular modeling allowed us to determine how they influence receptor structure and function. Amino acid sequence analysis shows how divergent T2Rs are from the well studied Class A GPCRs, raising questions regarding their mechanisms of activation and how T2Rs recognize natural bitter ligands with diverse chemical structures. Further structure-function analysis of the amino acids present in the extracellular and intracellular loops would give detailed insights into residues that contribute to ligand binding or G-protein coupling. Structure-function studies on T2Rs could result in the design of bitter blockers or antagonists for these bitter receptors, thereby increasing the palatability of health-promoting bitter foods.
Supplementary Material
Acknowledgment
We acknowledge use of the E-Tongue instrument at the Richardson Centre for Functional Foods and Nutraceuticals, University of Manitoba.
This work was supported by Discovery Grant RGPIN 356285 from the Natural Sciences and Engineering Research Council of Canada, an Establishment Grant from Manitoba Health Research Council, and a New Investigator Award from the Heart and Stroke Foundation of Canada (to P. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–6.
- GPCR
- G-protein-coupled receptor
- HEK293
- human embryonic kidney cells
- DXM
- dextromethorphan
- β2-AR
- β2-adrenergic receptor
- ICL
- intracellular loop
- ECL
- extracellular loop
- H-bond and H-bonding
- hydrogen bond and hydrogen bonding, respectively.
REFERENCES
- 1. Conte C., Ebeling M., Marcuz A., Nef P., Andres-Barquin P. J. (2002) Cytogenet. Genome Res. 98, 45–53 [DOI] [PubMed] [Google Scholar]
- 2. Lagerström M. C., Schiöth H. B. (2008) Nat. Rev. Drug Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 3. Shi P., Zhang J., Yang H., Zhang Y. P. (2003) Mol. Biol. Evol. 20, 805–814 [DOI] [PubMed] [Google Scholar]
- 4. Smith S. O. (2010) Annu. Rev. Biophys. 39, 309–328 [DOI] [PubMed] [Google Scholar]
- 5. Arakawa M., Chakraborty R., Upadhyaya J., Eilers M., Reeves P. J., Smith S. O., Chelikani P. (2011) Biochim. Biophys. Acta 1808, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Standfuss J., Edwards P. C., D'Antona A., Fransen M., Xie G., Oprian D. D., Schertler G. F. (2011) Nature 471, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V. P., Chien E. Y., Velasquez J., Kuhn P., Stevens R. C. (2008) Structure 16, 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chelikani P., Hornak V., Eilers M., Reeves P. J., Smith S. O., RajBhandary U. L., Khorana H. G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7027–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang M., Yao Y., Kuang D., Hampson D. R. (2006) J. Biol. Chem. 281, 8864–8870 [DOI] [PubMed] [Google Scholar]
- 10. Horn F., Bettler E., Oliveira L., Campagne F., Cohen F. E., Vriend G. (2003) Nucleic Acids Res. 31, 294–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fredriksson R., Lagerström M. C., Lundin L. G., Schiöth H. B. (2003) Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 12. Sakurai T., Misaka T., Ishiguro M., Masuda K., Sugawara T., Ito K., Kobayashi T., Matsuo S., Ishimaru Y., Asakura T., Abe K. (2010) J. Biol. Chem. 285, 28373–28378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Biarnés X., Marchiori A., Giorgetti A., Lanzara C., Gasparini P., Carloni P., Born S., Brockhoff A., Behrens M., Meyerhof W. (2010) PLoS ONE 5, e12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brockhoff A., Behrens M., Niv M. Y., Meyerhof W. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11110–11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Upadhyaya J., Pydi S. P., Singh N., Aluko R. E., Chelikani P. (2010) Biochem. Biophys. Res. Commun. 398, 331–335 [DOI] [PubMed] [Google Scholar]
- 16. Arakawa M., Yanamala N., Upadhyaya J., Halayko A., Klein-Seetharaman J., Chelikani P. (2010) Protein Sci. 19, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roy A., Kucukural A., Zhang Y. (2010) Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tusnády G. E., Simon I. (2001) Bioinformatics 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 19. Fiser A., Do R. K., Sali A. (2000) Protein Sci. 9, 1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krivov G. G., Shapovalov M. V., Dunbrack R. L., Jr. (2009) Proteins 77, 778–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedrichs M. S., Eastman P., Vaidyanathan V., Houston M., Legrand S., Beberg A. L., Ensign D. L., Bruns C. M., Pande V. S. (2009) J. Comput. Chem. 30, 864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 23. Trott O., Olson A. J. (2010) J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ballesteros J. A., Weinstein H. (1995) Methods Neurosci. 25, 366–428 [Google Scholar]
- 25. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 26. Anderson R. J., Weng Z., Campbell R. K., Jiang X. (2005) Proteins 60, 679–689 [DOI] [PubMed] [Google Scholar]
- 27. Meyerhof W., Batram C., Kuhn C., Brockhoff A., Chudoba E., Bufe B., Appendino G., Behrens M. (2010) Chem. Senses 35, 157–170 [DOI] [PubMed] [Google Scholar]
- 28. Flanagan C. A., Zhou W., Chi L., Yuen T., Rodic V., Robertson D., Johnson M., Holland P., Millar R. P., Weinstein H., Mitchell R., Sealfon S. C. (1999) J. Biol. Chem. 274, 28880–28886 [DOI] [PubMed] [Google Scholar]
- 29. Proulx C. D., Holleran B. J., Boucard A. A., Escher E., Guillemette G., Leduc R. (2008) Mol. Pharmacol. 74, 552–561 [DOI] [PubMed] [Google Scholar]
- 30. Sakmar T. P., Menon S. T., Marin E. P., Awad E. S. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 443–484 [DOI] [PubMed] [Google Scholar]
- 31. Angel T. E., Gupta S., Jastrzebska B., Palczewski K., Chance M. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chee M. J., Mörl K., Lindner D., Merten N., Zamponi G. W., Light P. E., Beck-Sickinger A. G., Colmers W. F. (2008) J. Biol. Chem. 283, 33337–33346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 34. Nygaard R., Valentin-Hansen L., Mokrosinski J., Frimurer T. M., Schwartz T. W. (2010) J. Biol. Chem. 285, 19625–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.