Abstract
The activation of cytosolic phospholipase A2α (cPLA2α) plays an important role in initiating the inflammatory response. The regulation of cPLA2α mRNA turnover has been proposed to control cPLA2α gene expression under cytokine and growth factor stimulation. However, the detailed mechanism is still unknown. In this report, we have demonstrated that the cPLA2α mRNA stability was increased under IL-1β treatment in A549 cells. By using EMSAs, HuR was identified as binding with the cPLA2α mRNA 3′-UTR, and the binding region was located at nucleotides 2716–2807, a fragment containing AUUUA flanked by U-rich sequences. IL-1β treatment enhanced the association of cPLA2α mRNA with cytosolic HuR. The reduction of HuR expression by RNA interference technology inhibited IL-1β-induced cPLA2α mRNA and protein expression. Furthermore, blocking the p38 MAPK signaling pathway with SB203580 abolished the effect of IL-1β-induced cPLA2α gene expression. Phosphorylation at residue Thr-118 of HuR is crucial in regulating the interaction between HuR and its target mRNAs. Mutation of HuR Thr-118 reduced the association between HuR and cPLA2α mRNA under IL-1β treatment. This inhibitory effect was also observed in binding with COX-2 mRNA. This result indicated that p38 MAPK-mediated Thr-118 phosphorylation may play a key role in regulating the interaction of HuR with its target mRNAs in inflammation.
Keywords: Eicosanoid, Gene Regulation, Inflammation, MAP Kinases (MAPKs), RNA-binding Protein, RNA Turnover, HuR, RNA Regulon, mRNA Stability
Introduction
Cytosolic phospholipase A2α (cPLA2α)2 is a key enzyme that catalyzes the hydrolysis of membrane glycerophospholipids at the sn-2 position to form arachidonic acid and lysophospholipids (1). These bioactive lipids initiate the production of eicosanoids, which play important roles in inflammation, cancer, and other cellular processes. Therefore, cPLA2α has been implicated in the pathogenesis of multiple diseases (2). For example, the up-regulation of cPLA2α activity and arachidonic acid release in spinal cord tissue was observed within hours after neurotrauma. Treatment with the cPLA2α inhibitor arachidonyl-trifluoromethyl ketone (AACOCF3) increases the survival of neurons and oligodendrocytes in rats that received compression spinal cord injuries (3). Moreover, it has been shown that reduced cPLA2α expression contributes to a remarkable resistance to acute respiratory distress syndrome (ARDS), asthma, and collagen-induced arthritis in the cPLA2α-deficient mouse model (4–6). In addition, dysregulation of cPLA2α has been documented in carcinogenesis. Depletion of cPLA2α in bone marrow-derived macrophages prevents spontaneous lung metastasis from primary tumors (7). Mice that carry APCMin/+, a germ line mutation in the adenomatous polyposis coli gene, and cPLA2α−/−, which lacks cPLA2α expression, have an 83% reduction in intestinal tumorigenesis compared with wild type mice (8).
The expression levels of cPLA2α are tightly regulated to maintain the optimum balance of eicosanoid metabolites (9). Proinflammatory cytokines and growth factors have been demonstrated to activate the cPLA2α expression, and the transcriptional regulation mechanism has been widely studied (10, 11). Sp1 and c-Jun cooperatively activate the cPLA2α promoter in lung epithelial cells and non-small cell lung cancer (12). Signal transducers and activators of transcription 3 (STAT-3)-dependent cPLA2α expression play an important role in thrombin-induced vascular smooth muscle cell motility (13). The minimal promoter required for basal transcriptional activity of cPLA2α has also been defined (14). Post-transcriptional regulation of mRNA stability has also been reported to regulate the cPLA2α expression. Tay et al. (15) found that the cPLA2α mRNA half-life is increased under treatment with EGF, PDGF, serum, and phorbol 12-myristate 13-acetate; and the AUUUA sequences in the 3′-untranslated region (3′-UTR) are responsible for the instability of cPLA2α mRNA. However, until now, no further report has been dedicated to deciphering the post-transcriptional regulation mechanism.
The regulation of gene expression in eukaryotic cells comprises multiple steps, from transcription to translation. Up to now, more and more studies have emphasized that post-transcriptional regulation, including mRNA stability and translation, is an important way that organisms use to control their expression of genetic information being transformed into proteome (16–18). The turnover rate of mRNA varies in cells under different extra stimulus (19), and the aberrant control of mRNA metabolism is implicated in diseases including cancer, chronic inflammation, and coronary disease (20, 21). The most well known determinant sequence in controlling mRNA stability consists of AU-rich elements (AREs) located within the 3′-UTR of short-lived mRNAs (22). An AU-rich element is composed of several copies of AUUUA pentamer or just a U-rich domain (23). A broad survey of the occurrence of AREs in mRNA sequences has revealed that ARE-containing mRNAs cover a large repertoire of protein-encoding genes implicated in a variety of biological processes (24).
The stability of ARE-containing mRNAs is thought to be controlled by trans-acting proteins that bind to AREs. Several proteins, such as HuR, AUF-1, and tristetraprolin (TTP), have been shown to interact with AREs specifically, playing a positive or negative role in regulating mRNA stability (25–27). HuR, a ubiquitously expressed RNA-binding protein belonging to the Drosophila embryonic lethal abnormal vision (ELAV) family, selectively binds to AREs and stabilizes the ARE-containing transcripts (28). Numerous inflammation-related genes, such as TNF-α, COX-2, and inducible NO synthase, have been demonstrated to bind with HuR and maintain their mRNA expression levels (29–31). Reducing the expression of HuR attenuates the inflammatory response followed by myocardial infarction and left ventricular dysfunction in IL-10-null mice. It suggests that HuR is a direct target of IL-10; depletion of HuR expression mimics the anti-inflammatory effects of IL-10 (32). Moreover, LPS-enhanced Toll-like receptor 4 mRNA expression in vascular smooth muscle cells is mediated by HuR expression, which implies that suppressing HuR activation is a promising means to prevent vascular inflammation (33).
We report here that IL-1β enhances cPLA2α mRNA and protein expression in A549 cells by increasing cPLA2α mRNA stability. HuR has been identified as binding with cPLA2α mRNA 3′-UTR to prevent its degradation. Moreover, our study also demonstrates that the p38 MAPK signaling pathway involved in IL-1β-induced cPLA2α expression and that phosphorylation at Thr-118 of HuR plays a key role in mediating the p38 MAPK signaling pathway in inflammation.
EXPERIMENTAL PROCEDURES
Materials
Recombinant human IL-1β and SB203580 were purchased from Calbiochem. Antibodies against HuR, cPLA2α, and actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). α-Tubulin antibody and actinomycin D were purchased from Sigma. F-12K medium, fetal bovine serum (FBS), Opti-MEM medium, and Lipofectamine 2000 were purchased from Invitrogen. The Dual-Luciferase assay system was purchased from Promega (Madison, WI). pSilencerTM 3.1-H1 neo vector was purchased from Ambion (Austin, TX).
Cell Culture and IL-1β Treatment
A549 cells (human lung adenoma cell line) were grown in F-12K nutrient mixture (Invitrogen) supplemented with 10% (v/v) fetal bovine serum, 100 μg/ml streptomycin, and 100 units/ml penicillin. Cells were propagated using standard culture techniques and maintained in a humidified 37 °C, 5% CO2 environment. Regarding the procedure of IL-1β treatment, the A549 cells were cultured in serum-free medium for 18 h and then treated with 5 ng/ml IL-1β (Calbiochem).
Reverse Transcription and PCR
Total RNA was isolated using the TRIzol (Invitrogen), and 3 μg of RNA was subjected to reverse transcription and PCR. The primers used are listed as follow: cPLA2α, forward (5′-GACTCTAGTCCTCCGTTCAAG-3′) and reverse (5′CCAGTCTCTCATGATCAGTACG-3′); COX-2 forward (5′-CCCACTTCAAGGGATTTT-3′) and reverse (5′-CCAGACCAAAGACCTCCT-3′); β-actin forward (5′-CCCAAGGCCAACCGCGAGAAG-3′) and reverse (5′-TCTTCATTGTGCTGGGTGCCA-3′); and GAPDH forward (5′-CCCACTCCTCCACCTTTGAC-3′) and reverse (5′-TCTCTCTTCCTCTTGTGCTCTTG-3′). The PCR products were separated in 1% agarose-gel with ethidium bromide.
Plasmid Construction
A 599-bp fragment of human cPLA2α promoter region was amplified from human genomic DNA and cloned into luciferase plasmid pXP1 via the KpnI and HindIII restriction sites. This construct was named as the PLA599 plasmid. Full-length cPLA2α 3′-UTR was amplified using specific primers and cloned into plasmid pGL3 promoter vector (Promega) using the XbaI and FseI sites. Various regions of the cPLA2α 3′-UTR were amplified by PCR and cloned into pGEM-T easy vector (Promega) for in vitro transcription to synthesize probes used in RNA electrophoretic mobility shift assays (EMSAs) and biotin-labeled probe pulldown assays. The HuR expression vectors were obtained by constructing the HuR coding region into the HA-pCDNA3.1vector, and the HuR T118A mutant was created by using a site-directed mutagenesis kit (Stratagene) to change the amino acid from threonine to alanine.
mRNA Turnover Analysis
A549 cells were grown in serum-free medium for 18 h and then treated with or without 5 ng/ml IL-1β for 3 h followed by the addition of 5 μg/ml actinomycin D (Sigma). Total RNA was isolated from the cells at different time points after the addition of actinomycin D and analyzed by Northern blot. The quantitative graph was obtained from five independent experiments.
Northern Blot Analysis
25 μg of total RNA/lane was separated in 1.2% (w/v) agarose/formaldehyde gels and transferred onto HybondTM-N nylon membrane. Hybridization was performed in ExpressHybTM hybridization solution (BD Biosciences) at 68°C for 2 h. The cPLA2α and GAPDH probes were labeled using a Rediprime II random prime labeling system (Amersham Biosciences). The blots were exposed to x-ray film overnight at −80°C.
Preparation of Cytosolic Cell Lysates
A549 cells from 10-cm plastic dishes were washed twice with phosphate-buffered saline and lysed with cytosolic lysis buffer (10 mm HEPES (pH 8.0), 40 mm KCl, 3 mm MgCl2, 5% glycerol, 2 mm DTT, and 0.5% Nonidet P-40) supplemented with protease inhibitors (Roche Applied Science). The total cell lysate was centrifuged at high speed (6000 rpm for 5 min at 4 °C), and then the supernatant was collected as cytosolic cell lysate and stored at −80 °C.
RNA EMSA
Radiolabeled probes were prepared by using the Riboprobe system (Promega) as per the manufacturer's instructions. Labeled probes were purified using a MicroSpinTM G-25 column. 20 μg of cytosolic protein was incubated with labeled probes in an RNA EMSA buffer (50 mm KCl, 5% (v/v) glycerol, 1 mm MgCl2, 1 mm dithiothreitol, 10 mm Tris/HCl (pH 8.0), 0.25 mg/ml heparin, and 0.2 mg/ml yeast tRNA) for 30 min at room temperature followed by the addition of RNase T1 for 15 min. For the competition assay, different concentrations of RNA homopolymers (poly(rA), poly(rU), and poly(rC); Amersham Biosciences) were added to the reaction mixture. If a supershift assay was carried out, 2 μg of antibody was adding to the binding reaction mixture. The gels were prerun for 30 min at 180 volts, and then the sample was loaded and complexes were separated on a 6% native acrylamide gel (acrylamide/bisacrylamide ratio 37.5:1) in 1× Tris borate-EDTA buffer. Gels were dried under vacuum and exposed to x-ray film.
Biotin-labeled Probe Pulldown Assay
800 μg of cytosolic protein from A549 cells was incubated with biotin-labeled RNA probes overnight at 4 °C in 50 mm KCl, 5% (v/v) glycerol, 2 mm MgCl2, 1 mm dithiothreitol, and 10 mm HEPES (pH 8.0) supplemented with 0.5 mg/ml tRNA, 0.7 mg/ml heparin, 0.5 unit/μl RNase inhibitor, and 1× protease inhibitor. After that, streptavidin-conjugated agarose beads (Sigma) were added, and the mixture was incubated for 2 h at 4 °C. The beads were pulled down by centrifugation and washed three times with wash buffer (10 mm HEPES (pH 8.0), 50 mm KCl, 2 mm MgCl2, 5% glycerol, 1 mm DTT, and 0.1% Nonidet P-40) supplemented with protease inhibitors. The RNA-protein complexes were analyzed by SDS-PAGE, and the protein of interest was detected by specific antibody.
RNA Immunoprecipitation Assay
800 μg of cytosolic proteins from A549 cells was incubated with 5 μg of HuR antibody in 10 mm HEPES (pH 8.0), 3 mm MgCl2, 40 mm KCl, 5% glycerol, 2 mm DTT, 1× protease inhibitor, and 0.5 unit/μl RNase inhibitor for 2 h at room temperature. Agarose-protein A/G beads saturated with tRNA were added to the reaction, which was then further incubated for 2 h. Immunoprecipitated complexes were washed three times with the binding buffer plus 0.5% Nonidet P-40. Finally, the RNA bound by HuR was extracted by TRIzol reagent, and cPLA2α mRNA was detected by RT-PCR.
RNA Interference Assay
the short interfering RNA targeted against HuR was synthesized by Dharmacon. The sequence was the same as reported previously (34) (HuR siRNA: 5′-AAGAGGCAAUUACCAGUUUCAtt-3′). To deliver the siRNA, A549 cells were plated in 6-well plate, and 50 nm siRNA was transfected into the cells using Lipofectamine 2000 (Invitrogen).
RESULTS
IL-1β Signaling Increases mRNA Stability of the cPLA2α Gene
Regulation of gene expression by increasing the mRNA stability was documented in many inflammation-related genes, such as COX-2, TNF-α, and CCL2. We were interested in determining whether the post-transcriptional regulation mechanism is involved in cPLA2α gene expression. Treatment of the non-small cell lung cancer cell line A549 with IL-1β (5 ng/ml) caused a time-dependent induction of cPLA2α mRNA and protein. Around 12 h after IL-1β treatment, the expression of cPLA2α mRNA and protein reached its maximum level (Fig. 1, A and B). Then, the luciferase expression vector bearing 599 bp of cPLA2α promoter was constructed to study the transcriptional effect. To our surprise, we found that the relative induction fold of cPLA2α promoter activity under IL-1β treatment was only slightly increased (∼1.4-fold) (Fig. 1C).
FIGURE 1.
IL-1β induces the expression of cPLA2α gene expression by increasing cPLA2α mRNA stability. A549 cells were starved for 18 h in serum-free medium and then treated with 5 ng/ml IL-1β for the indicated time. A, total RNA was isolated, and the expression of cPLA2α mRNA was examined by RT-PCR. The expression level of GAPDH was used as an internal control. The result was quantified by densitometry as shown in the lower panel. B, Western blot analysis of cPLA2α protein was performed, and β-actin was used as a control. The result was also quantified by densitometry. C, A549 cells were co-transfected with 1 μg of luciferase plasmid bearing the cPLA2α gene promoter and 0.1 μg of Renilla reporter. Luciferase activity was measured and normalized with the Renilla activity. The increased fold of promoter activity was determined by dividing the reporter value of pPLA599 with PXP vectors. The results are the mean ± S.D. of three independent experiments, each measured in triplicate. Statistical significance (*, p < 0.05) was analyzed by Student's t test. D, A549 cells were treated with or without 5 ng/ml IL-1β for 3 h, and then actinomycin D was added to block the newly synthesized RNA. Total RNA was isolated at the indicated time and examined by Northern blot analysis using cPLA2α and GAPDH cDNA probes. The quantitative results were plotted as a percentage of total cPLA2α mRNA remaining at different time points compared with time point 0 h.
Compared with the induction level of cPLA2α mRNA under IL-1β treatment, the contribution of IL-1β-induced cPLA2α promoter activation seemed to be insufficient to induce the large amount of cPLA2α mRNA expression. Therefore, we inferred that the mRNA turnover regulatory mechanism might also participate in inducing the expression of the cPLA2α gene. The A549 cells were treated with actinomycin D (5 μg/ml) to block ongoing transcription and to detect the degradation rate of cPLA2α mRNA under normal or IL-1β treatment. As shown in Fig. 1D, the mRNA stability of cPLA2α mRNA was increased when cells were treated with IL-1β, which indicates that the post-transcriptional regulation mechanism plays an important role in mediating the cPLA2α mRNA expression.
cPLA2α mRNA 3′-UTR Binds with Cytosolic Protein Complexes
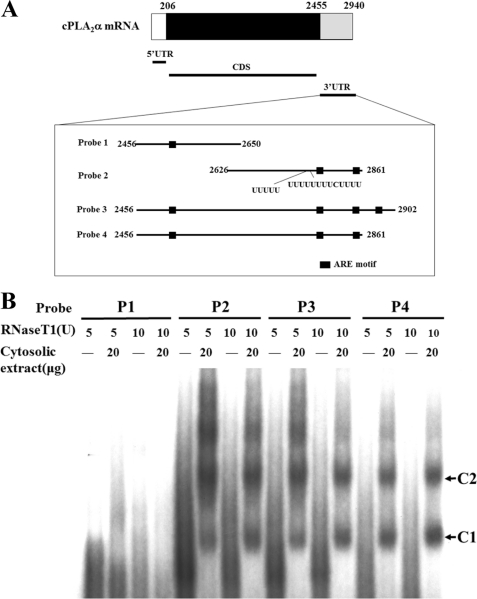
Because mRNA 3′-UTR participates in the regulation of mRNA stability, we tried to identify the cis- and trans- acting factors involved in cPLA2α mRNA expression. As the ARE sequence, which potentially mediates mRNA turnover regulation, has been identified in various genes (22), we also scanned the sequence of cPLA2α mRNA 3′-UTR and identified several AREs. Based on the location of AREs in the cPLA2α mRNA 3′-UTR, four different segments containing different lengths of cPLA2α 3′-UTR and AREs were constructed (Fig. 2A). To identify what kinds of RNA-binding proteins bound to these fragments, the in vitro transcribed RNAs were labeled with [α-32P]CTP to perform RNA EMSA experiments. As shown in Fig. 2B, probes P2–P4, but not P1, indicated that there were two kinds of protein-binding complexes. Although every fragment contained an ARE, the existence of AREs does not seem to be the only criterion for determining the region that interacts with protein complexes. Compared with the flanking sequence of ARE in probe P1 with other AREs, the ARE in P1 is flanked with a less U-rich sequence. Therefore, we propose that the protein complexes observed in the RNA EMSA may bind with the ARE flanked with a U-rich sequence on cPLA2α 3′-UTR.
FIGURE 2.
The 3′-UTR of cPLA2α mRNA binds with protein complexes. A, diagram of various probes used in the following RNA EMSA experiments. These probes span different regions of cPLA2α 3′-UTR, which contains different ARE and U-rich sequence. B, probes (P1–P4) were incubated with the cytosolic extracts prepared from A549 cells to perform an RNA EMSA under 5 units and 10 units of RNase T1 treatment. No protein-RNA complexes are formed on probe P1, but two protein complexes (C1 and C2) are indicated on probes P2–P4.
HuR Binds to the 3′-UTR of cPLA2α mRNA
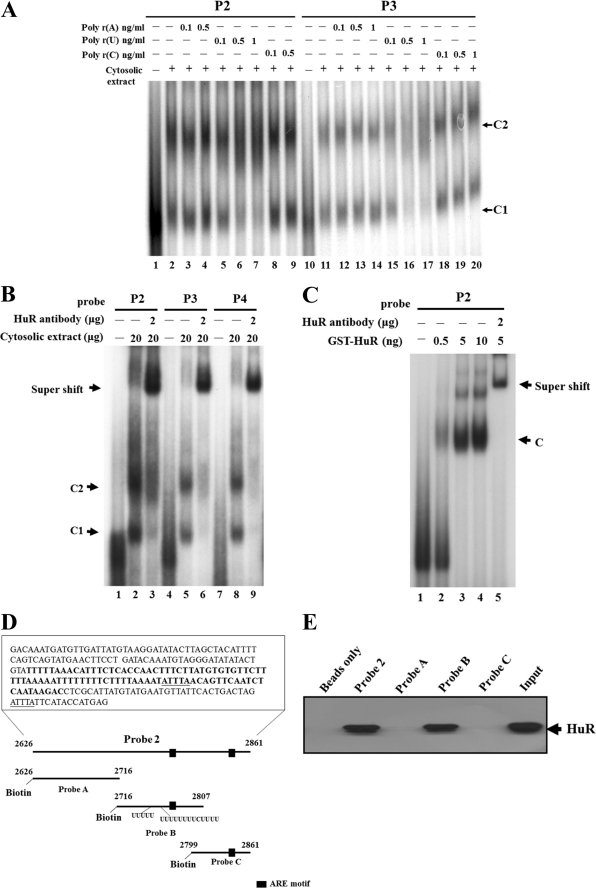
To further identify the kinds of proteins that exist in the protein-RNA complexes, the RNA homopolymers poly(rA), poly(rU), and poly(rC) were added to the mixture of RNA EMSA experiments to perform the competition assay (Fig. 3A). From the results, we found that RNA homopolymer(rA) and -(rC) did not have any effects on competing with the original binding, regardless of the increased concentration in probe P2 (Fig. 3A, lanes 3, 4, 8, and 9) or probe P3(lanes 12-14 and 18–20). However, RNA homopolymer(rU) could compete with the original binding complexes in a dose-dependent manner (lanes 5–7 and 15–17).This implies that the protein bound to the 3′-UTR of cPLA2α mRNA belongs to the U-rich sequence-binding proteins.
FIGURE 3.
HuR binds with cPLA2α mRNA 3′-UTR. A, the P1 and P2 probes were mixed with cytosolic extracts to perform a competitive RNA EMSA with different RNA homopolymers, poly(rA), poly(rU), and poly(rC), respectively. Only the poly(rU) homopolymer (lanes 5–7 and 15–17) can compete out the original binding complexes (C1 and C2). B, an antibody shift assay was performed to examine the composition of the protein complex. The shift-binding complex was found when antibody against HuR was added in the mixture (lanes 3, 6, and 9). C, GST-purified HuR protein was incubated with probe P2 to perform an RNA EMSA assay and antibody shift analysis. The binding complex was found when the probe was incubated with GST-HuR (lanes 2–4), and the addition of HuR antibody caused a supershift of the binding complex (lane 5). D, schematic diagram showing three divided segments of probe P2; these three segments were designated as probes A, B, and C. E, biotin-labeled probes were incubated with cytosolic extracts prepared from A549 cells and pulled down by streptavidin-agarose beads. The binding proteins were eluted and separated by SDS-PAGE, and the HuR antibody was used to do the Western blot analysis.
Among the RNA-binding proteins, HuR, a member of the Hu/ELAV family and a well known RNA-binding protein involved in regulating mRNA stability, prefers to bind to AREs and U-rich sequences (28). Therefore, we selected HuR as the candidate to demonstrate whether it would bind with the 3′-UTR of cPLA2α mRNA. As shown in Fig. 3B, probes P2–P4 were incubated with cytosolic lysates and 2 μg of HuR antibody to perform the antibody shift assay. With the addition of HuR antibody, we observed that a shift complex was formed in lanes 3, 6, and 9 (Fig. 3B), which indicated that HuR indeed bound to these probes. To further confirm this phenomenon, a GST-HuR fusion protein was produced to perform an additional RNA EMSA experiment. We found that the amount of binding complex was elevated as the amount of GST-HuR fusion protein was increased (Fig. 3C, compare lanes 2–4). In addition, the lysate mixed with the HuR antibody caused a supershift band from the original binding complex (Fig. 3C, lane 5). Based on these observations, we concluded that HuR binds to the 3′-UTR of cPLA2α mRNA.
According to the previous experiment, we found that HuR could bind to the 2626–2861-bp region (probe P2) on cPLA2α 3′-UTR mRNA. Next, we wanted to further identify the exact binding region between HuR and cPLA2α 3′-UTR mRNA. Three different fragments that spanned probe P2 were constructed. The RNA probes, designated as probes A, B, and C, were synthesized and labeled with biotin-CTP (Fig. 3D). These probes were incubated with cytosolic proteins to perform a biotin-pulldown assay combined with Western blot analysis. Only probe B was shown to associate with HuR protein (Fig. 3E). Probe P2, which bound HuR in an RNA EMSA experiment, is a positive control. In comparison with the sequences of probes A–C, the ARE in probe B flanks a U-rich sequence (Fig. 3D). This result is consistent with the competition assay showing that the protein complex binds with a U-rich sequence and also proves that HuR is the major protein of this complex.
IL-1β Signaling Increases Binding between HuR and the 3′-UTR of cPLA2α mRNA
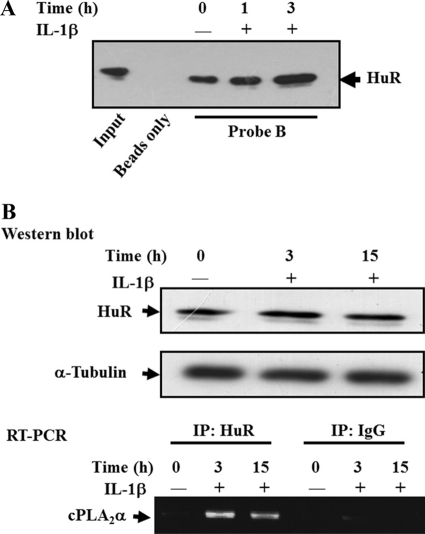
Because IL-1β signaling increases cPLA2α mRNA stability, we thought it would be interesting to study whether HuR contributes to this effect. Therefore, the A549 cells were treated with IL-1β (5 ng/ml) for 1 and 3 h, and the cytosolic proteins were collected and incubated with probe B to perform a biotin-pulldown assay. We found that binding between HuR and cPLA2α was increased under treatment with IL-1β for 1 and 3 h (Fig. 4A). To further demonstrate whether this phenomenon existed in vivo, the specific HuR antibody was used to immunoprecipitate the HuR-mRNA complex in cells with or without IL-1β treatment. The mRNAs were extracted from the complex to examine cPLA2α mRNA expression by RT-PCR. As shown in Fig. 4B, we found that the in vivo association between HuR and cPLA2α mRNA was increased under IL-1β treatment; and the Western blot shows that the protein used in RNA immunoprecipitation was equal. This result indicates that HuR may stabilize the cPLA2α mRNA under IL-1β treatment.
FIGURE 4.
IL-1β signaling increases the binding between HuR and the 3′-UTR of cPLA2α mRNA. A, cytosolic extracts from A549 cells treated with IL-1β for different time points were incubated with probe B, and the biotin-labeled probes were pulled down by streptavidin-agarose beads. The RNA-protein complex was subjected to Western blot and detected by HuR antibody. B, A549 cells were starved for 18 h in serum-free medium and then treated with 5 ng/ml IL-1β for 3 and 15 h. Cytosolic extract was incubated with HuR antibody, and the reaction mixture was pulled down by agarose-protein A/G. The RNAs associated with HuR were extracted by TRIzol, and the expression level of cPLA2α mRNA was further analyzed by RT-PCR. Western blot analysis showed that equal proteins were used in the RNA immunoprecipitation (IP) assay.
Reduction of HuR Protein Expression Decreases cPLA2α Gene Expression
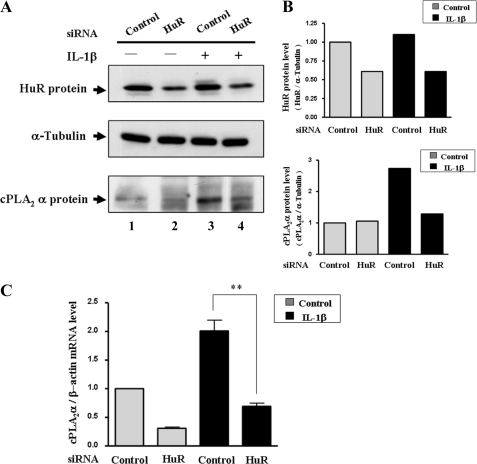
To clarify the functional role of HuR in regulating the expression of cPLA2α mRNA, RNA interference technology, which reduces HuR expression, was used. siRNA against HuR was transfected into the A549 cells with or without IL-1β treatment. From the results shown in Fig. 5A, we see that HuR can be down-regulated effectively around 50% (compare lanes 1 and 3 with 2 and 4). Under normal conditions, IL-1β can induce the expression of cPLA2α protein (Fig. 5A, compare lanes 1 and 3), similar to the previous results (Fig. 1A). However, the induction of cPLA2α protein by IL-1β was inhibited when the HuR expression level was reduced by siRNA technology (Fig. 5A, compare lanes 4 and 3). The expression of cPLA2α mRNA was also decreased under the reduction of HuR expression (Fig. 5C). This phenomenon indicates that IL-1β-induced expression of the cPLA2α gene is regulated by HuR through binding with cPLA2α mRNA to prevent its mRNA degradation.
FIGURE 5.
Reduction of HuR expression inhibits IL-1β-induced cPLA2α mRNA and protein expression. Control siRNA and HuR siRNA were transfected into A549 cells by Lipofectamine 2000. After transfection for 24 h, cells were cultured in serum-free medium for 18 h and then treated with or without IL-1β (5 ng/ml) for 3 h. A, total protein was extracted for analysis of the expression of HuR, tubulin, and cPLA2α. B, the expression level of HuR and cPLA2α protein was quantified by densitometry. C, the cPLA2α mRNA expression level was measure and quantified by quantitative RT-PCR (**, p < 0.01).
p38 MAPK Pathway Regulates cPLA2α mRNA Stability through HuR Phosphorylation
It has been reported that p38 MAPK plays an important role in mediating cytokine-induced mRNA stabilization. Thus, we speculated that IL-1β-stimulated cPLA2α mRNA turnover may act via the p38 MAPK pathway. To test this hypothesis, the p38 MAPK inhibitor SB203580 (20 μm) was used to block the pathway, and the effect on IL-1β-stimulated cPLA2α mRNA expression was monitored. As shown in Fig. 6A, p38 was activated by IL-1β treatment in A549 cells. Blockade of the p38 pathway by SB203580 inhibited the effect of IL-1β-induced cPLA2α mRNA expression (Fig. 6A, compare lane 4 with lanes 2 and 3). This result indicates that the p38 MAPK pathway plays a role in regulating cPLA2α mRNA turnover. Furthermore, we wanted to clarify the underlying mechanism of the p38 MAPK pathway on cPLA2α mRNA stability. HuR phosphorylation has been reported to regulate the formation of HuR-mRNA complexes (35, 36), and p38 MAPK has been reported to phosphorylate on HuR Thr-118 to mediate p21 mRNA stabilization under γ-radiation treatment (37). Therefore, we examined whether HuR Thr-118 is also critical for IL-1β-induced association between HuR and cPLA2α mRNA. The A549 cells were transfected with HA-HuR(wt) and phosphorylated mutant HA-HuR(T118A) and then treated with or without IL-1β. By immunoprecipitating these two forms of HuR-mRNA complex with HA antibody followed by RT-PCR, we found that IL-1β treatment increased the association of WT HA-HuR but not of the T118A mutant with cPLA2α mRNA (Fig. 6B). The result was also quantified by quantitative PCR (Fig. 6B, lower panel). This result implies that phosphorylation on Thr-118 by p38 MAPK regulates the binding of HuR to cPLA2α mRNA.
FIGURE 6.
The phosphorylation of HuR Thr-118 regulates the interaction between HuR and targeted mRNAs. A, A549 cells were pretreated with SB203580 for 30 min, and then IL-1β was added for 20 and 30 min. A Western blot assay of p38 phosphorylation status was used to verify the efficacy of SB203580. The RT-PCR analysis showed that pretreatment with SB203580 blocked the induction of cPLA2α mRNA by IL-1β treatment. B, A549 cells were transfected with plasmids encoding HA-tagged HuR and Thr-118 mutant (T118A), respectively. After transfection for 6 h, the Opti-MEM medium was replaced with serum-free medium for 18 h, and then the cells were treated with IL-1β for 3 h. Cytosolic extracts were then immunoprecipitated with HA antibody, RNA was purified from the pulled down complex, and the expression level of cPLA2α mRNA was determined by RT-PCR. The quantitative result of the RT-PCR is shown in the lower panel. C, the protein expression level of HA-HuR and Thr-118A mutant is shown to indicate the same amount of expressed proteins were used in Fig. 6B. D, an RNA immunoprecipitation (IP) assay was performed, and the RNAs extracted from the immunoprecipitation complexes by HuR antibody were analyzed for the expression of cPLA2α mRNA and COX-2 mRNA. Transfections of the A549 cells with plasmids encoding HA-tagged HuR and HuR Thr-118A mutant were performed as mentioned above. The RNAs immunoprecipitated by HA antibody were extracted to monitor COX-2 mRNA expression by RT-PCR. The quantitative result of the RT-PCR is shown in the lower panel (*, p < 0.05).
COX-2 is an inflammatory factor that also is reportedly regulated by HuR (38). We sought to test whether the phosphorylation on Thr-118 also regulates the association between HuR and COX-2 mRNA. First, the HuR complex was pulled down, and the mRNAs were extracted to determine the expression of COX-2 after IL-1β treatment. As shown in Fig. 6D, the increased interaction between COX-2 mRNA and HuR was found to be similar to that with cPLA2 mRNA. Also, the HuR T118A mutant reduced this effect (Fig. 6D, lower panel). Overall, these results indicate that HuR Thr-118 phosphorylation by p38 MAPK is very important for maintaining the mRNA stability of an inflammation-related gene under IL-1β treatment.
DISCUSSION
Inflammation is a complicated innate immune response that allows multicellular organisms to repair tissue damage. Many genes need to be expressed sequentially to produce an optimal response to the inciting events. Transcriptional regulation of inflammation-related genes has attracted much attention in this field. However, recent research has highlighted the importance of post-transcriptional regulation in inflammation (39, 40). Hao and Baltimore (40) report that the genes activated by tumor necrosis factor can be categorized into three groups with different induction kinetics. The differences in mRNA stability among these groups play an important role in influencing the temporal order of gene expression. Here, we found that the cPLA2α, a key player in the biosynthesis of eicosanoids that initiates inflammation, is also controlled through post-transcriptional regulation mechanism to modulate gene expression in A549 cells (Fig. 1). This phenomenon can also be found in HeLa cells (supplemental Fig. 1). The RNA-binding protein HuR was demonstrated to interact with the cPLA2α mRNA 3′-UTR to increase mRNA stability in vitro and in vivo (Figs. 3 and 6B and supplemental Fig. 3). Under IL-1β treatment, the association between HuR and cPLA2α mRNA was increased to prevent mRNA degradation (Figs. 4 and 5 and supplemental Fig. 2). These results provide an answer to the question of what kinds of mechanisms regulate cPLA2α mRNA half-life under treatment with EGF, PDGF, serum and phorbol 12-myristate 13-acetate (15). The important role of post-transcriptional regulation in inflammation cannot be overemphasized.
In inflammation, the p38 MAPK signaling pathway is well recognized as participating in post-transcriptional regulation. Activation of the p38 MAPK pathway has been implicated in stabilizing many ARE mRNAs such as COX-2, TNF-α, IL-6, macrophage inhibitory protein 1 (MIP 1), IL-8, and VEGF (41). Therefore, we were interested in identifying whether the phosphorylation of HuR by the p38 MAPK signaling pathway contributes to the enhancement of cPLA2α mRNA stability induced by IL-1β. By pretreating the cells with the p38 MAPK inhibitor SB203580, the increase of cPLA2α mRNA was inhibited under IL-1β treatment (Fig. 6A). This indicates that p38 MAPK is involved in the post-transcriptional regulation of cPLA2α mRNA expression.
The RNA-destabilizing factor TTP and the stabilizing factor HuR were shown to play important roles in regulating mRNA stability mediated by the p38 MAPK signaling pathway. Phosphorylation of TTP by the p38 MAPK downstream target MK2 released ARE-containing mRNAs from translational repression and inhibited the mRNA destabilizing activity of TTP (42). However, the detailed mechanism of how p38 MAPK regulates HuR to enhance the mRNA stability is still unknown. Recently, it has been indicated that post-translational modification of HuR could impair its subcellular localization and affect the mRNA stabilization outcome (43). For example, the cell cycle checkpoint kinase Chk2 phosphorylates HuR at residues Ser-88, Ser-100, and Thr-118 to regulate SIRT1 mRNA expression (35). Under angiotensin II treatment, PKCδ induces phosphorylation of HuR at Ser-221 and Ser-318, thereby promoting an increase in COX-2-derived PGE2 formation (44). Moreover, PKCα is also reported to phosphorylate HuR at Ser-158 and Ser-221 to enhance COX-2 mRNA stability after ATP stimulation (36). It seems that HuR can be phosphorylated on different sites by different signaling pathways in response to different stimuli. From a previous study, p38 MAPK has been demonstrated to phosphorylate HuR on Thr-118 to stabilize p21Cip1 mRNA under γ-radiation treatment (37). We wanted to study whether this modification was also functional in inflammation. Mutation at residue Thr-118 of HuR abolished the increased binding of HuR with cPLA2α mRNA after IL-1β stimulation (Fig. 6B). From the analysis of HuR protein expression in cytoplasm, the HuR T118A mutant slightly impaired the translocation ability of HuR under IL-1β treatment (Fig. 6C). Therefore, we infer that modification of the HuR Thr-118 phosphorylation site not only impairs the behavior of HuR translocation but is also responsible for the interaction between HuR and cPLA2α mRNA.
Recent findings demonstrate that multiple mRNAs are co-regulated by one or more sequence-specific RNA-binding proteins that orchestrate their splicing, export, stability, localization, and translation. These observations have given rise to the RNA regulon model in which genes encoding functionally related proteins are regulated in a coordinated manner by the association of specific subsets of mRNAs with RNA-binding proteins (45, 46). COX-2 is a key enzyme that mediates the inflammation response initiated by cPLA2α, and the increased expression of cPLA2α and COX-2 has been found to promote angiogenesis in several cancers (47). The involvement of HuR and p38 MAPK in maintaining COX-2 mRNA stability was reported under UVB and anticancer drug treatment (48, 49). From our results, COX-2 mRNA associated with HuR was increased under IL-1β treatment, and the HuR T118A mutant abolished this effect as observed in cPLA2α mRNA (Fig. 6D). This result highlights the concept of an RNA regulon in which phosphorylation on HuR Thr-118 may regulate a set of inflammation-related, ARE-containing mRNAs to fulfill their physiological functions in inflammation.
Overall, we have demonstrated that HuR and the p38 MAPK signaling pathway participate in the regulation of cPLA2α mRNA expression. Thus the phosphorylation of HuR Thr-118 is critical for HuR to interact with ARE mRNAs to conduct its functional role in inflammation.
Supplementary Material
This work was supported by National Science Council (Taiwan) Grants NSC 97-2320-B-006-026-MY3 and NSC 99-2923-B-006-002-MY3.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- cPLA2α
- cytosolic phospholipase A2α
- ARE
- AU-rich element
- TTP
- tristetraprolin.
REFERENCES
- 1. Leslie C. C. (2004) Prostaglandins Leukot. Essent. Fatty Acids 70, 373–376 [DOI] [PubMed] [Google Scholar]
- 2. Linkous A., Yazlovitskaya E. (2010) Cell. Microbiol. 12, 1369–1377 [DOI] [PubMed] [Google Scholar]
- 3. Huang W., Bhavsar A., Ward R. E., Hall J. C., Priestley J. V., Michael-Titus A. T. (2009) J. Neurotrauma 26, 1429–1434 [DOI] [PubMed] [Google Scholar]
- 4. Nagase T., Uozumi N., Ishii S., Kume K., Izumi T., Ouchi Y., Shimizu T. (2000) Nat. Immunol. 1, 42–46 [DOI] [PubMed] [Google Scholar]
- 5. Myou S., Sano H., Fujimura M., Zhu X., Kurashima K., Kita T., Nakao S., Nonomura A., Shioya T., Kim K. P., Munoz N. M., Cho W., Leff A. R. (2001) Nat. Immunol. 2, 145–149 [DOI] [PubMed] [Google Scholar]
- 6. Hegen M., Sun L., Uozumi N., Kume K., Goad M. E., Nickerson-Nutter C. L., Shimizu T., Clark J. D. (2003) J. Exp. Med. 197, 1297–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weiser-Evans M. C., Wang X. Q., Amin J., Van Putten V., Choudhary R., Winn R. A., Scheinman R., Simpson P., Geraci M. W., Nemenoff R. A. (2009) Cancer Res. 69, 1733–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hong K. H., Bonventre J. C., O'Leary E., Bonventre J. V., Lander E. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3935–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirabayashi T., Murayama T., Shimizu T. (2004) Biol. Pharm. Bull. 27, 1168–1173 [DOI] [PubMed] [Google Scholar]
- 10. Kudo I., Murakami M. (2002) Prostaglandins Other Lipid Mediat. 68–69, 3–58 [DOI] [PubMed] [Google Scholar]
- 11. Ghosh M., Tucker D. E., Burchett S. A., Leslie C. C. (2006) Prog. Lipid Res. 45, 487–510 [DOI] [PubMed] [Google Scholar]
- 12. Blaine S. A., Wick M., Dessev C., Nemenoff R. A. (2001) J. Biol. Chem. 276, 42737–42743 [DOI] [PubMed] [Google Scholar]
- 13. Dronadula N., Liu Z., Wang C., Cao H., Rao G. N. (2005) J. Biol. Chem. 280, 3112–3120 [DOI] [PubMed] [Google Scholar]
- 14. Cowan M. J., Yao X. L., Pawliczak R., Huang X., Logun C., Madara P., Alsaaty S., Wu T., Shelhamer J. H. (2004) Biochim. Biophys. Acta 1680, 145–157 [DOI] [PubMed] [Google Scholar]
- 15. Tay A., Maxwell P., Li Z. G., Goldberg H., Skorecki K. (1994) Biochem. J. 304, 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ing N. H. (2005) Biol. Reprod. 72, 1290–1296 [DOI] [PubMed] [Google Scholar]
- 17. Keene J. D., Tenenbaum S. A. (2002) Mol. Cell 9, 1161–1167 [DOI] [PubMed] [Google Scholar]
- 18. Wilusz C. J., Wilusz J. (2004) Trends Genet. 20, 491–497 [DOI] [PubMed] [Google Scholar]
- 19. van Hoof A., Parker R. (2002) Curr. Biol. 12, R285–287 [DOI] [PubMed] [Google Scholar]
- 20. Hollams E. M., Giles K. M., Thomson A. M., Leedman P. J. (2002) Neurochem. Res. 27, 957–980 [DOI] [PubMed] [Google Scholar]
- 21. Audic Y., Hartley R. S. (2004) Biol. Cell 96, 479–498 [DOI] [PubMed] [Google Scholar]
- 22. Chen C. Y., Shyu A. B. (1995) Trends Biochem. Sci. 20, 465–470 [DOI] [PubMed] [Google Scholar]
- 23. Kracht M., Saklatvala J. (2002) Cytokine 20, 91–106 [DOI] [PubMed] [Google Scholar]
- 24. Bakheet T., Williams B. R., Khabar K. S. (2006) Nucleic Acids Res. 34, D111–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma W. J., Cheng S., Campbell C., Wright A., Furneaux H. (1996) J. Biol. Chem. 271, 8144–8151 [DOI] [PubMed] [Google Scholar]
- 26. Brewer G. (1991) Mol. Cell. Biol. 11, 2460–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carballo E., Lai W. S., Blackshear P. J. (1998) Science 281, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 28. Brennan C. M., Steitz J. A. (2001) Cell. Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rajasingh J., Bord E., Luedemann C., Asai J., Hamada H., Thorne T., Qin G., Goukassian D., Zhu Y., Losordo D. W., Kishore R. (2006) FASEB J. 20, 2112–2114 [DOI] [PubMed] [Google Scholar]
- 30. Sureban S. M., Murmu N., Rodriguez P., May R., Maheshwari R., Dieckgraefe B. K., Houchen C. W., Anant S. (2007) Gastroenterology 132, 1055–1065 [DOI] [PubMed] [Google Scholar]
- 31. Linker K., Pautz A., Fechir M., Hubrich T., Greeve J., Kleinert H. (2005) Nucleic Acids Res. 33, 4813–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krishnamurthy P., Lambers E., Verma S., Thorne T., Qin G., Losordo D. W., Kishore R. (2010) FASEB J. 24, 2484–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin F. Y., Chen Y. H., Lin Y. W., Tsai J. S., Chen J. W., Wang H. J., Chen Y. L., Li C. Y., Lin S. J. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 2622–2629 [DOI] [PubMed] [Google Scholar]
- 34. Yeh C. H., Hung L. Y., Hsu C., Le S. Y., Lee P. T., Liao W. L., Lin Y. T., Chang W. C., Tseng J. T. (2008) Mol. Biol. Cell 19, 3812–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abdelmohsen K., Pullmann R., Jr., Lal A., Kim H. H., Galban S., Yang X., Blethrow J. D., Walker M., Shubert J., Gillespie D. A., Furneaux H., Gorospe M. (2007) Mol. Cell 25, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Doller A., Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. (2007) Mol. Biol. Cell 18, 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lafarga V., Cuadrado A., Lopez de Silanes I., Bengoechea R., Fernandez-Capetillo O., Nebreda A. R. (2009) Mol. Cell. Biol. 29, 4341–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sengupta S., Jang B. C., Wu M. T., Paik J. H., Furneaux H., Hla T. (2003) J. Biol. Chem. 278, 25227–25233 [DOI] [PubMed] [Google Scholar]
- 39. Anderson P. (2010) Nat. Rev. Immunol. 10, 24–35 [DOI] [PubMed] [Google Scholar]
- 40. Hao S., Baltimore D. (2009) Nat. Immunol. 10, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khabar K. S. (2010) Cell. Mol. Life Sci. 67, 2937–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ronkina N., Menon M. B., Schwermann J., Tiedje C., Hitti E., Kotlyarov A., Gaestel M. (2010) Biochem. Pharmacol. 80, 1915–1920 [DOI] [PubMed] [Google Scholar]
- 43. Doller A., Pfeilschifter J., Eberhardt W. (2008) Cell. Signal. 20, 2165–2173 [DOI] [PubMed] [Google Scholar]
- 44. Doller A., Akool el-S., Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. (2008) Mol. Cell. Biol. 28, 2608–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keene J. D. (2007) Nat. Rev. Genet. 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 46. Morris A. R., Mukherjee N., Keene J. D. (2010) Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 162–180 [DOI] [PubMed] [Google Scholar]
- 47. Nakanishi M., Rosenberg D. W. (2006) Biochim. Biophys. Acta 1761, 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fernau N. S., Fugmann D., Leyendecker M., Reimann K., Grether-Beck S., Galban S., Ale-Agha N., Krutmann J., Klotz L. O. (2010) J. Biol. Chem. 285, 3896–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Subbaramaiah K., Marmo T. P., Dixon D. A., Dannenberg A. J. (2003) J. Biol. Chem. 278, 37637–37647 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.