Abstract
Trehalose synthase (TreS) catalyzes the reversible interconversion of maltose and trehalose and has been shown recently to function primarily in the mobilization of trehalose as a glycogen precursor. Consequently, the mechanism of this intriguing isomerase is of both academic and potential pharmacological interest. TreS catalyzes the hydrolytic cleavage of α-aryl glucosides as well as α-glucosyl fluoride, thereby allowing facile, continuous assays. Reaction of TreS with 5-fluoroglycosyl fluorides results in the trapping of a covalent glycosyl-enzyme intermediate consistent with TreS being a member of the retaining glycoside hydrolase family 13 enzyme family, thus likely following a two-step, double displacement mechanism. This trapped intermediate was subjected to protease digestion followed by LC-MS/MS analysis, and Asp230 was thereby identified as the catalytic nucleophile. The isomerization reaction was shown to be an intramolecular process by demonstration of the inability of TreS to incorporate isotope-labeled exogenous glucose into maltose or trehalose consistent with previous studies on other TreS enzymes. The absence of a secondary deuterium kinetic isotope effect and the general independence of kcat upon leaving group ability both point to a rate-determining conformational change, likely the opening and closing of the enzyme active site.
Keywords: Carbohydrate Metabolism, Enzyme Inactivation, Enzyme Mechanisms, Glycosidases, Mycobacterium tuberculosis, Intermediate Trapping, Kinetic Isotope Effects, Linear Free Energy, Sugar Isomerases
Introduction
Trehalose is a non-reducing disaccharide in which two glucose molecules are linked via an α,α-(1,1) glycosidic bond. This disaccharide has been found in a wide spectrum of living organisms ranging from mycobacteria, bacteria, and fungi to higher orders of the plant kingdom and lower orders of the animal kingdom (1). The amount of trehalose found in some of these organisms can be very significant, implying important biological roles for this simple disaccharide with the early view being that trehalose served as the storehouse of glucose similar to that of starch or glycogen (1–3). Although this is still the case in some organisms (3, 4), studies in the past three decades have revealed many other important biological functions for trehalose. For example, trehalose has been convincingly shown in many cases to stabilize membranes and cellular proteins against various environmental stresses such as dehydration, oxygen radicals, heat, and cold (1, 5) because high concentrations are found in a number of microorganisms after exposure to heat shock, oxygen stress, or lowered temperature (6–8). Besides playing a role as a protectant, trehalose is also an important component of the cell walls of many mycobacteria and corynebacteria in the form of glycolipids. The most well studied example is trehalose dimycolate (or cord factor), which consists of a trehalose core with the unusual fatty acid mycolic acid esterified at the 6-OH and 6′-OH positions (9). The cord factor is the most toxic lipid produced by Mycobacterium tuberculosis and significantly increases the impermeability of the cell wall to various antibiotics and thus was identified as a virulence factor (10, 11).
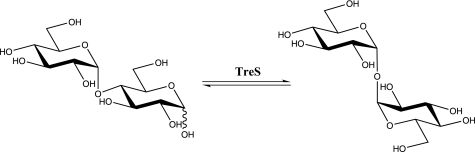
The important biological functions of trehalose combined with the fact that humans do not biosynthesize this sugar initially raised interest in its biosynthetic enzymes as targets for the development of novel antituberculosis drugs (12). However, the realization that there are three biosynthetic pathways for trehalose in M. tuberculosis has tempered enthusiasm for this approach (13). The major biosynthetic pathway involves a two-step enzymatic process. The first step is a glucosyl transfer onto glucose 6-phosphate catalyzed by trehalose-6-phosphate synthase (or OtsA in Escherichia coli) to produce trehalose 6-phosphate (14). This is followed by dephosphorylation to generate the free trehalose catalyzed by trehalose-6-phosphate phosphatase (or OtsB in E. coli.) (12). The second pathway (TreY-TreZ) utilizes glycogen or maltooligosaccharides as the starting material. The maltosyl moiety at the reducing end is first isomerized into a trehalosyl moiety by maltooligosyltrehalose synthase (TreY). Hydrolytic release of the trehalose is then catalyzed by a second enzyme, maltooligosyltrehalose trehalohydrolase (TreZ) (15–17). Interestingly, the combined use of the TreY-TreZ enzymes has allowed facile, inexpensive industrial production of trehalose (5, 18). The third biosynthetic pathway utilizes only one enzyme, trehalose synthase (TreS),3 which catalyzes the reversible interconversion of maltose and trehalose (Fig. 1) with an equilibrium lying in favor of trehalose by a factor of 4.1:1 (19). Importantly, the equilibria of the first two processes are driven in the synthetic direction by coupled hydrolytic processes, whereas that for TreS relies upon inherent thermodynamics and can be more readily displaced in the opposite direction.
FIGURE 1.
Reversible transformation of maltose and trehalose catalyzed by TreS.
TreS activity was first demonstrated in Pimelobacter sp. R48 and Pseudomonas putida through an extensive screening of 2,500 strains of soil bacteria (20). Subsequently, TreS has been purified and characterized from many other microorganisms such as Thermus aquaticus (21), Thermus caldophilus (22), Thermus thermophilus (23), Picrophilus torridus (24), and Mycobacterium smegmatis (13). The interest in TreS initially stemmed from its promise as a low cost way to produce trehalose, which has found a range of applications in the food, cosmetics, and pharmaceutical industries. This was complemented by interest in the mycobacterial enzyme as an antibiotic target until the two other pathways were uncovered. However, very recently, a new α-glucan biosynthetic pathway was discovered in M. tuberculosis in which trehalose is converted into branched α-1,4-glucan by the sequential action of four enzymes: TreS, Pep2 (maltokinase), GlgE (maltosyltransferase), and GlgB (branching enzyme) (25). Inactivation of GlgE has been demonstrated to cause rapid death of M. tuberculosis through a self-poisoning accumulation of maltose 1-phosphate. Inhibitors of GlgE therefore hold promise as novel antituberculosis drugs. However, uptake of maltose by M. tuberculosis is very poor relative to trehalose (26); thus, the development of maltose analogues that can be converted to potent GlgE inhibitors by TreS and Pep2 would represent an ideal approach. Mechanistic characterization of TreS including its substrate specificity would therefore be a valuable step toward implementing this strategy.
TreS is a retaining α-transglycosidase in the α-amylase family (GH13) (27). Sequence alignment of several TreS enzymes with other GH13 glycosidases has already identified several conserved regions (21, 24, 28–30). Consequently, a classical double displacement mechanism for the interconversion of maltose and trehalose (31, 32) seems probable but remains unproven. Likewise, the nature of the aglycone rearrangement process is unclear with some studies suggesting an intramolecular process within the confines of the active site (33, 34) and others suggesting release of the glucose into the medium and rebinding (13). In the present study, we designed and synthesized a series of compounds and evaluated these as substrates or inhibitors with TreS from M. smegmatis. A convenient spectrophotometric assay was developed and used to determine the pH dependence of the enzyme. 5-Fluoroglycosyl fluorides were utilized to trap the covalent glycosyl-enzyme intermediate, and the catalytic nucleophile was identified through LC-MS/MS analysis of peptic digests. Isotope exchange studies were used to demonstrate an intramolecular mechanism within an enclosed active site, and further kinetic studies suggested that protein conformational changes are rate-limiting.
EXPERIMENTAL PROCEDURES
General Methods
Recombinant wild type TreS enzyme was produced and purified according to literature procedures (13). An extinction coefficient of 1.2 × 105 m−1 cm−1 at 280 nm (obtained by entering the primary sequence of TreS into the ExPASy website using ProtParam software) was used to determine enzyme concentrations. All α-aryl glucosides (including 2,4-dinitrophenyl 1-{2H}-α-glucoside), α-glucosyl fluoride (αGlcF), 5-fluoro α-d-glucosyl fluoride (5FGlcF), and 5-fluoro-β-l-idosyl fluoride (5FIdoF) were synthesized as described previously (35–38). Deoxynojirimycin (DNJ) was purchased from Carbosynth, Berkshire, UK. Casuarine was a generous gift from Prof. Andrea Goti from the Department of Organic Chemistry, University of Florence, Florence, Italy. Isofagomine and xylo-DNJ were provided by Tara Hill and Dr. Ethan D. Goddard-Borger, respectively, from the Department of Chemistry, University of British Columbia.
Kinetic Evaluation of Aryl α-Glucosides as Substrates
All kinetic studies were carried out at 37 °C in 40 mm potassium phosphate buffer, pH 6.8 unless otherwise noted. Michaelis-Menten kinetic parameters for aryl α-glucosides were measured by monitoring the increase of absorbance at 400 nm using a Varian CARY 300 spectrophotometer equipped with a circulating water bath. Quartz cuvettes (200 μl) with a path length of 1 cm were used. An approximate Km value for each substrate was first determined by measuring initial rates at three widely different concentrations of the substrate. The accurate values of kcat and Km were then determined by using six to eight different substrate concentrations ranging from 0.3 × Km to 5 × Km (depending on the availability or solubility of the substrate). All enzyme kinetic data were processed using the program GraFit 5.0.13 (Erithacus Software Limited, 2006). The extinction coefficients for phenols and corresponding aryl glycosides were determined by measuring the absorbances of carefully prepared stock solutions of each compound in the enzyme buffer at 37 °C.
Kinetic Isotope Effect Measurements for 2,4-Dinitrophenyl 1-{2H}-α-Glucoside (d-DNPGlc)
For (kcat)H/(kcat)D measurements, either DNPGlc or d-DNPGlc was added to the TreS reaction mixture in 200-μl quartz cuvettes to a final concentration of 30 mm (∼10 × Km), and their initial rates were measured alternately seven times over. The corresponding kcat values were calculated by dividing initial rates by the enzyme concentrations. For (kcat/Km)H/(kcat/Km)D measurements, the substrate depletion method was used in which the complete hydrolysis of a low substrate concentration (100 μm DNPGlc or d-DNPGlc; ∼0.03 × Km) by TreS was monitored. The increase of absorbance at 400 nm was monitored until the reactions were complete (within 25 min in this study), and the kcat/Km value was calculated by dividing the pseudo first-order rate constant derived by curve fitting, by the enzyme concentration. Each substrate pair (protio and deuterio) was studied six times in alternation, and the (kcat/Km)H/(kcat/Km)D value was calculated by dividing the first-order rate constant for the protio substrate by the first-order rate constant for the deuterio substrate in each case.
αGlcF Kinetics
Reaction rates were determined by monitoring the release of fluoride using a fluoride ion electrode. Glass vials containing αGlcF (0.1–1 mm) were incubated at 37 °C and monitored to establish a steady-state spontaneous hydrolysis rate before 10 μl of the stock solution of TreS enzyme (0.47 μm) was added to bring the final volume to 300 μl. The initial rates determined (corrected for the spontaneous hydrolysis rate) were then analyzed using the GraFit program in the normal manner.
pH Profile Studies
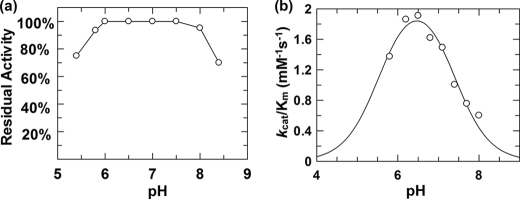
The stability of TreS at a range of pH values was first studied by incubating TreS for 1 h in buffers of different pH values ranging from 5.4 to 8.4. Aliquots of the mixtures were then assayed using 2 mm DNPGlc in standard assay buffer (40 mm potassium phosphate, pH 6.8), which was preincubated at 37 °C. The following buffers were used: 40 mm citrate-phosphate, pH 5.4–5.8; 40 mm sodium phosphate, pH 5.8–8.0; and 40 mm sodium borate, pH 8.0–8.4. This study revealed that TreS was stable from pH 5.8 to 8.0 for 1 h, and this range was selected for pH profile studies.
The pH dependence of kcat/Km was measured using the substrate depletion method at a low concentration (50 μm) of DNPGlc ([S] ≪ Km). TreS was added to the preincubated substrate solution at 37 °C (final enzyme concentration, 0.7 μm), and the increase of absorbance at 400 nm was followed for 60 min until the reaction was complete. The data were fitted to a first-order curve using GraFit 5.0 software to yield pseudo first-order rate constants. The kcat/Km values were obtained from the values by dividing by the enzyme concentration.
Fluorosugar Inactivation Studies
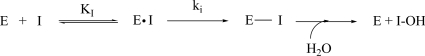
Samples of TreS (0.28 μm) were incubated in buffer in the presence of a range of concentrations of the inhibitors (1–10 mm 5FGlcF or 5–30 mm 5FIdoF) at 37 °C. Aliquots (10 μl) of these inactivation mixtures were removed at time intervals and diluted into assay cells containing 190 μl of DNPGlc substrate ([S] = 3 mm) preincubated at 37 °C. The residual enzymatic activity was determined from the rate of hydrolysis of the substrate, which is directly proportional to the amount of active enzyme. The process was monitored until 80–90% inactivation. Pseudo first-order rate constants (kobs) for each inactivator concentration were calculated by fitting plots of the residual activity versus time to a single exponential equation using GraFit. These values of kobs were then fit to the following equation (Equation 1) describing the inactivation process shown in Reaction 1 to obtain values of ki and KI using GraFit software.
REACTION 1.
 |
Determination of Ki Values for Azasugars and Apparent Ki for 5FGlcF
The apparent Ki values of 5FGlcF were determined by measuring the rates of reaction at differing 5FGlcF concentrations (0–150 μm) for a single DNPGlc concentration (3 mm) in the presence of 23 nm TreS enzyme. The same concentration of enzyme was also used in reactions with different concentrations (1–10 mm) of DNPGlc to determine the Vmax value. The residual enzymatic rates in the presence of 5FGlcF were then plotted in the form of a Dixon plot (1/rate versus [5FGlcF]), and the intersection of the line with y = 1/Vmax yields the inverse value of Ki. Similar experiments were performed to determine the Ki values for azasugars as reversible inhibitors of TreS except that multiple DNPGlc concentrations (2–4 mm) were used.
Reactivation Experiments
TreS that had been fully inactivated by 5FIdoF was freed of excess inactivator by 5-fold dilution with buffer into a total volume of 500 μl and then concentrated at 4 °C using a 10-kDa-nominal cutoff centrifugal concentrator to a volume of ∼50 μl a total of eight times. The resultant solution was then diluted to 100 μl with either buffer alone or with glucose solution (final [glucose], 100 mm) and then incubated at 37 °C. Reactivation was monitored by removal of aliquots (10 μl) at appropriate time intervals and assaying using 3 mm DNPGlc substrate at 37 °C.
RESULTS AND DISCUSSION
Aryl α-Glucosides and α-Glucosyl Fluoride as TreS Substrates
The lack of a convenient assay for TreS has hampered mechanistic studies. However, many hydrolytic α-glucosidases in GH13 family can be conveniently assayed spectrophotometrically by using aryl α-glucoside substrates; thus, it seemed possible that these might also work with TreS, although it was not clear whether transglycosylation or hydrolysis would occur. Previous reports on TreS have indicated that this enzyme does have weak hydrolytic activity along with its isomerization activity when interconverting its natural substrates, producing small amounts (≤10%) of glucose along with isomerized disaccharide products (13, 22, 33). Indeed, incubation of the first five aryl α-glucoside substrates (shown in Table 1) with TreS resulted in a continuous increase of absorbance at 400 nm over time, allowing the determination of Michaelis-Menten parameters. Surprisingly, the enzymatic turnover of 3-nitrophenyl α-glucoside was too slow to be measured. TLC analysis of reaction mixtures containing TreS and aryl α-glucoside substrates indicated that only hydrolysis reactions occurred because no transglycosylation products were observed (data not shown).
TABLE 1.
Kinetic parameters of TreS substrates
N.D., no turnover detected; N/A, not applicable, since not UV-Vis absorbing.
| Substrate | pKa | Km | kcat | kcat/Km | Δϵ |
|---|---|---|---|---|---|
| mm | s−1 | mm−1s−1 | m−1cm−1 | ||
| DNPGlc | 3.96 | 2.9 ± 0.1 | 4.7 ± 0.1 | 1.6 | 9,800 |
| 34DNPGlc | 5.36 | 2.5 ± 0.1 | (4.3 ± 0.1) × 10−2 | 1.7 × 10−2 | 14,220 |
| 4C2NPGlc | 6.45 | 2.2 ± 0.1 | 8.3 ± 0.2 | 3.8 | 1,300 |
| 4NPGlc | 7.18 | 5.8 ± 0.2 | 0.10 ± 0.01 | 1.7 × 10−2 | 6,464 |
| 2NPGlc | 7.22 | 0.7 ± 0.1 | 8.1 ± 0.3 | 12 | 1,650 |
| 3NPGlc | 8.39 | N.D.a | N.D. | N.D. | 167 |
| aGlcF | 3.17 | 0.15 ± 0.03 | 5.3 ± 0.3 | 35 | N/A |
| Maltose | ∼16 | 8.0 ± 0.9 | 19 ± 1 | 2.4 | N/A |
| Trehalose | ∼16 | 87 ± 6 | 66 ± 2 | 0.75 | N/A |
Glycosyl fluorides have also been utilized as substrates to analyze a wide spectrum of glycosidases and generate useful mechanistic insights (39). They were of particular interest in this case because the small size of the aglycone might allow two molecules to bind at once and hence transglycosylation. Incubation of αGlcF with TreS enzyme resulted in a continuous release of fluoride ion as monitored by a fluoride electrode, allowing the determination of kinetic parameters for αGlcF (Table 1). However, TLC analysis of such reaction mixtures confirmed that glucose was the sole carbohydrate product (data not shown).
Interestingly, very similar turnover numbers were seen for DNPGlc, 4C2NPGlc, 2NPGlc, and αGlcF, suggesting that some common step in the mechanisms is rate-limiting (Table 1). By contrast, turnover numbers for 34DNPGlc and 4NPGlc were some 50–100 times lower, whereas no turnover whatsoever was seen for 3NPGlc. The lack of any defensible dependence of rate constants upon leaving group ability indicates that initial bond cleavage is unlikely to be rate-limiting. The possibility that the deglycosylation step is a common rate-limiting step for the “good” substrates is made less likely by the fact that Km values did not decrease with the increase of aglycone leaving group ability (with the notable exception of α-glucosyl fluoride) as would be expected were this to be the case. A common test for rate-limiting deglycosylation step is to examine the effects of exogenously added neutral nucleophiles such as methanol or ethylene glycol on the steady-state rate. This has been demonstrated for several retaining β-glycosidases wherein kcat for deglycosylation-rate-limiting substrates increases proportionally to the nucleophile concentration, whereas no increase is seen for glycosylation-rate-limiting substrates (40–42). Methanol was selected for the study of TreS because of its small size. However, the addition of methanol at concentrations ranging from 100 to 800 mm did not result in any rate increase when using DNPGlc, 4C2NPGlc, or 2NPGlc as TreS substrates (data not shown), rendering it unlikely that the deglycosylation step is rate-limiting, although this experiment must be interpreted with caution because access of the nucleophile to the active site may well be limited. Further evidence that neither chemical step is rate-limiting was obtained by kinetic isotope effect measurements. 2,4-Dinitrophenyl 1-{2H}-α-glucoside was synthesized according to literature procedures (37, 38), and kinetic isotope effects for TreS were determined. Values of (kcat)H/(kcat)D = 1.0 ± 0.02 and (kcat/Km)H/(kcat/Km)D = 1.0 ± 0.02 were determined, strongly suggesting that non-chemical steps are rate-limiting for these hydrolytic reactions.
For the purpose of comparison, kinetic parameters of TreS-catalyzed turnover of its natural substrates, maltose and trehalose, were determined using a stopped assay as described previously (results shown in Table 1) (13). These kcat values (66 and 19 s−1 for trehalose and maltose, respectively) were significantly greater than those of even the good hydrolytic substrates, indicating that the step limiting hydrolytic turnover is not limiting in the transglycosylation process. Indeed, it is interesting to note that previous studies have shown that ∼10% hydrolysis accompanies the isomerization reaction. This accompanying hydrolytic reaction would then presumably exhibit kcat values of approximately 1.9 and 6.6 s−1 for maltose and trehalose, respectively. These values are right in the range measured for hydrolysis of the aryl glucosides, suggesting that the hydrolytic process is common to both sets of substrates.
The values of kcat/Km derived from these parameters are 2.4 and 0.75 mm−1 s−1, respectively, for maltose and trehalose. It is not clear whether the general similarity of these values to those of the aryl glucosides has any significance given the disparity in structural types of the aglycone. However, the availability of this pair of values does allow an equilibrium constant for the overall reaction of 3.2 in favor of trehalose to be determined using the Haldane relationship (43). This compares well with the value of 4.1 calculated using thermodynamic parameters determined previously by calorimetric methods (19).
pH Stability and Profile
pH dependence studies generate important mechanistic insights into the ionization states of residues that are essential for enzymatic catalysis. Stability tests were first carried out in which the enzyme was incubated in buffers at a series of pH values, and aliquots removed at different time periods were assayed under standard kinetic conditions. This enzyme was found to be stable from pH 5.8 to 8.0 for 60 min, and this region was therefore selected for pH dependence studies (shown in Fig. 2a). The pH dependence of kcat/Km for TreS was measured using the substrate depletion method at low substrate concentrations ([S] ≪ Km). Assays were performed with 50 μm DNPGlc because its Km is 2.9 ± 0.1 mm. The obtained kcat/Km values were then plotted versus pH as shown in Fig. 2b. This bell-shaped curve suggests that two ionizable groups are involved in catalysis with assigned pKa values of 5.6 ± 0.2 and 7.4 ± 0.1, although only the higher pKa value is reliable because instability at lower pH values precluded collection of further low pH data points. Such a profile is consistent with the classical double displacement mechanism adopted by retaining glycosidases and transglycosidases with the lower pKa value corresponding to deprotonation of the catalytic nucleophile and the higher value (7.4 ± 0.1) arising from the general acid/base residue.
FIGURE 2.
a, residual enzyme activity of TreS after incubation at 37 °C for 1 h at different pH values. b, dependence of kcat/Km upon pH for hydrolysis of DNPGlc by wild type TreS. The data were fit to a double ionization pH curve by nonlinear regression using the program GraFit 5.0.13 (Erithacus Software Limited, 2006).
Inhibition Studies
Inhibition of TreS by a range of known α-glucosidase inhibitors was assayed, and the results are shown in Table 2. All behaved as competitive inhibitors with the tightest binder being DNJ (Ki = 250 nm), whereas isofagomine bound TreS with ∼500-fold lower affinity. This inhibition behavior is characteristic of a retaining α-glycosidase (31, 44) and thus is consistent with the mechanism proposed. Very interestingly, the absence of the C5 hydroxymethyl substituent dramatically decreased the affinity of DNJ some 1,000-fold as seen in the relative Ki values of xylo-DNJ and DNJ.
TABLE 2.
Ki values for inhibition of TreS by several α-glucosidase inhibitors
| Compound name | Ki |
|---|---|
| μm | |
| Isofagomine | 140 |
| DNJ | 0.25 |
| Xylodeoxynojirimycin | 300 |
| d-Gluconohydroximino-1,5-lactam | 2.1 |
| Casuarine | 2.5 |
Mechanism-based Inhibitors of TreS
5-Fluoroglycosyl fluorides function as effective mechanism-based inhibitors of retaining α-glycosidases by forming long lived covalent glycosyl-enzyme intermediates (32, 35, 45) as a consequence of the substantial inductive effect of the C5 fluorine on the oxocarbenium ion-like transition state. However, incubation of TreS enzyme with 5FGlcF resulted in no time-dependent inactivation. Instead, 5FGlcF was found to behave like a reversible inhibitor with an apparent Ki′ value of 16 μm as shown in supplemental Fig. 1. This very low Ki value is not consistent with normal ground state binding for such a compound given the Km values of 150 μm and 2.9 mm for α-glucosyl fluoride and DNPGlc, respectively. Rather, as seen previously (46, 47), it suggests substantial steady-state accumulation of a covalent glycosyl-enzyme intermediate but one whose turnover is too fast to appear as inactivation. Indeed, incubation of 5FGlcF and TreS resulted in the continuous release of fluoride anion as monitored using a fluoride electrode (supplemental Fig. 2b). By using a large excess of substrate ([5FGlcF] ≫ Ki′), the turnover number of 5FGlcF by TreS was determined to be kcat = 2.9 × 10−3 s−1, which is roughly 2,000-fold lower than that of αGlcF (Table 1).
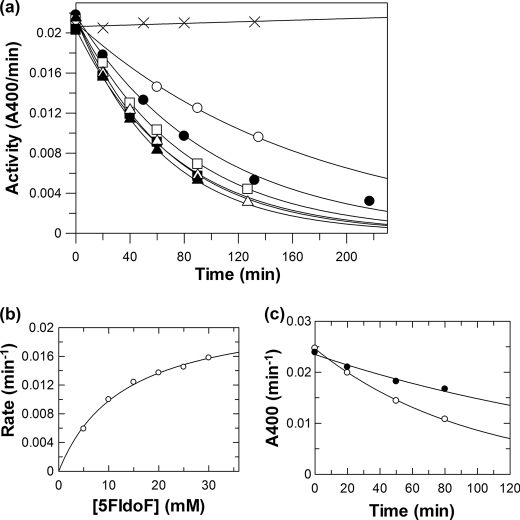
In such circumstances, lower deglycosylation turnover rates and thus trapping on a useful time scale can be achieved by use of the C5 epimer of 5FGlcF, 5FIdoF, as the inactivator because the bulky axial hydroxymethyl group results in further transition state destabilization (46, 47). Incubation of TreS with 5FIdoF indeed resulted in time-dependent decay of enzymatic activity as shown in Fig. 3a. Pseudo first-order kinetics of inactivation were seen at each concentration of 5FIdoF, allowing the determination of a series of rate constants for inactivation (kobs) at each inactivator concentration. A replot of these inactivation rate constants versus inactivator concentration allowed the determination of inactivation parameters of ki = 0.022 ± 0.001 min−1 and KI = 13 ± 1 mm (Fig. 3b) and thus a second-order rate constant for inactivation of ki/KI = 0.0017 min−1 mm−1, respectively. This corresponds to a half-life for inactivation at saturating ligand of t½ = 31 min. Demonstration that inactivation was an active site event was provided by the determination that the rate constant for inactivation by 10 mm 5FIdoF was reduced the expected amount from 0.010 to 0.005 min−1 by co-incubation with 5 μm casuarine (Ki = 2.5 μm) (Fig. 3c).
FIGURE 3.
Inactivation of TreS by 5FIdoF at 37 °C. a, plot of residual enzymatic activity versus time at different inhibitor concentrations: ×, 0 mm; ○, 5 mm; ●, 10 mm; □, 15 mm; ■, 20 mm; △, 25 mm; and ▴, 30 mm. b, replot of inactivation rate constants versus concentration of 5FIdoF. c, inactivation of TreS by 10 mm 5FIdoF in the absence (○) and the presence (●) of 5 μm casuarine at 37 °C.
Reactivation experiments were attempted to demonstrate the catalytic competence of the covalent 5-fluoroidosyl-enzyme intermediate of TreS. Inactivated enzyme was freed from excess inactivator by ultrafiltration and then incubated in buffer at 37 °C. Aliquots of the reactivation mixture were taken at time intervals and assayed, but even after 4 days of incubation, there was no significant recovery of enzymatic activity. The hydrolytic stability of the trapped intermediate is not too surprising because the inactivation rate (ki) of 0.022 ± 0.001 min−1 (t½ = 31 min) with 5FIdoF was relatively low; thus, reactivation (turnover) must be even slower for the intermediate to be trapped. Denaturation of the enzyme may well be occurring faster than reactivation because control experiments showed that free TreS lost roughly 50% of its activity after incubation at 37 °C for 4 days. Attempts to accelerate reactivation via transglycosylation to 100 mm glucose were likewise unsuccessful; no recovery of enzymatic activity was seen over 4 days at 37 °C. This lack of reactivation could well be a consequence of a conformationally closed active site formed when the covalent intermediate was trapped so that the exogenously added glucose could not enter.
Identification of Catalytic Nucleophile by MS/MS
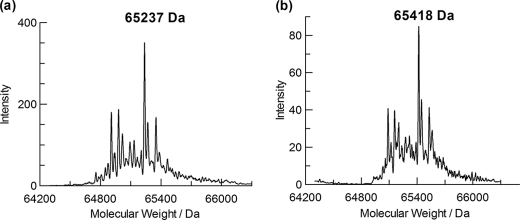
The complete inactivation of TreS by 5FIdoF opened up the possibility of identifying the amino acid to which it is attached by LC-MS/MS studies of proteolytic digests (48). The stoichiometry of inactivation was first shown by comparing the masses of the intact TreS and 5FIdoF-labeled TreS using a quadrupole TOF instrument. The reconstructed mass spectrum of the unlabeled wild type TreS [M + H+] (Fig. 4a) revealed a range of masses with a principal peak at a mass of 65,237 ± 2 Da. The predicted mass of the intact 593-amino acid enzyme is 68,202 Da. Because TreS has been reported to undergo limited proteolytic degradation from its original 68-kDa form during long term storage on ice (49), the detected masses presumably reflect these proteolytic products. Upon incubation of the enzyme with 10 mm 5FIdoF at 37 °C for 6 h, the whole manifold of peaks shifted up in mass with the major peak in the mass spectrum now found at 65,418 ± 2 Da, increased by 181 Da (Fig. 4b). This difference correlates very well with the incorporation of a single 5-fluoro-α-l-idopyranosyl moiety (181 Da) into the protein. The fact that all peaks shifted indicates that all are active species.
FIGURE 4.
Mass spectra of intact TreS enzyme (a) and a mixture of TreS enzyme with 10 mm 5FIdoF (b) incubated at 37 °C for 6 h.
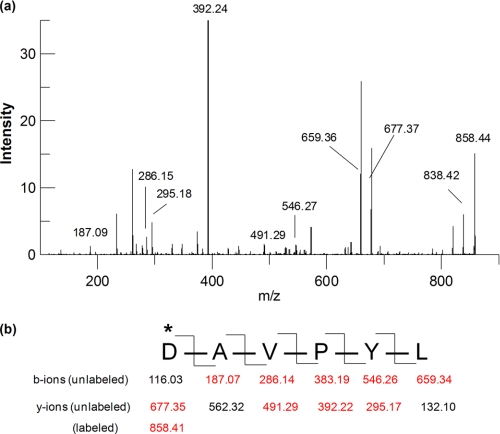
Samples of 5FIdoF-labeled TreS and non-labeled enzyme control were subjected to pepsin digestion at pH 2.0 at room temperature for 2 h, and the two resultant peptide mixtures were individually subjected to HPLC separation with analysis by electrospray ionization MS. Comparative mapping studies of elution profiles of the two samples allowed identification of a short peptide, P2 (m/z 858.4), that was present in the labeled TreS mixture but not in the control sample. Instead, another peptide, P1 (m/z 677.3), was found in the non-labeled TreS mixture. The difference in m/z of these two peptides (181.1) is consistent with P2 being a singly charged peptide to which a label of mass 181.1 (the mass of the 5-fluoro-α-l-idopyranosyl moiety) is attached. P1 and P2 were therefore good candidates for the peptides of interest and were subjected to sequencing by collision-induced fragmentation.
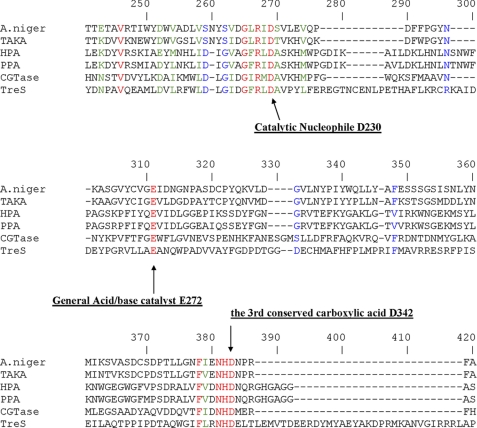
The sequence of peptide P1 was determined to be 230DAVPYL235 as evidenced by the fragment ion spectrum (data not shown). Analysis of the fragmentation of the labeled peptide P2 confirmed that it was the same peptide as P1 except that the peak at m/z 858.4 corresponded to the 5-fluorosugar-labeled peptide 230DAVPYL235 and its HF elimination product (m/z 838.4) as shown in Fig. 5, a and b. These results clearly confirmed that the hexapeptide starting from Asp230 was indeed the site of labeling by 5FIdoF. Alignment of the primary sequence of TreS with those of other GH13 enzymes (Fig. 6) revealed that Asp230 is fully conserved in this family and indeed corresponds to the residue identified as the nucleophile in other enzymes of GH13. The sequence alignment also suggested that Glu272 is the best candidate as the general acid/base catalyst, whereas the third highly conserved active site carboxylic residue was identified as Asp342.
FIGURE 5.
Electrospray ionization MS/MS analysis of labeled peptide P2. a, MS/MS fragment ion spectrum of peptide P2. b, fragmentation pattern of peptide P2 and corresponding m/z of singly charged b-ions and y-ions. Red-colored fragments can be found in the spectrum. *, residue labeled.
FIGURE 6.
Partial sequence alignment of several GH13 enzymes including TreS. The three conserved carboxylic acid residues are labeled with arrows and the corresponding TreS numbering. The Swiss-Prot accession numbers are indicated in parentheses. The sequences shown are Aspergillus niger α-amylase (P56271), Aspergillus oryzae α-amylase (TAKA) (P0C1B3), human pancreatic α-amylase (HPA) (P04746), porcine pancreatic α-amylase (PPA) (P00690), Bacillus circulans cyclomaltodextrin glucanotransferase (CGTase) (P43379), and M. smegmatis trehalose synthase (A0R6E0).
Rearrangement Mechanism
A key remaining question concerning the mechanism of this enzyme is whether the glucose molecule released upon formation of the glucosyl-enzyme intermediate reorients within the active site and reattacks or whether it is released into the medium and rebinds. The literature contains conflicting reports on this question with different TreS homologues with two reports suggesting a completely intramolecular (no release) process (33, 34) and one suggesting some level of release and rebinding (13). Two approaches to this question were adopted in this study. In the first approach, TreS was incubated with isotopically labeled glucose and unlabeled disaccharide (either maltose or trehalose), and disaccharide products were analyzed by MS. To facilitate the TLC separation of maltose and trehalose, the enzymatic reaction mixtures were lyophilized and acetylated (acetic anhydride/pyridine) before analysis. Two reactions containing 10 mm glucose (either d-glucose-6,6′-d2 or d-[U-13C]glucose), 2 mm maltose, and TreS were incubated at 37 °C for 24 h along with a control sample to which no enzyme had been added. After lyophilization, acetylation, and aqueous workup, the three samples were analyzed by TLC and MS. As shown in supplemental Fig. 2a, TLC analysis of the reaction of d-[U-13C]glucose, maltose, and TreS clearly demonstrated enzymatic conversion of maltose into trehalose, whereas no such conversion had occurred in the control sample (supplemental Fig. 2b). The mass spectrum of the mixture (supplemental Fig. 2d) revealed that the two major peaks, 419.2 and 701.3, correspond to [per-O-acetylated (6 × 13C) glucose + Na+] and [per-O-acetylated maltose/trehalose + Na+], respectively. No incorporation of exogenous [13C]glucose into disaccharides was observed as this would have shifted the mass of the disaccharide products by 6 Da to 707 Da. Similar results were obtained for the reaction mixture containing d-glucose-6,6′-d2, maltose, and TreS. These results unambiguously demonstrate that TreS does not catalyze incorporation of exogenous glucose into disaccharide reactants under these conditions. It is difficult to evaluate the single previous claim that TreS could incorporate exogenous glucose into disaccharides because few experimental details were provided. However, their use of a radioactive incorporation assay may have allowed the detection of very small amounts of incorporation.
In a second approach, α-glucosyl fluoride was used as a glycosyl donor with a sufficiently small aglycone, fluoride, that it might be possible to accommodate two sugar molecules in the active site at once and catalyze the assembly of maltose/trehalose especially in the presence of relatively high concentrations of added glucose. A sample containing TreS plus 5 mm αGlcF and 15 mm glucose was incubated at room temperature alongside a control from which TreS had been omitted. TLC analysis of these mixtures (supplemental Fig. 2c) clearly showed essentially complete hydrolysis of the αGlcF and no evidence of transglycosylation to a disaccharide. The control showed no reaction. This lack of transglycosylation again supports an intramolecular rearrangement mechanism.
Substrate Scope Studies
The recent revelation of the new α-glucan biosynthetic pathway in M. tuberculosis has opened up the possibility that fluorinated maltose 1-phosphate analogues might serve as inhibitors of the maltosyltransferase/phosphorylase GlgE and serve as novel therapeutic agents to treat tuberculosis (25). Theoretically, 2-fluoromaltose might be used in the expectation that the mycobacterial maltose kinase would convert it to the inhibitory form. However, because M. tuberculosis has a poor maltose uptake system but a good trehalose transporter, it was of interest to see whether 2-fluorotrehalose could be synthesized by TreS. This in turn would reveal whether the endogenous TreS could indeed convert this back to 2-fluoromaltose “in situ.”
Three fluorinated maltose compounds, 2-fluoromaltose, 2-fluoro-“manno” maltose (4-O-α-d-glucopyranosyl-2-deoxy-2-fluoromannose), and 2-deoxy-2,2-difluoromaltose (50), were incubated with TreS enzyme, and reactions were monitored by TLC and 19F NMR. Satisfyingly, both of the monofluorinated maltose analogues were isomerized into their corresponding trehalose analogues in over 60% yield consistent with the equilibrium constant lying in favor of trehalose. Unfortunately, the isomerization of 2-deoxy-2,2-difluoromaltose was extremely slow and thus was not pursued further. These two monofluoromaltose analogues therefore have potential for control of tuberculosis and will be evaluated as novel antituberculosis agents in the future.
Conclusion
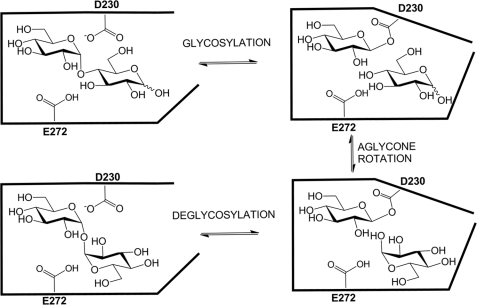
All the data collected within this study are consistent with TreS acting as a GH13 transglucosylase in which a covalent β-glucosyl-enzyme intermediate is formed via attack of Asp230 on the sugar anomeric center (Fig. 7). The glucose thus released is retained within the active site and reorients within this protected environment such that either the 1-hydroxyl or the 4-hydroxyl are positioned for reattack in a productive complex. Although protected within this enclosed site, the glycosyl-enzyme intermediate nonetheless undergoes some transfer to water such that ≈10% hydrolysis accompanies isomerization. Simple monosaccharide derivatives such as aryl α-glucosides or α-glucosyl fluoride can also act as substrates but undergo only the hydrolytic reaction as revealed by the reaction products and as reflected in their kcat values relative to that of maltose. The absence of any defensible dependence of kcat or kcat/Km for aryl glucosides upon aglycone reactivity and the absence of an α-deuterium kinetic isotope on either parameter for the substrate 2,4-dinitrophenyl α-glucoside suggest that a non-chemical step is rate-limiting for these substrates. These findings in conjunction with the strictly intramolecular nature of the rearrangement process suggest that a protein conformational change is the rate-limiting step for the majority of the aryl glucosides studied as well as αGlcF. Significantly slower turnover of the substrates lacking an ortho-aryl substituent is surprising but possibly reflects a slower conformational change in such enzyme-substrate complexes. Further evidence in support of a classical “α-glycosidase” mechanism is derived from the potent inhibition exhibited by deoxynojirimycin, a classical α-glucosidase inhibitor, and much weaker inhibition by isofagomine.
FIGURE 7.
Proposed catalytic mechanism of TreS.
Although no three-dimensional structures have been reported to date on any TreS enzymes, there are several structures available for another similar class of GH13 enzymes, the sucrose mutases. These enzymes catalyze the isomerization of sucrose (α-d-glucopyranosyl-1,2-β-d-fructofuranoside) to generate isomaltulose (α-d-glucopyranosyl-1,6-d-fructofuranoside) and trehalulose (α-d-glucopyranosyl-1,1-β-d-fructofuranoside) as the major products. For this reason, these enzymes are sometimes named as trehalulose synthases or isomaltulose synthases. Earlier studies have demonstrated that sucrose mutase also operates by an intramolecular rearrangement mechanism (51). Although in general the active sites are very similar to those of GH13 glycosidases (52–54), a key difference is that two aromatic residues, Phe256 and Phe280 (numbering of trehalulose synthase from Pseudomonas mesoacidophila MX-45), have been found to form a clamp at the entrance to the enzymatic active site (53). Mutation of either one or both of the clamping phenylalanines results in greatly enhanced enzymatic hydrolysis with the double mutant being an exclusive hydrolase. Presumably, these two hydrophobic clamps shield the active site during catalysis and suppress hydrolysis of the glucosyl-enzyme while the fructosyl residue rotates. Indeed, very few interactions are seen between the fructosyl moiety and the enzyme active site consistent with such mobility (54).
Another related enzyme is the maltooligosyltrehalose synthase (TreY) referred to earlier. This enzyme catalyzes an isomerization reaction similar to that of TreS but with a longer version of the substrate. TreY has also been shown to carry out an intramolecular rearrangement (16), although a small degree of intermolecular rearrangement was reported in one study as was also seen with TreS (55). Two crystal structures of TreY are available, and both reveal a classical GH13 −1 subsite and a “pocket-like” +1 subsite (56, 57) as seen in the sucrose mutases. A similarly enclosed active site can be anticipated for TreS.
Supplementary Material
This work was supported in part by the Canadian Institutes of Health Research.
This paper is dedicated to Dr. Alan D. Elbein who passed away on November 30, 2009.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- TreS
- trehalose synthase
- GH13
- glycoside hydrolase family 13
- DNPGlc
- 2,4-dinitrophenyl α-d-glucoside
- 4C2NPGlc
- 4-chloro-2-nitrophenyl α-d-glucoside
- 2NPGlc
- 2-nitrophenyl α-d-glucoside
- 34DNPGlc
- 3,4-dinitrophenyl α-d-glucoside
- 4NPGlc
- 4-nitrophenyl α-d-glucoside
- 3NPGlc
- 3-nitrophenyl α-d-glucoside
- αGlcF
- α-glucosyl fluoride
- d-DNPGlc
- 2,4-dinitrophenyl 1-{2H}-α-glucoside
- 5FGlcF
- 5-fluoro α-d-glucosyl fluoride
- 5FIdoF
- 5-fluoro-β-l-idosyl fluoride
- DNJ
- deoxynojirimycin.
REFERENCES
- 1. Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. (2003) Glycobiology 13, 17R–27R [DOI] [PubMed] [Google Scholar]
- 2. Clegg J. S., Filosa M. F. (1961) Nature 192, 1077–107814478157 [Google Scholar]
- 3. Becker A., Schlöder P., Steele J. E., Wegener G. (1996) Experientia 52, 433–439 [DOI] [PubMed] [Google Scholar]
- 4. Thevelein J. M. (1984) Microbiol. Rev. 48, 42–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schiraldi C., Di Lernia I., De Rosa M. (2002) Trends Biotechnol. 20, 420–425 [DOI] [PubMed] [Google Scholar]
- 6. Madin K. A., Crowe J. H. (1975) J. Exp. Zool. 193, 335–342 [Google Scholar]
- 7. Singer M. A., Lindquist S. (1998) Trends Biotechnol. 16, 460–468 [DOI] [PubMed] [Google Scholar]
- 8. Kandror O., DeLeon A., Goldberg A. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9727–9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hunter R. L., Armitige L., Jagannath C., Actor J. K. (2009) Tuberculosis 89, S18-S25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hunter R. L., Olsen M. R., Jagannath C., Actor J. K. (2006) Ann. Clin. Lab. Sci. 36, 371–386 [PubMed] [Google Scholar]
- 11. Brennan P. J., Nikaido H. (1995) Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 12. Edavana V. K., Pastuszak I., Carroll J. D., Thampi P., Abraham E. C., Elbein A. D. (2004) Arch. Biochem. Biophys. 426, 250–257 [DOI] [PubMed] [Google Scholar]
- 13. Pan Y. T., Koroth Edavana V., Jourdian W. J., Edmondson R., Carroll J. D., Pastuszak I., Elbein A. D. (2004) Eur. J. Biochem. 271, 4259–4269 [DOI] [PubMed] [Google Scholar]
- 14. Gibson R. P., Tarling C. A., Roberts S., Withers S. G., Davies G. J. (2004) J. Biol. Chem. 279, 1950–1955 [DOI] [PubMed] [Google Scholar]
- 15. Maruta K., Nakada T., Kubota M., Chaen H., Sugimoto T., Kurimoto M., Tsujisaka Y. (1995) Biosci. Biotechnol. Biochem. 59, 1829–1834 [DOI] [PubMed] [Google Scholar]
- 16. Nakada T., Maruta K., Tsusaki K., Kubota M., Chaen H., Sugimoto T., Kurimoto M., Tsujisaka Y. (1995) Biosci. Biotechnol. Biochem. 59, 2210–2214 [DOI] [PubMed] [Google Scholar]
- 17. Nakada T., Maruta K., Mitsuzumi H., Kubota M., Chaen H., Sugimoto T., Kurimoto M., Tsujisaka Y. (1995) Biosci. Biotechnol. Biochem. 59, 2215–2218 [DOI] [PubMed] [Google Scholar]
- 18. Higashiyama T. (2002) Pure Appl. Chem. 74, 1263–1269 [Google Scholar]
- 19. Tewari Y. B., Goldberg R. N. (1991) Biophys. Chem. 40, 59–67 [DOI] [PubMed] [Google Scholar]
- 20. Nishimoto T., Nakano M., Ikegami S., Chaen H., Fukuda S., Sugimoto T., Kurimoto M., Tsujisaka Y. (1995) Biosci. Biotechnol. Biochem. 59, 2189–2190 [DOI] [PubMed] [Google Scholar]
- 21. Tsusaki K., Nishimoto T., Nakada T., Kubota M., Chaen H., Fukuda S., Sugimoto T., Kurimoto M. (1997) Biochim. Biophys. Acta 1334, 28–32 [DOI] [PubMed] [Google Scholar]
- 22. Koh S., Shin H. J., Kim J. S., Lee D. S., Lee S. Y. (1998) Biotechnol. Lett. 20, 757–761 [Google Scholar]
- 23. Wang J. H., Tsai M. Y., Lee G. C., Shaw J. F. (2007) J. Agric. Food Chem. 55, 1256–1263 [DOI] [PubMed] [Google Scholar]
- 24. Chen Y. S., Lee G. C., Shaw J. F. (2006) J. Agric. Food Chem. 54, 7098–7104 [DOI] [PubMed] [Google Scholar]
- 25. Kalscheuer R., Syson K., Veeraraghavan U., Weinrick B., Biermann K. E., Liu Z., Sacchettini J. C., Besra G., Bornemann S., Jacobs W. R., Jr. (2010) Nat. Chem. Biol. 6, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Titgemeyer F., Amon J., Parche S., Mahfoud M., Bail J., Schlicht M., Rehm N., Hillmann D., Stephan J., Walter B., Burkovski A., Niederweis M. (2007) J. Bacteriol. 189, 5903–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yue M., Wu X. L., Gong W. N., Ding H. B. (2009) Microb. Cell Fact. 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Y., Wei D., Zhang J., Wang Y., Xu H., Xing L., Li M. (2010) Extremophiles 14, 1–8 [DOI] [PubMed] [Google Scholar]
- 30. Tsusaki K., Nishimoto T., Nakada T., Kubota M., Chaen H., Sugimoto T., Kurimoto M. (1996) Biochim. Biophys. Acta 1290, 1–3 [DOI] [PubMed] [Google Scholar]
- 31. Zechel D. L., Withers S. G. (2000) Acc. Chem. Res. 33, 11–18 [DOI] [PubMed] [Google Scholar]
- 32. Zhang R., Li C., Williams L. K., Rempel B. P., Brayer G. D., Withers S. G. (2009) Biochemistry 48, 10752–10764 [DOI] [PubMed] [Google Scholar]
- 33. Nishimoto T., Nakano M., Nakada T., Chaen H., Fukuda S., Sugimoto T., Kurimoto M., Tsujisaka Y. (1996) Biosci. Biotechnol. Biochem. 60, 640–644 [DOI] [PubMed] [Google Scholar]
- 34. Koh S., Kim J., Shin H. J., Lee D., Bae J., Kim D., Lee D. S. (2003) Carbohydr. Res. 338, 1339–1343 [DOI] [PubMed] [Google Scholar]
- 35. McCarter J. D., Withers S. G. (1996) J. Am. Chem. Soc. 118, 241–242 [Google Scholar]
- 36. Lee S. S., Yu S., Withers S. G. (2003) Biochemistry 42, 13081–13090 [DOI] [PubMed] [Google Scholar]
- 37. Berven L. A., Withers S. G. (1986) Carbohydr. Res. 156, 282–285 [Google Scholar]
- 38. Chen H. M., Withers S. G. (2007) Chembiochem 8, 719–722 [DOI] [PubMed] [Google Scholar]
- 39. Williams S. J., Withers S. G. (2000) Carbohydr. Res. 327, 27–46 [DOI] [PubMed] [Google Scholar]
- 40. Zechel D. L., Reid S. P., Stoll D., Nashiru O., Warren R. A., Withers S. G. (2003) Biochemistry 42, 7195–7204 [DOI] [PubMed] [Google Scholar]
- 41. Kempton J. B., Withers S. G. (1992) Biochemistry 31, 9961–9969 [DOI] [PubMed] [Google Scholar]
- 42. Vocadlo D. J., Wicki J., Rupitz K., Withers S. G. (2002) Biochemistry 41, 9727–9735 [DOI] [PubMed] [Google Scholar]
- 43. Garces E., Cleland W. W. (1969) Biochemistry 8, 633–640 [DOI] [PubMed] [Google Scholar]
- 44. Lillelund V. H., Jensen H. H., Liang X., Bols M. (2002) Chem. Rev. 102, 515–553 [DOI] [PubMed] [Google Scholar]
- 45. Rempel B. P., Withers S. G. (2008) Glycobiology 18, 570–586 [DOI] [PubMed] [Google Scholar]
- 46. McCarter J. D., Withers S. G. (1996) J. Biol. Chem. 271, 6889–6894 [DOI] [PubMed] [Google Scholar]
- 47. Numao S., Kuntz D. A., Withers S. G., Rose D. R. (2003) J. Biol. Chem. 278, 48074–48083 [DOI] [PubMed] [Google Scholar]
- 48. Withers S. G., Aebersold R. (1995) Protein Sci. 4, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pan Y. T., Carroll J. D., Asano N., Pastuszak I., Edavana V. K., Elbein A. D. (2008) FEBS J. 275, 3408–3420 [DOI] [PubMed] [Google Scholar]
- 50. Zhang R., McCarter J. D., Braun C., Yeung W., Brayer G. D., Withers S. G. (2008) J. Org. Chem. 73, 3070–3077 [DOI] [PubMed] [Google Scholar]
- 51. Cheetham P. S. (1984) Biochem. J. 220, 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang D., Li N., Lok S. M., Zhang L. H., Swaminathan K. (2003) J. Biol. Chem. 278, 35428–35434 [DOI] [PubMed] [Google Scholar]
- 53. Ravaud S., Robert X., Watzlawick H., Haser R., Mattes R., Aghajari N. (2007) J. Biol. Chem. 282, 28126–28136 [DOI] [PubMed] [Google Scholar]
- 54. Rhimi M., Haser R., Aghajari N. (2008) Biologia 63, 1020–1027 [Google Scholar]
- 55. Kato M., Takehara K., Kettoku M., Kobayashi K., Shimizu T. (2000) Biosci. Biotechnol. Biochem. 64, 319–326 [DOI] [PubMed] [Google Scholar]
- 56. Kobayashi M., Kubota M., Matsuura Y. (2003) J. Appl. Glycosci. 50, 1–8 [Google Scholar]
- 57. Cielo C. B., Okazaki S., Suzuki A., Mizushima T., Masui R., Kuramitsu S., Yamane T. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 397–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.