Background: ADP/ATP carrier (Ancp) is a model of mitochondrial carriers that mediates the transport of metabolic intermediates.
Results: Yeast Ancp exhibits inhibitor-dependent solvent accessibility. Ancp signature sequence is involved in the ADP/ATP binding step.
Conclusion: Ancp has a highly dynamic structure, with different protein parts acting in synergy.
Significance: Learning the functional dynamics of Ancp is crucial for understanding ADP/ATP transport mechanism.
Keywords: Bioenergetics, Membrane Proteins, Membrane Transport, Mitochondrial Transport, Protein Dynamics, ADP/ATP Carrier, Bongkrekic Acid, Carboxyatractyloside, Conformational Dynamics, Hydrogen/Deuterium Exchange
Abstract
The mitochondrial ADP/ATP carrier, or Ancp, is a member of the mitochondrial carrier family responsible for exchanging ADP and ATP across the mitochondrial inner membrane. ADP/ATP transport involves Ancp switching between two conformational states. These can be analyzed using specific inhibitors, carboxyatractyloside (CATR) and bongkrekic acid (BA). The high resolution three-dimensional structure of bovine Anc1p (bAnc1p), as a CATR-carrier complex, has been solved. However, because the structure of the BA-carrier complex has not yet been determined, the detailed mechanism of transport remains unknown. Recently, sample processing for hydrogen/deuterium exchange experiments coupled to mass spectrometry was improved, providing novel insights into bAnc1p conformational transitions due to inhibitor binding. In this work we performed both hydrogen/deuterium exchange-mass spectrometry experiments and genetic manipulations. Because these are very difficult to apply with bovine Anc1p, we used Saccharomyces cerevisiae Anc isoform 2 (ScAnc2p). Significant differences in solvent accessibility were observed throughout the amino acid sequence for ScAnc2p complexed to either CATR or BA. Interestingly, in detergent solution, the conformational dynamics of ScAnc2p were dissimilar to those of bAnc1p, in particular for the upper half of the cavity, toward the intermembrane space, and the m2 loop, which is thought to be easily accessible to the solvent from the matrix in bAnc1p. Our study then focused on the methionyl residues of the Ancp signature sequence, RRRMMM. All our results indicate that the methionine cluster is involved in the ADP/ATP transport mechanism and confirm that the Ancp cavity is a highly dynamic structure.
Introduction
The mitochondrial ADP/ATP carrier (Ancp)5 plays a key role in supplying energy to aerobic eukaryotic cells. It is the last element of the oxidative phosphorylation system, allowing ATP to be exported into the cytosol in exchange for ADP. It catalyzes one of the most active transport activities in eukaryotic cells, and in many higher organisms it is the most abundant mitochondrial membrane protein. The mitochondrial ADP/ATP exchange process can be blocked by two very specific inhibitors: carboxyatractyloside (CATR) and bongkrekic acid (BA). CATR and BA bind with high affinity to two distinct pre-existing conformations of the carrier referred to as CATR and BA conformations, respectively. Well defined regions in the resulting CATR- and BA-carrier complexes show different reactivity to chemical, enzymatic, and immunochemical reagents (1). It has been suggested that the transition between the CATR and BA conformations is involved in ADP/ATP transport across the mitochondrial inner membrane (1).
Ancp belongs to the mitochondrial carrier family (MCF), all members of which share common features, i.e. similar molecular masses of about 30 kDa, a so-called tripartite organization consisting in three sequence repeats of about 100 amino acids, each containing a conserved PX(D/E)XX(K/R) motif (2). In addition, all Ancp share a common signature sequence, RRRMMM, that is not found in other MCF members (3). The intrinsic mechanisms of ADP/ATP transport, nucleotide recognition, and Ancp conformational changes have been widely investigated, mainly using bovine isoform 1 (bAnc1p) and Saccharomyces cerevisiae isoform 2 (ScAnc2p) (4).
Bovine Anc1p crystallized in complex with CATR was the first mitochondrial carrier for which high resolution structural data were obtained (5). The 2.2-Å resolution structure of the CATR conformation showed six tilted transmembrane α-helices forming a wide cavity open toward the intermembrane space (IMS) and closed on the matrix side (5). The high resolution three-dimensional structure of the bovine carrier in complex with BA, however, has not yet been described. Recently, both complexes were compared in detergent solution by hydrogen/deuterium exchange coupled to mass spectrometry (HDX-MS). These analyses provided new information on the BA conformation and, thus, on the conformational dynamics of bAnc1p during ADP/ATP transport (6). However, the bovine model remains limited to biochemical studies of the native form because its overexpression in bacteria or in yeast is difficult (7). In contrast, the yeast carrier, ScAnc2p, has been widely studied by site-directed mutagenesis. It has been suggested that, despite extensive homology and similar activity, bAnc1p and ScAnc2p present some distinct biochemical properties (7) while sharing similar structural features, i.e. a common two-dimensional structure formed by six transmembrane helices and three large loops.
In this study we investigated the conformational dynamics of ScAnc2p by HDX-MS to decipher the role of specific amino acid residues involved in the structural transitions that Ancp undergoes during ADP/ATP transport. The results are discussed with respect to the biochemical data available on ScAnc2p and on Ancp in general. We then focused our studies on the Ancp signature sequence, RRRMMM, specifically on the triplet of methionyl residues. Our results provide new insights into the biochemical role of this motif, which is located at the bottom of the cavity (toward the matrix) and may contribute to the ADP/ATP binding step. After mutation of the first methionyl residue of the triplet, spontaneous intragenic suppressor mutants were identified. These were located at the top of the cavity, indicating that the conserved sequence may also play a role in the structural complementarity between both sides of the cavity formed by the carrier and in the nucleotide translocation process.
Taken together, our results indicate that ScAnc2p has a highly dynamic structure, with different protein parts acting in synergy. Moreover, the first and the third methionyl residues within the Ancp signature sequence appear to be crucial for nucleotide transport.
EXPERIMENTAL PROCEDURES
Chemicals and Immunochemicals
BA and [3H]atractyloside ([3H]ATR) were prepared as previously described (8, 9). Hydroxylapatite (HTP) was from Bio-Rad. Triton X-100 and n-dodecyl-β-d-maltoside were purchased from Anatrace. CATR, pepsin, and bovine serum albumin were obtained from Sigma. Recombinant type XVIII protease was prepared as described (10). Trifluoroacetic acid, acetonitrile, and dichloromethane were purchased from Sigma, Carlo Erba Reagenti, and Riedel de Haën, respectively. Rabbit polyclonal antibodies directed against yeast SDS-treated VDAC were generously provided by Guy Lauquin (Bordeaux 2 University). The polyclonal ScAnc2p antibodies used were produced in rabbits inoculated with SDS-treated, HTP-purified ScAnc2p.
Bacterial and Yeast Strains and Media
The Escherichia coli strain used in this work was JM109: recA1, endA1, gyrA96, thi, hsdR17 (rK-, mK+), relA1, supE44, Δ(lac-proAB), [F′, traD36, proAB, lacIqZΔM15] (Promega). Bacteria were grown on LB medium (Difco) supplemented with 100 μg/ml ampicillin when necessary. Bacteria were transformed according to standard methods using CaCl2 (11). The following S. cerevisiae strains were used in this study: W303-1B (MATα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1 can1-100) and its derivatives JL1-3 (W303-1B anc1::LEU2 anc2::HIS3 anc3::URA3) (12), JL-1Δ2Δ3u− (JL1–3 Δanc2::HIS3 Δ[269–975]anc3) (13), JL1–3ANC2 and JL1–3ANC2(His6) (W303-1B anc1::LEU2 anc3::URA3) (14), and JL1–3ANC2Δpor1 (W303–1B anc1::LEU2 anc3::URA3 por1::HIS3).6 Strains were cultivated and transformed as described (15). A full description of the compositions of all media used, YPL (lactate-containing rich medium) and YNB Glc W− and YNB Gal W− (tryptophan-free minimal medium containing glucose or galactose, respectively) can be found in Clémençon et al. (15).
Site-directed Mutagenesis and Plasmid Construction
Site-directed mutagenesis of ScANC2 was performed using the Transformer site-directed mutagenesis kit (Roche Applied Science) with the mutagenic primers presented in Table 1. The mutated scanc2 genes were subcloned into a centromeric plasmid, pRS314, under the control of ScANC2 regulatory sequences as described (16). The resulting plasmids were used to transform the JL1Δ2Δ3u− strain using the LiCl procedure (17). Their capacity to complement the ScANC2 deletion was assessed on solid YPL medium.
TABLE 1.
Site-directed mutagenesis strategy
Fw, forward; Rv, reverse.
| Amino acid change | Oligonucleotides useda | Strain nameb |
|---|---|---|
| M254A; M255A; M256A | AAA-Fw, gttagaagaagagcggcggcgacctccggtcaagct | scanc2/AAA |
| AAA-Rv, agcttgaccggaggtcgccgccgctcttcttctaac | ||
| M254A; M255A | AAM-Fw, gttagaagaagagcggcgatgacctccggtcaagct | scanc2/AAM |
| AAM-Rv, agcttgaccggaggtcatcgccgctcttcttctaac | ||
| M254A; M256A | AMA-Fw, gttagaagaagagcgatggcgacctccggtcaagct | scanc2/AMA |
| AMA-Rv, agcttgaccggaggtcgccatcgctcttcttctaac | ||
| M255A; M256A | MAA-Fw, gttagaagaagaatggcggcgacctccggtcaagct | scanc2/MAA |
| MAA-Rv, agcttgaccggaggtcgccgccattcttcttctaac | ||
| M254A | AMM-Fw, gttagaagaagagcgatgatgacctccggtcaagct | scanc2/AMM |
| AMM-Rv, agcttgaccggaggtcatcatcgctcttcttctaac | ||
| M255A | MAM-Fw, gttagaagaagaatggcgatgacctccggtcaagct | scanc2/MAM |
| MAM-Rv, agcttgaccggaggtcatcgccattcttcttctaac | ||
| M256A | MMA-Fw, gttagaagaagaatgatggcgacctccggtcaagct | scanc2/MMA |
| MMA-Rv, agcttgaccggaggtcgccatcattcttcttctaac |
a The altered bases are underlined.
b Nomenclature for JL1Δ2Δ3u− strains carrying pRS314 harboring a wild type or a mutated ScANC2 gene under the control of ScANC2 regulatory sequences.
Isolation of Mitochondria
Yeast cells grown on YPL or YNB Gal W− were harvested during the late log phase (A600 nm near 5 in YPL or near 3 in YNB Gal W−). Mitochondria were prepared as previously described (18).
ADP/ATP Transport Assay with Isolated Mitochondria
ADP/ATP transport was measured by a luminescence assay, as detailed in Dassa et al. (19). Briefly, freshly prepared mitochondria were incubated for 5 min at 25 °C in 10 mm Tris-HCl, pH 7.4, 10 mm KH2PO4, 0.6 m mannitol, 0.1 mm EGTA, 2 mm MgCl2, 10 μm Ap5A (an adenylate kinase inhibitor), and 1 mm α-ketoglutarate in the presence of 0.1% (w/v) luciferin and 0.1% (w/v) luciferase.
[3H]ATR Binding Assays
[3H]ATR binding assays on isolated mitochondria were carried out as described (3). Briefly, mitochondria were diluted (0.5–1 mg of protein/ml) in 1 ml of standard medium (0.12 m KCl, 10 mm Mops, pH 6.8, 1 mm EDTA), and [3H]ATR was added at concentrations up to 3 μm. After incubation for 45 min at 0 °C, mitochondria were sedimented, and radioactivity associated with the pellet was determined by scintillation counting (Ready Value, Beckman). Assays were also performed in the presence of 20 μm CATR to correct for minor nonspecific [3H]ATR binding.
ADP/ATP Carrier Purification
ADP/ATP carrier proteins were isolated from yeast mitochondria as a CATR-carrier complex or a BA-carrier complex. ScAnc2p was enriched from JL1–3ANC2 or JL1–3ANC2Δpor1 strains by HTP in the presence of Triton X-100 (1% (w/v) final concentration) according to the described method (3). C-terminal histidine-tagged carrier was purified from strain JL1–3ANC2(His6) in the presence of n-dodecyl-β-d-maltoside (0.05% (w/v) final concentration) according to the described procedure (14). The final carrier protein concentration in each purified fraction ranged between 0.05 and 0.1 mg/ml, depending on the method used.
ADP/ATP Carrier Digestion
All in-solution protein digestions were performed in an ice bath at 0 °C. Protease solutions were prepared in 20 mm glycine, pH 2.5, and cooled to 0 °C. The Ancp was digested in 20 mm glycine, pH 2.5, for 2 min using a substrate/protease ratio between 1:0.25 and 1:2 (w/w) for pepsin or between 1:1.5 and 1:24 (w/w) for recombinant type XVIII protease. On-line digestion of the ADP/ATP carrier in the presence of 1 m guanidinium chloride was performed in an ice-bath at 0 °C using a column packed with pepsin immobilized on a POROS-20AL resin, as described (10).
SDS-PAGE and Western Blotting
Protein concentrations were determined using a BCA protein assay kit (Sigma) with BSA as a standard. For SDS-PAGE, samples were prepared as described (18). Antibodies directed against SDS-treated ScAnc2p or yeast SDS-treated VDAC were used at a 1:1500 and 1:1000 final dilution, respectively. Antibody binding was revealed using horseradish peroxidase-coupled protein A and the enhanced chemiluminescence (ECL) system (Amersham Biosciences).
H/D Exchange on ADP/ATP Carrier Complexes
The ADP/ATP carrier-inhibitor complexes were purified and concentrated up to 5 mg of protein/ml. The HDX reaction was initiated by diluting protein 10-fold in a 4 °C deuterated buffer containing 10 mm MOPS-NaOH, pH 6.8, 10 mm NaCl, 1 mm EDTA. The kinetics of H/D exchange was followed over a 10,000-s period by sequentially withdrawing 80-μl samples. Aliquots were immediately added to 20 μl of quenching buffer (1 m glycine-HCl, pH 2.5), rapidly mixed, and flash-frozen in liquid nitrogen. Samples were stored in liquid nitrogen until analysis.
High Pressure Liquid Chromatography
Peptides obtained by on-line protein digestion were loaded onto a Peptide MicroTrap column (Michrom Bioressources). Samples were desalted, and detergent and lipids were removed, as previously described (20). Peptides were separated on a reversed phase C18 column (1 × 100 mm, Jupiter; Phenomenex), as previously described (20). To minimize back-exchange, the valves, trap column, and analytical column were cooled to 0 °C by immersion in an ice-water bath. Protein mass was measured as follows. 200 pmol of carrier solubilized in 0.05% n-dodecyl-β-d-maltoside were loaded onto a Protein MicroTrap column (Michrom Bioressources). The sample was desalted for 1 min with 200 μl of water. Protein and detergent were then eluted onto a C8 RP-column (1 × 100 mm, Jupiter, Phenomenex) for separation.
MS Analyses
Liquid chromatography tandem MS (LC ESI-MS/MS) analyses were performed on an ion trap mass spectrometer (Esquire 3000+, Bruker Daltonics). The HPLC system was connected directly to the spectrometer's ESI source through a T-piece splitting device. The settings used were as previously described (20). Data were processed using DataAnalysis 3.0 (Bruker), and MASCOT was used to match MS/MS spectra to a single-protein data base containing the ScAnc2p sequence. A liquid chromatography electrospray time-of-flight MS (LC ESI-ToF-MS) 6210 system (Agilent Technologies) was used to analyze deuterated samples and for accurate mass measurements. Isotope envelopes were extracted using Mass Hunter Qualitative Analyses software (Agilent Technologies), and the centroid of the isotopic distribution was measured using MagTran software (Zhongqi Zhang, Amgen, Thousand Oaks, CA) (21). Carrier mapping was represented using DrawMap (MS Tools (36)).
RESULTS
Molecular Weight of ScAnc2p
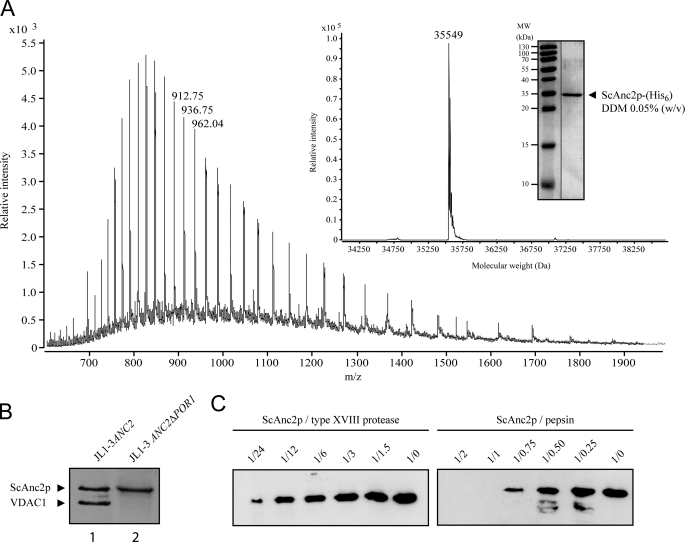
The exact number of amino acids and whether any post-translational modifications are present on the mature ScAnc2p have not yet been determined. LC ESI-ToF-MS was used to determine the accurate molecular weight of ScAnc2p according to the method described under “Experimental Procedures.” HTP-purified ScAnc2p preparations contain several contaminant proteins, in particular mitochondrial porin isoform one (VDAC1) and large amounts of ergosterol and detergent (22). We, therefore, purified a modified form of the yeast carrier to which a six-histidine extension had been added at the C terminus (ScAnc2p-His6). This allowed contaminants to be eliminated and decreased the amount of detergent present in the final purified protein solution, thus facilitating MS analysis. From the deconvoluted mass spectrum, purified ScAnc2p-His6 exhibits a molecular mass of 35,549 Da (Fig. 1A), which is in agreement with the calculated mass of 35,547 Da for ScAnc2p-His6, where the N-terminal methionyl residue has been removed and the new N-terminal residue acetylated, as previously suggested (23).
FIGURE 1.
Analysis of the ScAnc2p primary amino acid sequence and proteolysis in HDX-compatible conditions. A, LC ESI-ToF-MS analysis of ScAnc2p-His6 in 0.05% (w/v) n-dodecyl-β-d-maltoside (DDM) solution is shown. Inset, shown is a deconvoluted spectrum of the carrier purified from crude mitochondrial extracts from JL1–3 ANC2(His6) strain. In the gel, proteins were analyzed by SDS-PAGE (12.5% acrylamide) and revealed by Coomassie Blue staining. B, ScAnc2p purification from crude mitochondrial extracts of JL1–3ANC2 (lane 1) or JL1–3Δpor1-ANC2 (lane 2) strains were analyzed by SDS-PAGE as for A. C, ScAnc2p proteolysis is shown. Lysates from pepsin and type XVIII digestion were analyzed by SDS-PAGE as for A. Fragments were revealed by Western blot with anti-SDS-ScAnc2p. Digestion was performed in acidic buffer for 2 min at 4 °C, with 2 μg of ScAnc2p at a substrate/protease ratio between 1:0.25 and 1:2 (w/w) for pepsin or between 1:1.5 and 1:24 (w/w) for type XVIII protease.
Purification and Digestion of ScAnc2p
As stated above, ScAnc2p, purified by a single chromatographic step on HTP, contained VDAC1 as a major contaminant (Fig. 1B, lane 1). For structural and conformational analyses, a highly purified, unmodified ScAnc2p (without His6 tag) form is necessary. To produce this, we used the JL1–3ANC2Δpor1 strain in which the genes encoding VDAC1 and Ancp isoforms 1 and 3 (ScANC1 and ScANC3) were disrupted (see “Experimental Procedures”). ScAnc2p in complex with CATR or BA was isolated in Triton X-100 from this yeast strain as a pure protein (Fig. 1B, lane 2). The detergent used is relevant for further HDX-MS studies, as shown in Rey et al. (20). Two different acidic proteases were tested for in-solution digestion of ScAnc2p as a CATR complex. The corresponding lysates were analyzed by Western blot using antibodies raised against SDS-treated ScAnc2p (Fig. 1C). Proteases were used at an enzyme/ScAnc2p ratio (w/w) of between 0.25 and 2 for commercial pepsin and between 1.5 and 24 for the home-produced recombinant type XVIII protease (10). The results clearly showed pepsin to be more efficient for ScAnc2p digestion, as no residual fragment was immunodetected from an enzyme/ScAnc2p ratio (w/w) of 1 (Fig. 1C). Type XVIII protease was also able to almost completely digest ScAnc2p, although only at an enzyme/ScAnc2p ratio (w/w) of 24. Similar results were obtained with the BA-carrier complex (not shown). Because of its more efficient digestion of ScAnc2p, pepsin was used in subsequent experiments. Our previous studies of soluble and membrane-bound proteins showed immobilized proteases to offer several advantages in HDX-MS experiments (6, 10, 20). In addition to higher protease stability and the absence of auto-digestion, immobilized proteases provide a much higher local protein concentration. This leads to a better digestion efficiency and makes it possible to use chaotropic reagents such as guanidinium chloride. Thus, for fast, efficient, and highly reproducible digestion of ScAnc2p in complex with CATR or BA, we performed on-line proteolysis using a column packed with immobilized pepsin.
Peptide Mapping of ScAnc2p Covers Half the Carrier Sequence
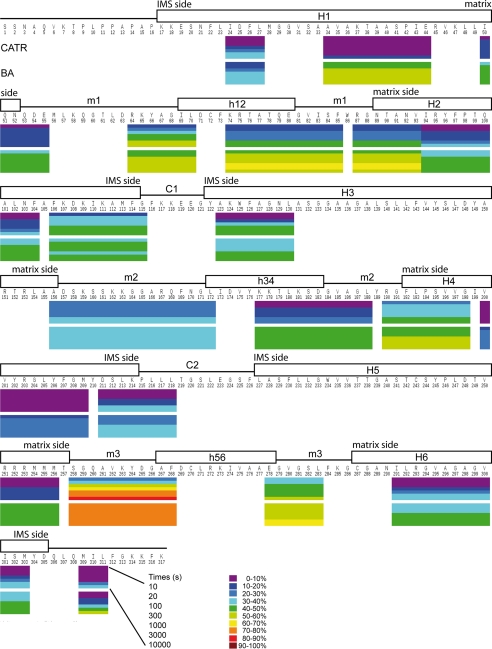
ScAnc2p as a CATR- or BA-carrier complex was digested as described above. Lysates were analyzed by LC ESI-MS/MS. The MS/MS spectra allowed identification of 55 peptides, homogeneously covering 76% of the protein sequence (data not shown). Only the N-terminal region of ScAnc2p was not covered by any peptides. This is probably due to the presence of numerous prolyl residues that hamper carrier digestion. In contrast, in HDX-MS experiments, protein coverage fell to 51% (20 peptides). This was due to a combination of low signal-to-noise ratio and overlapping signals (Fig. 2). Because of this, helices H3, H5, and H6 could only be analyzed based on one peptide per region. We, therefore, attempted to improve protein coverage by adapting our digestion process. Combinations of pepsin and type XVIII protease were tested for in-solution digestion of deuterated proteins, but no improvement in coverage was noted (data not shown). Similarly, a column packed with immobilized recombinant type XVIII protease did not improve protein coverage in HDX-MS experiments (data not shown).
FIGURE 2.
HDX profile of ScAnc2p in complex with CATR or BA mapped to its sequence after exchange for 10, 30, 100, 300, 1000, 3000, and 10000 s. The deuterium levels of each segment at each time point are shown by color-coding, from purple (≤10% deuteration) through to maroon (>90%) as indicated by the key. Putative secondary structures for ScAnc2p were transmembrane helices (H1 to H6), matrix loops (m1, m2, and m3) encompassing matrix helices (h12, h34, and h56), and IMS loops (C1 and C2), deduced from the three-dimensional structure of the bovine carrier bound to CATR (5). These are indicated above the sequence.
HDX-MS Analysis of ScAnc2p Reveals Typical Conformational Dynamics
The conformational dynamics of Triton X-100- extracted ScAnc2p were investigated by HDX-MS. To monitor local HDX kinetics for individual parts of ScAnc2p in complex with CATR or BA, the protein was digested after between 10 and 10,000 s of incubation in a deuterium-containing buffer. Results for each peptide region are presented in Fig. 2. All the regions analyzed exhibited inhibitor-sensitive deuterium accessibility. Moreover, deuteration was always higher and/or faster for the ScAnc2p-BA complex than for the ScAnc2p-CATR complex (Fig. 2), suggesting that overall and throughout the protein sequence, the amidic protons of the BA conformer were more exchangeable with deuterium than those of the CATR conformer. For both complexes analyzed, deuteration data recorded at the end of the experiment are presented in Fig. 3.
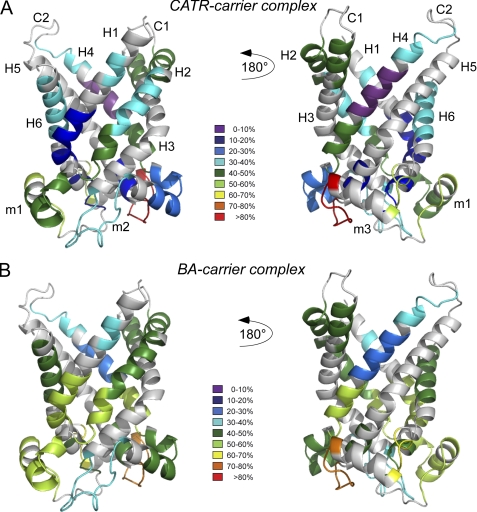
FIGURE 3.
Deuteration data after exchange for 10000 s mapped on a three-dimensional model of ScAnc2p. A, shown is a ribbon diagram of the CATR-carrier complex. B, shown is a ribbon diagram of the BA-carrier complex. The three-dimensional model was generated using the Protein Homology/analogY Recognition Engine (PHYRE, Version 0.2) server (35) with the structure of bAnc1p in complex with CATR (PDB ID 1OKC) as a template. Modeling of the ScAnc2p three-dimensional structure covers residues 2–308. The deuterium levels of each segment are shown by color-coding, from purple (≤10% deuteration) through to red (>80%) as indicated by the key. For the putative secondary structures for ScAnc2p, see the Fig. 2 legend.
Membrane Topography of the Matrix Loops in ScAnc2p Complexed Either to CATR or BA as Assessed by HDX-MS Experiments
In the three-dimensional structure of bovine Anc1p, loops m1 to m3 are positioned parallel to the membrane surface, where they appear to strengthen the carrier closed conformation on the matrix side when bound to CATR (5). In our studies of ScAnc2p, loop m1 was covered mainly by regions 64–70 and 74–85, loop m2 by regions 156–172 and 177–188, and loop m3 by regions 258–268 and 278–283 (Fig. 2). When complexed to CATR, the amide protons of loops m1 and m3 were generally more readily exchangeable with deuterium than those of loop m2 (Fig. 2). Region 258–268, located within loop m3, was the most extensively deuterated (81% of HDX at the end of the experiment). In contrast, region 177–188, from loop m2, was the most protected from deuteration (27% of HDX at the end of the experiment). In the BA-carrier complex, amidic protons of the matrix loops exhibited a greater level of deuterium exchange than in the CATR-carrier complex (Figs. 2 and 3). Interestingly, regions 64–70, 156–172, and 258–268 (located mainly at the N-terminal coil of matrix helices) were deuterated faster in the BA-carrier complex than in the CATR-carrier complex (17, 25, and 27% of HDX in presence of CATR versus 41, 38, and 76% of HDX in the presence of BA, respectively, after 10 s of incubation). However, the level of deuteration recorded at the end of the experiment was inhibitor-insensitive (52 versus 51, 36 versus 38, and 81 versus 80% of HDX, respectively) (Figs. 2 and 3). In contrast, regions 74–85, 177–188, and 278–283, which encompass a large part of the C-terminal coil of the matrix helices, did not behave in the same way. These regions were more rapidly and extensively deuterated in the presence of BA at the end of the experiment (42, 27, and 53% of HDX in presence of CATR versus 59, 45, and 63% of HDX in the presence of BA) (Figs. 2 and 3). Overall, these results were consistent with better exposure of the matrix loops to the solvent in the BA conformer than in the CATR conformer. This behavior may indicate that the carrier opens toward the matrix, as previously hypothesized for bAnc1p (6). However, the lower level of deuteration measured for loop m2 in the CATR-carrier complex is difficult to interpret when considered alongside the other two loops (Fig. 3). Indeed, all these loops are predicted to be symmetrically positioned within the carrier, directed toward the matrix.
Conformation-dependent Accessibility of the Carrier Cavity Analyzed by Local HDX in the Presence of CATR or BA
From the structural prediction based on bovine Anc1p, helix H1 of ScAnc2p is covered by regions 24–27, 34–44, and 50–55; helix H2 is covered by regions 87–93, 94–104, and 106–115; helices H3, H5, and H6 are covered only by regions 123–130, 251–256, and 291–303, respectively, whereas regions 190–198, 200–209, and 211–218 fully cover helix H4 (Fig. 2). In the CATR-carrier complex, helix H1 initially exhibited a very low level of deuterium incorporation (4–4% of HDX after 300 s of incubation, depending on the region analyzed). Moreover, the regions located in the C-terminal half of helix H1 (regions 34–44 and 50–55) were strongly protected from deuteration throughout the experiment (14 and 16% of HDX at the end, respectively), with the intrahelical region 34–44, encompassing one part of the first MCF motif, being the most protected. Thus, these results suggest that a major part of helix H1 was weakly accessible to solvent in the CATR conformation. This behavior was also observed with amide protons from region 251–256, encompassing one part of the third MCF motif (helix H5). Indeed, these protons were also less exchangeable, with rates ranging from 7 to 12% of HDX over the duration of the experiment. Amidic protons from intrahelical regions 94–104 (helix H2), 200–209 (helix H4), and 291–303 (helix H6) also allowed weak exchange at the beginning of the HDX experiment (17, 7, and 14% of HDX after 300 s of incubation, respectively). Moreover, the 200–209 region remained strongly protected from deuterium exchange, showing only 6–10% incorporation over the course of the experiment. In the CATR-carrier complex, regions 87–93 and 190–198 (corresponding to the N-terminal extremities of helices H2 and H4) are thought to face the matrix. These regions were more rapidly deuterated and presented higher incorporation levels at the end of the experiment (42 and 41% of HDX, respectively) than the corresponding intrahelical regions 94–104 and 200–209 (33 and 10% of HDX, respectively), located deeper inside the cavity. Similar behavior was observed for regions 106–115 and 211–218 (48 and 40% of HDX at the end of the experiment, respectively). These regions correspond to the C-terminal extremities of helices H2 and H4, respectively. All these results are consistent with better exposure of these regions to solvent. However, amidic protons from region 211–218 were significantly protected from deuteration at the beginning of the HDX experiment (only 13% of HDX after 300 s of incubation), and those from region 309–311, covering the C terminus, which is predicted to be outside the mitochondrial inner membrane, were surprisingly also less exchangeable at the beginning of the experiment (7% of HDX after 300 s of incubation). Taken together, all these results suggest that the major part of the carrier cavity was not very accessible to solvent in the CATR-carrier complex; this is in contrast to observations with the bovine carrier (6).
The largest differences (relative to the CATR-carrier complex) were observed for amidic protons from regions 34–44 and 251–256 (see below), from helices H1 and H5, and from regions 50–55, 87–93, 94–104, 190–198, covering the lower part of the cavity toward the matrix (Figs. 2 and 3). These protons were more exchangeable in the BA-carrier complex (56, 47, 48, 59, 43, and 57% of HDX at the end of the experiment, respectively) than in the CATR-carrier complex (14, 7, 16, 42, 33, and 41% of HDX at the end of the experiment, respectively). These results would appear to indicate better accessibility of the BA conformation to the solvent from the matrix side (see above). Surprisingly, some regions from the upper part of the cavity were more rapidly deuterated in the BA-carrier complex than in the CATR-carrier complex (regions 106–115, 123–130, and 211–218); however, overall the final level of deuterium incorporation was inhibitor-insensitive (Figs. 2 and 3). Other regions in the same area were rapidly and slightly more extensively deuterated (regions 200–209, 291–303, and 309–311) in the BA-carrier complex than in the CATR-carrier complex (Figs. 2 and 3). These results are discussed below.
Deuterium Accessibility Changes within the Ancp Signature Sequence
In the three-dimensional -structure of the bovine carrier bound to CATR, the methionine cluster of the Ancp signature sequence (RRRMMM) occupies a bulky volume that could control access to the cavity from the matrix side, acting as a plug during the transport mechanism (5). The corresponding region in ScAnc2p (251–256) was more rapidly and extensively deuterated in the presence of BA than CATR (7% of HDX with CATR versus 47% of HDX with BA at 10,000 s). The role of the arginine triplet has been investigated in ScAnc2p (24, 25); we, therefore, decided to further investigate the functional role of the triplet of methionyl residues in this sequence. Variant proteins with the three methionyl residues replaced by alanyl residues, simultaneously, in pairs or singly were produced and biochemically characterized.
Growth Phenotypes on a Non-fermentable Carbon Source Show Different Roles for Each Methionyl Residue in the MMM Cluster
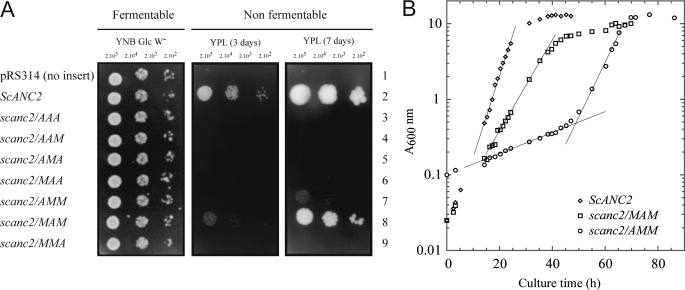
When the three methionyl residues were simultaneously replaced by alanyl residues, the resulting carrier, ScAnc2pAAA, could not rescue JL1Δ2Δ3u− growth on medium containing a non-fermentable carbon source (YPL) (Fig. 4A). Thus, assuming that there is no impairment in the import of the carrier into the mitochondrial inner membrane, one, two, or all three of these residues may play a crucial role in nucleotide transport. When only two methionyl residues were replaced at a time (ScAnc2pAAM, ScAnc2pAMA, and ScAnc2pMAA) JL1Δ2Δ3u− growth on YPL was not restored either (Fig. 4A). Mutation of one methionyl residue at a time (ScAnc2pAMM, ScAnc2pMAM, and ScAnc2pMMA) clearly demonstrated that each of these residues has a different role in the transport mechanism (Fig. 4A). Indeed, scanc2/MMA was unable to develop on YPL, although scanc2/MAM displayed a slow growth phenotype with a doubling time of 5 h and A600 nm = 10 for the saturation phase, whereas for ScANC2 the doubling time was 2.5 h, with A600 nm = 13 for the saturation phase (Fig. 4B and Table 2). In the case of scanc2/AMM, growth on solid YPL could only be measured after 7 days. Furthermore, in liquid YPL, the 2-phase growth curve was striking; for 50–70 h, the doubling time was ∼30 h; then A600 nm increased rapidly, corresponding to a doubling time of 4 h (Fig. 4B). Interestingly, the A600 nm value at the growth plateau was 12, which equates to only a minor decrease in the biomass yield compared with the wild types strain (Fig. 4B). These results suggested that the mitochondrial metabolism in scanc2/AMM could reach the same efficiency as in the wild type strain but only after significant metabolic adaptation. Strikingly, this adaptation completely disappeared when the strain was inoculated into fresh liquid YPL from the stationary phase (results not shown). Therefore, we looked for occurrence of intragenic or extragenic suppressors of the M254A mutation.
FIGURE 4.
Yeast growth phenotype for strains carrying mutations of the methionyl residues of the MMM triplet. Strain JL1Δ2Δ3u− was transformed with plasmid pRS314 containing no gene (line 1), wild type ScANC2 gene (line 2), or scanc2 genes carrying mutations in the MMM cluster (lines 3–9). Mutations replaced methionyl residues with alanyl. The three positions were mutated simultaneously (scanc2/AAA), in pairs (scanc2/AAM, scanc2/AMA, scanc2/MAA), or singly (scanc2/AMM, scanc2/MAM, scanc2/MMA) under the control of ScANC2 regulatory sequences. A, yeast transformants were isolated and inoculated in complete, tryptophan-free liquid minimal medium containing 2% glucose as carbon source (YNB Glc W−). When cultures reached the log phase, cells were diluted to obtain 2 ×105 to 2 ×103 cells per 4 μl before spotting onto YNB Glc W− (glucose) or rich lactate-containing medium (YPL) plates. Plates were incubated for 3 days (YNB Glc W−) or between 3 and 7 days (YPL) at 28 °C. B, shown are growth curves for strains ScANC2, scanc2/MAM, and scanc2/AMM in rich liquid medium (YPL). Cultures were inoculated with cells previously grown on YNB Glc W− medium to obtain an initial A600 nm = 0.1. Growth was monitored by measuring absorbance at 600 nm (A600 nm) for ∼45, 65, and 85 h, depending on the yeast strain. Except for scanc2/AMM, which was a revertant strain, values are the means of at least three independent experiments.
TABLE 2.
scanc2/AMM mutants growing on a non-fermentable carbon source due to the emergence of compensatory second-site mutations in the scanc2/AMM ORF
TMS, transmembrane segment.
| Revertant strains | ORF position | Condon changes | Amino acid changes | Location |
|---|---|---|---|---|
| scanc2/AMM, G29C | 88 | GGT → TGT | G29C | TMS1 |
| scanc2/AMM, S32N | 98 | AGT → AAT | S32N | TMS 1 |
| scanc2/AMM, A101T | 304 | GCT → ACT | A101T | TMS 2 |
| scanc2/AMM, F104L | 315 | TTC → TTG | F104L | TMS 2 |
| scanc2/AMM, A105T | 316 | GCC → ACC | A105T | TMS 2 |
| scanc2/AMM, A105V | 317 | GCC → GTC | A105V | TMS 2 |
| scanc2/AMM, Y206H | 619 | TAC → CAC | Y206H | TMS 4 |
| scanc2/AMM, G208S | 625 | GGT → AGT | G208S | TMS 4 |
| scanc2/AMM, G297S | 892 | GGT → AGT | G297S | TMS 4 |
| scanc2/AMM, A298S | 895 | GCT → TCT | A298S | TMS 6 |
| scanc2/AMM, I301T | 905 | ATC → ACC | I301T | TMS 6 |
| scanc2/AMM, L307P | 923 | CTG → CCG | L307P | C terminus |
Compensatory Mutations in ScAnc2pAMM Can Rescue scanc2/AMM Growth on a Non-fermentable Carbon Source
After overnight culture in a selective synthetic liquid minimal medium, scanc2/AMM cells were plated onto YPL and incubated for 10 days at 28 °C to select for regain-of-function revertants. We isolated 30 clones and sequenced their scanc2 genes. None of them corresponded to true revertants, but 17 contained an intragenic second-site mutation, corresponding to 12 unique mutations in 11 different codons (Table 2). Their locations on the hypothetical three-dimensional structure of ScAnc2p are shown in Fig. 5. All the mutants are on the same side of the membrane (IMS) and are distant from their parent mutation both in terms of amino acid sequence and the putative three-dimensional structure. To confirm that the observed phenotypes resulted specifically from the mutations identified in the scanc2/AMM gene, all the corresponding scanc2/AMM genes were subcloned into the original expression vector and used to back-transform JL1Δ2Δ3u−. The back-transformed cells exhibited similar growth properties on YPL to ScANC2 (not shown). Therefore, we conclude that the phenotypes observed are due solely to the occurrence of compensatory mutations in the scanc2/AMM gene when cells were grown on a non-fermentable carbon source. The scanc2/MMA mutant exhibited a permanent growth defect in YPL, indicating that the M256A mutation is too deleterious to be compensated for by a single compensatory amino acid change.
FIGURE 5.
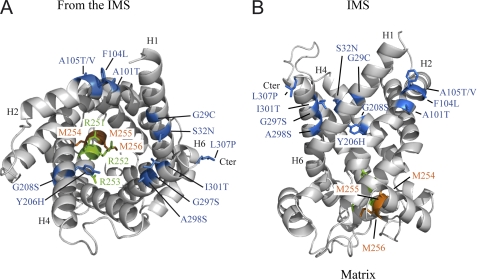
Location of second-site mutations on a three-dimensional model of ScAnc2p generated as described in the legend of the Fig. 3. Shown is a ribbon diagram of the carrier viewed from the IMS (A) and viewed from the side (B). The Ancp signature sequence is highlighted, with methionyl residues 254 (parental mutation), 255, 256 indicated in orange and arginyl residues 251–253 indicated in green. The 12-s-site mutations detected after selection of revertant strains from yeast bearing the M254A mutation are shown in blue. The putative transmembrane segments, H1, H2, H4, H6, and the C-terminal extremity (Cter) encompassing the second-site mutations are indicated. Ribbon diagrams were drawn using PyMOL v0.99 (DeLano Scientific LLC).
Mutations in the MMM Cluster Do Not Prevent ScAnc2p Translocation to Mitochondria
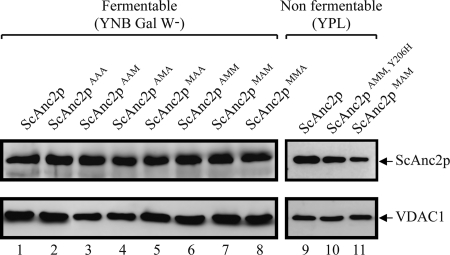
Growth impairment on YPL associated with mutations in the MMM cluster may be related to lower ScAnc2p levels in the MIM. Therefore, we evaluated carrier content in mitochondrial lysates by Western blot. Yeast VDAC1 was used as a control of mitochondrial protein loading (Fig. 6). All strains were grown in a medium containing galactose, a fermentable carbon source. ScAnc2p could be immunodetected in every transformant, and, globally, relative amounts of all variants were similar to levels of the wild type carrier (Fig. 6, lanes 1–8). Thus, the growth defect observed on YPL does not appear to be correlated to a defect in importing the mutated carriers but, rather, to impairment of ADP/ATP exchange activity. The mitochondrial content of ScAnc2pMAM was also evaluated after growing yeast on YPL. In these conditions, ScAnc2pMAM is less imported and/or less stable in the mitochondria than the wild type carrier (Fig. 6, lanes 9 and 11). The regain-of-function revertants showed a normal level of ScAnc2p production, as observed for the double mutant M254A/Y206H (Fig. 6, lane 10).
FIGURE 6.
Relative levels of mitochondrial wild type and variant ScAnc2p. Mitochondrial protein extracts (20 μg/lane) were prepared from wild type JL-ScANC2 (lanes 1 and 9) and variant strains (lanes 2–8, 10, and 11). Strains were cultured on either fermentable (lanes 1–8) or non-fermentable (lanes 9–11) carbon sources. SDS-PAGE (12.5% acrylamide) and Western blotting were performed. Proteins were detected using polyclonal antibodies directed against SDS-treated Anc2p (1:1500 final dilution). Yeast VDAC1, revealed using polyclonal anti-VDAC1 antibodies (1:1000 final dilution), was used as a loading control. Immune complexes were detected with ECL. Identified polypeptides are indicated by arrows.
Each Residue of the Methionine Cluster Is Involved in ATR Binding
ATR is a very specific inhibitor of ADP/ATP transport, which binds with high affinity to Ancp. Determining the maximum number of ATR binding sites allows quantification of the amount of functional Ancp present in the membrane. Because ATR blocks Ancp in a conformation thought to be involved in adenine nucleotide transport, a high affinity for ATR reflects functional protein folding. We performed [3H]ATR binding experiments with mitochondria isolated from the strains expressing the methionine cluster variants (Table 3). When cultivated in the presence of a fermentable carbon source such as galactose, mitochondria from ScAnc2 and scanc2/MAM strains present similar numbers of ATR binding sites at saturation (BmaxATR), binding ∼120 pmol·mg−1. However, when cells were cultivated on a non-fermentable carbon source, the BmaxATR value for the scanc2/MAM mitochondria was about one-third that of wild type (205 ± 30 versus 680 ± 80 pmol/mg of proteins, respectively), although cytochrome aa3 content did not vary significantly (data not shown). Similar data were obtained using 3′-O-naphthoyl-ADP or 6-O-naphthoyl-ATR as probes to quantify mitochondrial ScAnc2p (data not shown). Thus, the amount of ScAnc2pMAM variant in mitochondria could account for the phenotype observed with scanc2/MAM in YPL. In addition, compared with the wild type carrier, the ATR binding properties were dramatically modified for all variants. Indeed, the ATR dissociation constant (KDATR) for ScAnc2pMAM is 3-fold that of the wild type carrier (1130 ± 155 versus 375 ± 44 nm, respectively). All other variants were unable to bind ATR, suggesting that they may be incorrectly folded in the mitochondria, making ATR and probably nucleotide binding impossible. Interestingly, compensatory second-site mutations in ScAnc2pAMM can rescue ATR binding (Table 4).
TABLE 3.
Ligand binding data for carriers and kinetic parameters of ADP/ATP transport activity measured in mitochondria isolated from strains ScANC2 or scanc2/MAM
Cells were cultivated on liquid YPL medium.
| Strains | ScANC2 | scanc2/MAM |
|---|---|---|
| [3H]ATR bindinga | ||
| Bmax pmol/mg of proteins | 680 ± 80 | 205 ± 36 |
| Kd (nm) | 375 ± 44 | 1130 ± 155 |
| ADP/ATP exchangeb | ||
| Vmax (nmol ATP/min/mg if protein) | 1080 ± 127 | 247 ± 135 |
| kcat (min−1) | 1538 ± 214 | 1205 ± 497 |
| Km external ADP (μm) | 3.7 ± 2 | 12 ± 1 |
a [3H]ATR binding parameters are given as the mean of three independent experiments. Number of binding sites, Bmax, and Kd values were calculated from Scatchard plots of [3H]ATR binding data.
b Vmax and apparent Km for external free ADP were calculated from kinetic data using the Michaelis-Menten equation. kcat refers to carrier turnover, calculated based on the carrier content determined by [3H]ATR binding experiments, assuming that 1 mol of ATR binds to 1 mol of transport unit. Values are the means of three independent experiments.
TABLE 4.
Ligand binding data for carriers and kinetic parameters of ADP/ATP transport activity measured in mitochondria isolated from scanc2/AMM mutants growing on a non-fermentable carbon source due to the emergence of compensatory second-site mutations in the scanc2/AMM ORF
| Revertant strains | [3H]ATR bindinga |
ADP/ATP exchangeb |
|||
|---|---|---|---|---|---|
| Bmax | Kd | Vmax | Kcat | Km external ADP | |
| pmol/mg proteins | nm | nmol ATP/min/mg protein | min−1 | μm | |
| Scanc2/AMM, G29C | 498 ± 69 | 398 ± 49 | 640 ± 154 | 1258 ± 265 | 3.9 ± 3 |
| scanc2/AMM, S32N | 689 ± 55 | 458 ± 42 | 1150 ± 148 | 1685 ± 358 | 5.6 ± 2 |
| scanc2/AMM, A101T | 510 ± 78 | 259 ± 55 | 525 ± 89 | 1015 ± 198 | 3.5 ± 3 |
| scanc2/AMM, F104L | 480 ± 95 | 335 ± 72 | 656 ± 245 | 1352 ± 272 | 2.8 ± 4 |
| scanc2/AMM, A105T | 720 ± 89 | 468 ± 61 | 1025 ± 108 | 1459 ± 298 | 3.9 ± 3 |
| scanc2/AMM, A105V | 569 ± 45 | 325 ± 87 | 705 ± 105 | 1252 ± 165 | 3.7 ± 1 |
| scanc2/AMM, Y206H | 523 ± 95 | 550 ± 55 | 689 ± 78 | 1298 ± 309 | 4.8 ± 2 |
| scanc2/AMM, G208S | 563 ± 74 | 490 ± 69 | 925 ± 258 | 1608 ± 158 | 4.6 ± 3 |
| scanc2/AMM, G297S | 630 ± 35 | 295 ± 94 | 1089 ± 115 | 1751 ± 254 | 2.9 ± 2 |
| scanc2/AMM, A298S | 496 ± 86 | 369 ± 67 | 645 ± 103 | 1285 ± 304 | 5.3 ± 3 |
| scanc2/AMM, I301T | 587 ± 83 | 520 ± 45 | 630 ± 145 | 1052 ± 156 | 4.9 ± 3 |
| scanc2/AMM, L307P | 690 ± 36 | 430 ± 71 | 1115 ± 258 | 1598 ± 389 | 3.1 ± 4 |
a [3H]ATR binding parameters are given as the mean of three independent experiments. Number of binding sites, Bmax, and Kd values were calculated from Scatchard plots of [3H]ATR binding data.
b Vmax and apparent Km for external free ADP were calculated from kinetic data using the Michaelis-Menten equation. kcat refers to carrier turnover, calculated based on the carrier content determined by [3H]ATR binding experiments assuming that 1 mol of ATR binds to 1 mol of transport unit. Values are the means of three independent experiments.
Residues 254 and 256 Are Crucial to Nucleotide Translocation or Binding
The ADP/ATP transport rate for wild type ScAnc2p and the ScAnc2pMAM variant was measured using the luciferase/luciferin system with mitochondria isolated from the corresponding strains grown on YPL. As shown in Table 3, the Vmax value for ScAnc2pMAM was about 4.4 times lower than that of ScAnc2p (247 ± 135 versus 1080 ± 127 nmol ATP/min/mg of proteins, respectively). However, given the unchanged kcat value, we suggest that Met-255 does not participate in the translocation process per se. The apparent Km value of external free ADP for ScAnc2pMAM was 3.2-fold higher than for ScAnc2p (12 ± 1 versus 3.7 ± 2 μm, respectively), suggesting that Met-255 is involved in the ADP binding step. All these results could account for the reduced growth observed on YPL for scanc2/MAM. In contrast, Met-254 and Met-256 were crucial to either nucleotide translocation or binding as we were unable to measure any ADP/ATP transport activity on mitochondria freshly isolated from the corresponding strains (scanc2/AMM and scanc2/MMA) grown under fermentable conditions. But compensatory second-site mutations in ScAnc2pAMM can rescue nucleotide translocation or binding (Table 4).
DISCUSSION
The physiological role of Ancp is to export ATP from the mitochondria in exchange for cytosolic ADP. During the transport process, the carrier switches between two different conformations that can be blocked by two specific inhibitors: CATR and BA. We recently used HDX-MS to demonstrate that, on the matrix side, the bAnc1p-BA complex exhibits greater solvent accessibility than the bAnc1p-CATR complex. This corroborates data obtained using different approaches (6). To explain our results, we hypothesized that the cavity was closed on the IMS side and open toward the matrix in the bAnc1p-BA complex. The yeast Anc2p affords the opportunity to use site-directed mutagenesis to study the bAnc1p peptide regions presenting the highest levels of variability in terms of deuterium exchange as a function of the inhibitor used. As both proteins share a significant level of amino acid sequence identity, we also decided to analyze the conformational dynamics of the corresponding peptide regions in ScAnc2p by HDX-MS. This was a starting point before going on to further mutagenesis experiments to highlight the functional role of some residues in the ADP/ATP transport mechanism. The results were analyzed in the light of a predicted ScAnc2p three-dimensional structure model, based on the bAnc1p structure.
Like the bovine carrier, ScAnc2p exhibited inhibitor-dependent deuterium accessibility. These results corroborated those using chemical, enzymatic, and immunochemical reagents to label inhibitor-carrier complexes (18, 26–31). The matrix loops were more extensively deuterated in the presence of BA than in the presence of CATR (Figs. 2 and 3). This is a sign of their rearrangement, probably improving solvent accessibility of regions located in the lower part of the cavity. Indeed, the largest differences in deuteration levels were observed for regions encompassing parts of the MCF motifs located at the bottom of the cavity (Figs. 2 and 3). This behavior was similar to that observed with bAnc1p (6) and once again underlined the pivotal role played by the MCF motifs in the conformational dynamics of Ancp. Among other common features, deuterium exchange within the major part of helix H4 (regions 200–209 and 211–218), comprising the aromatic ladder, which is conserved in all Ancp sequences, was only weakly sensitive to the inhibitor used (this work and Ref. 6).
However, our results also highlight some differences between the yeast and bovine Ancp in detergent solution. Interestingly, during HDX-MS experiments with ScAnc2p in complex with CATR, the majority of loop m2 was protected from deuterium exchange, in contrast with the other two loops. Moreover, its N-terminal region (region 156–172) remained sparsely deuterated with both inhibitors (Figs. 2 and 3). This behavior was not observed with the bovine carrier (6). A role for the m2 loop in substrate binding has already been postulated by other authors for the yeast carrier in the membrane (23). They found it possible to photolabel peptides 182–190 with 2-azido-3′-O-naphthoyl-[β-32P]ADP. This is consistent with an intrusion of this loop into the cavity, as 2-azido-3′-O-naphthoyl-[β-32P]ADP cannot penetrate the mitochondrial inner membrane (23). It has also been shown that region 156–192 protruded into the cavity in both inhibitor complexes (18). All these results could explain how the m2 loop is protected against deuteration.
Besides, the deuterium incorporation levels for the upper part of the cavity were also different in yeast and bovine Ancp. Indeed, unlike what was observed with bAnc1p, this region was (i) overall weakly deuterated in the ScAnc2p-CATR complex (Figs. 2 and 3), whereas it was highly deuterated in the bAnc1p-CATR complex, in line with the carrier structure (6) and (ii) not so extensively deuterated in the ScAnc2p-BA complex (Figs. 2 and 3) versus weakly deuterated in the bAnc1p-BA complex (6). Thus, in detergent solution this part of the cavity remained globally better protected from deuteration in ScAnc2p than in bAnc1p. These observations could be linked to the intrusion of loop m2 into the cavity in both complexes, as demonstrated by Dahout-Gonzalez et al. (18). Therefore, it is conceivable that loop m2 protects the upper part of the cavity from deuteration whatever the complex analyzed. However, in the bAnc1p-CATR crystal structure, all matrix loops present similar folding, and none protrudes into the membrane or cavity (5). Taken together, these results suggest conformational differences between the m2 matrix loop in bAnc1p and ScAnc2p.
The RRRMMM motif is highly conserved in Ancp sequences (32), and deuterium exchange for this sequence was significantly affected by the inhibitor used. This observation supports a role for this motif in the conformational dynamics of ScAnc2p. The three-dimensional structure of bAnc1p revealed this sequence to be involved in both CATR binding inside the cavity and probably in the nucleotide transport mechanism (5). The role of the arginine triplet was previously investigated in ScAnc2p (24, 25). Arginyl residues are essential to transporter function, as for the three single R → I mutants, no intracellular oxidative ATP production was detected. However, the corresponding variants were correctly imported into mitochondria and conserved their ability to bind inhibitors (25). In this article we have investigated the function of the methionine triplet (Met-254–Met-255–Met-256), the second part of this signature sequence, by mutating each of the methionyl residues into alanine. We showed that Met-255 was not essential to nucleotide transport, as ScAnc2p and ScAnc2pMAM exhibited very similar kcat values. However, ScAnc2pMAM presented a significant decrease in ATR affinity, and the value of the apparent Km for external free ADP was 3 times higher than for the wild type carrier (Table 3). Because of a partial overlap in the ATR and ADP binding sites (7), our results suggest that Met-255 is involved in ADP binding, but the exact mechanism remains to be determined. Although there is no evidence of direct interaction between CATR and the methionine triplet in the bovine three-dimensional structure (5), we can note that Met-238 of bAnc1p (Met-255 of ScAnc2p) is oriented toward the cavity in the CATR conformation. Thus, this residue could contribute to stabilization of ADP in the cavity, assuming that the CATR binding site resembles that of the ADP. Mutating the methionyl residues simultaneously or in pairs prevented yeast growth on YPL. We showed that Met-254 and Met-256 are essential not only to nucleotide transport activity but also to ATR binding. The side chains of Met-237 and Met-239 in bAnc1p (Met-254 and Met-256 in ScAnc2p) are perpendicular to the vertical axis of the cavity in the CATR-carrier complex, probably acting as a steric plug toward the matrix as previously hypothesized (5). We suggest that mutations M254A or M256A modify the size of this plug, thus altering the cavity geometry and inhibiting ATR and probably adenylic nucleotide binding.
Variants of ScAnc2p that could not grow on YPL were subjected to selection for spontaneous suppressors, which were obtained only with ScAnc2pAMM. This suggests a major structural and/or functional alteration in mutants M256A. This also implies that the M254A mutation located on the matrix side propagates a structural effect that can be suppressed by mutations of amino acids on the other side of the carrier cavity. Those second site mutations are distant from the parental mutation by as many as 225 amino acids (G29C) and follow a discernable pattern. Nine of the changes occur in amino acid residues that are very well conserved in Ancp (Gly-29, Ala-101, Phe-104, Ala-105, Tyr-206, Gly-208, Gly-297, Ala-298) (4). Analysis of the mutations stresses the importance of particular residues with -OH groups (A101T, A105T, G208S, G297S, A298S, and I301T) or -SH groups (G29C) in restoring the ADP/ATP transport activity (Table 2). R251I mutation could not be reversed, whereas R252I and R253I led to a large number of second site suppressors (24). Nine of these changes (G29C, S32N, F104L, A105T, Y206H, G208S, A298S, I301T, and L307P) also occur in ScAnc2pAMM revertants. Consequently, mutations of Arg-252, Arg-253, and Met-254 of ScAnc2p into hydrophobic residues led to the appearance of the same second site suppressors, suggesting that these residues play a similar biochemical role in ADP/ATP transport.
Among the second site mutations, L307P is notable. Indeed, it is localized within the C-terminal extremity of ScAnc2p, which we have recently shown to be involved in nucleotide recognition and binding steps (15). Thus, mutation L307P, localized outside the cavity, would affect the conformational state of the C-terminal extremity of the carrier by offsetting the negative perturbation induced by the M254A mutation, making nucleotide binding possible. This phenomenon could also be an indicator of cooperation between different, non-neighboring regions of the carrier, which could be involved in ADP/ATP binding. The A105T second-site mutant is also very interesting. Indeed, Ala-105 is strictly conserved in all the Ancp sequences (4), and its mutation into aspartic acid is involved in autosomal dominant progressive external ophthalmoplegia, a human neuromuscular degenerative mitochondrial disease (33). Recently, we demonstrated in yeast that this mutation leads to complete inactivation of carrier activity (34). However, the A → D mutation can be suppressed by secondary mutation of the first methionyl residue of the RRRMMM motif, replacing it with a lysyl residue (34). These results corroborate those presented above and confirm the key role of Met-254 in the ADP/ATP transport mechanism.
As a conclusion of our studies, the yeast and bovine Ancp present different conformational dynamics in detergent solution, although they share significant sequence identity (47%). First, unlike bAnc1p, ScAnc2p exhibits a lower level of deuterium exchange in the m2 loop and in the upper part of the cavity with both inhibitors. This part of the cavity is oriented through the IMS. The reason for this difference between yeast and bovine Ancp is not clear. But the yeast results are in support of the postulated intrusion of the m2 loop into the cavity. Nonetheless, the conformational changes occurring between both conformations affect the yeast carrier as a whole, as shown for bAnc1p. Second, focusing our studies on the methionine triplet of the RRRMMM motif, which is common to all Ancp, we suggest that the differences in deuterium exchange caused by either CATR or BA binding could be linked to the functional dynamics of ScAnc2p. Indeed, we have shown that the methionine triplet contributes mainly to the ADP/ATP binding step. Finally, the fact that a mutation located at the bottom of the cavity on the matrix side can be compensated for by a second mutation on top of the cavity on the IMS side and vice versa (this work and Ref. 34) shows a real synergy between the two sides of the carrier during nucleotide transport. Moreover, the occurrence of numerous second site mutations restoring the protein function highlights the dynamic structure of the Ancp cavity and strengthens the results obtained by HDX-MS experiment.
Acknowledgments
We thank Dr. Gérard Klein and Dr. Gérard Brandolin for constructive discussions.
This work was supported by grants from the Université Joseph Fourier, the French CNRS, and the Commissariat à l'Energie Atomique.
A. Le Saux, unpublished data.
- Ancp
- ADP/ATP carrier
- bAnc1p
- bovine isoform 1 Ancp
- ScAnc2p
- yeast isoform 2 Ancp
- CATR
- carboxyatractyloside
- ATR
- atractyloside
- BA
- bongkrekic acid
- IMS
- intermembrane space
- HTP
- hydroxylapatite
- VDAC1
- isoform one of the yeast mitochondrial porin
- HDX
- hydrogen/deuterium exchange
- ESI
- electrospray ionization
- MCF
- mitochondrial carrier family
- YPL
- yeast peptone liver.
REFERENCES
- 1. Klingenberg M. (2008) Biochim. Biophys. Acta 1778, 1978–2021 [DOI] [PubMed] [Google Scholar]
- 2. Walker J. E., Runswick M. J. (1993) J. Bioenerg. Biomembr. 25, 435–446 [DOI] [PubMed] [Google Scholar]
- 3. Brandolin G., Le Saux A., Trézéguet V., Vignais P. V., Lauquin G. J. M. (1993) Biochem. Biophys. Res. Commun. 192, 143–150 [DOI] [PubMed] [Google Scholar]
- 4. Dahout-Gonzalez C., Nury H., Trézéguet V., Lauquin G. J. M., Pebay-Peyroula E., Brandolin G. (2006) Physiology 21, 242–249 [DOI] [PubMed] [Google Scholar]
- 5. Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G. J. M., Brandolin G. (2003) Nature 426, 39–44 [DOI] [PubMed] [Google Scholar]
- 6. Rey M., Man P., Clémençon B., Trézéguet V., Brandolin G., Forest E., Pelosi L. (2010) J. Biol. Chem. 285, 34981–34990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trézéguet V., Pélosi L., Lauquin G. J. M., Brandolin G. (2008) J. Bioenerg. Biomembr. 40, 435–443 [DOI] [PubMed] [Google Scholar]
- 8. Brandolin G., Meyer C., Defaye G., Vignais P. M., Vignais P. V. (1974) FEBS Lett. 46, 149–153 [DOI] [PubMed] [Google Scholar]
- 9. Lauquin G. J., Vignais P. V. (1976) Biochemistry 15, 2316–2322 [DOI] [PubMed] [Google Scholar]
- 10. Rey M., Man P., Brandolin G., Forest E., Pelosi L. (2009) Rapid Commun. Mass Spectrom. 23, 3431–3438 [DOI] [PubMed] [Google Scholar]
- 11. Cohen S. N., Chang A. C., Hsu L. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 2110–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drgon T., Sabová L., Nelson N., Kolarov J. (1991) FEBS Lett. 289, 159–162 [DOI] [PubMed] [Google Scholar]
- 13. De Marcos Lousa C., Trézéguet V., Dianoux A. C., Brandolin G., Lauquin G. J. M. (2002) Biochemistry 41, 14412–14420 [DOI] [PubMed] [Google Scholar]
- 14. Fiore C., Trézéguet V., Roux P., Le Saux A., Noël F., Schwimmer C., Arlot D., Dianoux A. C., Lauquin G. J. M., Brandolin G. (2000) Protein Expr. Purif. 19, 57–65 [DOI] [PubMed] [Google Scholar]
- 15. Clémençon B., Rey M., Dianoux A. C., Trézéguet V., Lauquin G. J. M., Brandolin G., Pelosi L. (2008) J. Biol. Chem. 283, 11218–11225 [DOI] [PubMed] [Google Scholar]
- 16. Le Saux A., Roux P., Trézéguet V., Fiore C., Schwimmer C., Dianoux A. C., Vignais P. V., Brandolin G., Lauquin G. J. M. (1996) Biochemistry 35, 16116–16124 [DOI] [PubMed] [Google Scholar]
- 17. Gietz D., St Jean A., Woods R. A., Schiestl R. H. (1992) Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dahout-Gonzalez C., Ramus C., Dassa E. P., Dianoux A. C., Brandolin G. (2005) Biochemistry 44, 16310–16320 [DOI] [PubMed] [Google Scholar]
- 19. Dassa E. P., Dahout-Gonzalez C., Dianoux A. C., Brandolin G. (2005) Protein Expr. Purif. 40, 358–369 [DOI] [PubMed] [Google Scholar]
- 20. Rey M., Mrázek H., Pompach P., Novák P., Pelosi L., Brandolin G., Forest E., Havlícek V., Man P. (2010) Anal. Chem. 82, 5107–5116 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Z., Marshall A. G. (1998) J. Am. Soc. Mass Spectrom 9, 225–233 [DOI] [PubMed] [Google Scholar]
- 22. Knirsch M., Gawaz M. P., Klingenberg M. (1989) FEBS Lett. 244, 427–432 [DOI] [PubMed] [Google Scholar]
- 23. Dianoux A. C., Noël F., Fiore C., Trézéguet V., Kieffer S., Jaquinod M., Lauquin G. J. M., Brandolin G. (2000) Biochemistry 39, 11477–11487 [DOI] [PubMed] [Google Scholar]
- 24. Nelson D. R., Douglas M. G. (1993) J. Mol. Biol. 230, 1171–1182 [DOI] [PubMed] [Google Scholar]
- 25. Nelson D. R., Lawson J. E., Klingenberg M., Douglas M. G. (1993) J. Mol. Biol. 230, 1159–1170 [DOI] [PubMed] [Google Scholar]
- 26. Iwahashi A., Kihira Y., Majima E., Terada H., Yamazaki N., Kataoka M., Shinohara Y. (2006) Mitochondrion 6, 245–251 [DOI] [PubMed] [Google Scholar]
- 27. Kihira Y., Majima E., Shinohara Y., Terada H. (2005) Biochemistry 44, 184–192 [DOI] [PubMed] [Google Scholar]
- 28. Kihira Y., Ueno M., Terada H. (2007) Biol. Pharm. Bull. 30, 885–890 [DOI] [PubMed] [Google Scholar]
- 29. Boulay F., Vignais P. V. (1984) Biochemistry 23, 4807–4812 [DOI] [PubMed] [Google Scholar]
- 30. Brandolin G., Boulay F., Dalbon P., Vignais P. V. (1989) Biochemistry 28, 1093–1100 [DOI] [PubMed] [Google Scholar]
- 31. Marty I., Brandolin G., Gagnon J., Brasseur R., Vignais P. V. (1992) Biochemistry 31, 4058–4065 [DOI] [PubMed] [Google Scholar]
- 32. Brandolin G., Le Saux A., Trézéguet V., Lauquin G. J. M., Vignais P. V. (1993) J. Bioenerg. Biomembr. 25, 459–472 [DOI] [PubMed] [Google Scholar]
- 33. Deschauer M., Hudson G., Müller T., Taylor R. W., Chinnery P. F., Zierz S. (2005) Neuromuscul. Disord. 15, 311–315 [DOI] [PubMed] [Google Scholar]
- 34. Thomas A., Rey M., Aubry L., Pelosi L. (2011) Biochimie 93, 1415–1423 [DOI] [PubMed] [Google Scholar]
- 35. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 36. Kavan D., Man P. (2011) Int. J. Mass Spectrom. 302, 53–58 [Google Scholar]