Background: The role of transferrin receptor-1 (TfR1) in cell signaling remains unknown.
Results: Using gambogic acid, we found that TfR1 is bound to Src and its phosphorylation at Tyr20 potentiates breast cancer cell survival.
Conclusion: In addition to its iron uptake function, TfR1 is involved in cell survival.
Significance: High expression of TfR1 in cancer cells presents a therapeutic target for gambogic acid and Src inhibitor.
Keywords: Breast Cancer, Iron, Iron Metabolism, Src, Transferrin Receptor
Abstract
Transferrin receptor 1 (TfR1) is a ubiquitous type II membrane receptor with 61 amino acids in the N-terminal cytoplasmic region. TfR1 is highly expressed in cancer cells, particularly under iron deficient conditions. Overexpression of TfR1 is thought to meet the increased requirement of iron uptake necessary for cell growth. In the present study, we used transferrin (Tf), a known ligand of TfR1, and gambogic acid (GA), an apoptosis-inducing agent and newly identified TfR1 ligand to investigate the signaling role of TfR1 in breast cancer cells. We found that GA but not Tf induced apoptosis in a TfR1-dependent manner in breast cancer MDA-MB-231 cells. Estrogen receptor-positive MCF-7 cells lack caspase-3 and were not responsive to GA treatment. GA activated the three major signaling pathways of the MAPK family, as well as caspase-3, -8, and Poly(ADP-ribose)polymerase apoptotic pathway. Interestingly, only Src inhibitor PP2 greatly sensitized the cells to GA-mediated apoptosis. Further investigations by confocal fluorescence microscopy and immunoprecipitation revealed that Src and TfR1 are constitutively bound. Using TfR1-deficient CHO TRVB cells, point mutation studies showed that Tyr20 within the 20YTRF23 motif of the cytoplasmic region of TfR1 is the phosphorylation site by Src. TfR1 Tyr20 phosphomutants were more sensitive to GA-mediated apoptosis. Our results indicate that, albeit its iron uptake function, TfR1 is a signaling molecule and tyrosine phosphorylation at position 20 by Src enhances anti-apoptosis and potentiates breast cancer cell survival.
Introduction
Transferrin receptor-1 (TfR1,2 also known as CD71) is an essential membrane protein involved in iron uptake and the regulation of cell growth (1). TfR1 knock-out embryos die before embryonic day 12.5 due to defects in erythropoiesis and neurological development (2). Delivery and uptake of iron from transferrin (Tf) into cells occur through the internalization of the iron-loaded Tf-TfR1 complex. TfR1 is a type II transmembrane glycoprotein, which contains a large extracellular C-terminal domain (671 amino acids), a single-pass transmembrane domain (28 amino acids), and a short intracellular N-terminal domain (61 amino acids) (3). The tetrapeptide 20YTRF23 motif within the cytoplasmic region of TfR1 has been shown as the structural recognition motif to TfR1 endocytosis (4).
TfR1 is a key molecule controlling cellular iron homeostasis, and its level is mainly regulated posttranscriptionally through the iron-responsive element/iron regulatory protein system. Under iron-deficient conditions, iron regulatory proteins binding to the iron-responsive elements in the 3′-untranslated region of TfR1 mRNA prevents its degradation and enhances its translation (5). However, TfR1 expression is also regulated at the transcriptional level. TfR1 expression is up-regulated by the hypoxia-inducible factor (6), which is typically activated under hypoxic conditions (7), but can also be turned on by a number of non-hypoxic stimuli, including inflammatory signals such as nitric oxide and l/opolysaccharide in macrophages (8) and 17β-estradiol in estrogen receptor-positive cells (9).
Numerous studies have shown elevated levels of TfR1 expression in cancer cells when compared with their normal counterparts. A comparison of benign and malignant breast epithelia in the same section shows that TfR1 expression could be up to 4–5-fold higher in malignant breast cells than non-neoplastic breast cells (10, 11). On a multivariate analysis, TfR1 was found to be an independent prognostic factor in breast cancer outcome, and high levels of TfR1 expression could be used to define patients less likely to respond to endocrine therapy such as tamoxifen treatment (12). Thus far, the most accepted explanation for high expression of TfR1 in cancer cells is that they require more iron as a nutrient and a co-factor of the ribonucleotide reductase enzyme involved in DNA synthesis of rapidly dividing cells (3).
Iron is mostly bound to Tf and delivered to cells using an endocytic pathway involving TfR1. The mechanism by which cells regulate clathrin-mediated endocytosis of iron-bound Tf was viewed as a constitutive process that occurs continuously without regulatory constraints. A recent study indicates that endocytosis of TfR1 is a regulated process requires activated Src kinase, and subsequent phosphorylation of two important components of the endocytosis machinery, the large GTPase dynamin 2 and its associated actin-binding protein, cortactin (13). Significant attention has been paid toward understanding the basic endocytic machine and the structure of the human TfR1-Tf complex has been solved (14). More recently, several novel TfR1 ligands have been identified. These include ferritin H chain (15), New World hemorrhagic fever arenaviruses (16), and gambogic acid (GA), an apoptosis-inducing agent (17). Tumor cells overexpressing TfR1 treated with GA underwent significant apoptosis as compared with that observed for primary normal cells (18, 19). These results indicate that TfR1 has unidentified functions other than iron uptake. However, it is unclear whether high expressions of TfR1 in cancer cells are associated with novel functions, and conformational changes, such as phosphorylation, might occur in the short cytoplasmic tail of TfR1 to initiate downstream signaling cascades.
To gain better understanding of the role of TfR1 in cancer biology, we tested a hypothesis that greater expression of TfR1 in cancer cells plays an important role in cell signaling, contributing to cancer cell survival. In the present study, we used Tf, a known TfR1 ligand, and GA, a newly identified TfR1 ligand that is independent of the Tf binding site (17). We demonstrated that TfR1 is bound to Src, a non-receptor tyrosine kinase, in breast cancer MDA-MB-231 cells and phosphorylation of tyrosine at position Tyr20 within the cytoplasmic region promotes cancer cell survival. Thus, our findings provide new insight into the high expression of TfR1 in cancer cells not only for enhancing iron uptake but also for increasing survival.
EXPERIMENTAL PROCEDURES
Reagents
GA was purchased from Gaia Chemical Corp. (Gaylordsville, CT). Holo-Tf (its two iron binding sites are 100% saturated by iron), kinase inhibitors, such as PP2, PP3, SB202190, PD98059, U0126, SP600125, wortmannin, and propidium iodide (PI) were purchased from Sigma. Antibodies against caspase-3 and -8, PARP, Bcl-2, Bcl-xL, Src, Bax, GAPDH and phospho-ERK, JNK, and p38 were purchased from Cell Signaling (Danvers, MA). Antibodies against phospho-Tyr and TfR1 were from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 555-labeled donkey anti-rabbit antibody and Alexa Fluor 488-labeled donkey anti-mouse antibody were purchased from Invitrogen. pcDNA3-TfR1 vector is a kindly gift from Dr. J. E. Pessin (Albert Einstein College of Medicine New York) (20).
Site-directed Point Mutation
Point mutants of Y20L and Y20F at the YTRF motif were generated by using a site-directed mutagenesis kit (Stratagene, La Jolla, CA) with a pcDNA3-TfR1 vector of the following primers: Y20L, 5′-TTGGTGGAGAACCATTGTCATTAACCCGGTTCAGCCTGGC-3′ (sense) and 5′-GCCAGGCTGAACCGGGTTAATGACAATGGTTCTCCACCAA-3′ (antisense); Y20F, 5′-TTGGTGGAGAACCATTGTCATTTACCCGGTTCAGCCTGGC-3′ (sense) and 5′-GCCAGGCTGAACCGGGTAAATGACAATGGTTCTCCACCAA-3′ (antisense). Briefly, primers containing point mutation were used for PCR amplifications in a pcDNA3-TfR1 vector as the template. After PCR reactions, the DNA templates were digested with DpnI for 1 h, followed by transformation of Escherichia coli. Five clones were randomly chosen for sequencing, and three clones from each mutation were used to establish stable cell lines.
Cells
Human breast cancer MCF-7 and MDA-MB-231 cells were purchased from the American Tissue Culture Collection (Manassas, VA) and were maintained in DMEM with 10% FBS. CHO-TRVB cells (a TfR1-deficient CHO cell line) and TRVB1 (CHO cells expressing human TfR1) were kindly provided by Dr. T. E. McGraw (21) and were cultured in F-12 medium with 10% FBS. pcDNA3-TfR1-Y20L and pcDNA3-TfR1-Y20F were stably transfected into TRVB cells. All of the cells were cultured in 5% CO2 incubator at 37 °C.
Western Blot
Cells were seeded into a six-well plate and treated with GA or holo-Tf for indicated doses and time points. The cell extracts were collected in a boiling buffer consisting of 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm glycerophosphate, 1 mm Na3VO4, and 1 μg/ml leupeptin. After sonication to break genomic DNA, proteins were quantified, and equal amounts of protein were loaded in 10% SDS-polyacrylamide gels. After transferring to PVDF membrane, the membranes were probed with the primary antibodies overnight. After washing, the HRP-labeled secondary antibody was added for 1 h, and the bands were visualized by enhanced chemiluminescence kit (PerkinElmer Life Sciences).
Immunoprecipitation (IP)
Cells were seeded into 10-cm dishes with or without GA treatments for indicated time points. After treatment, cells were collected on ice using radioimmune precipitation assay lysis buffer (Santa Cruz Biotechnology). The cell lysates were sonicated to break DNA and centrifuged at 10,000 rpm for 5 min to pellet debris. The supernatants were mixed with 1 μg/ml normal mouse IgG and 20 μl of protein A/G-labeled agarose and incubated for 1 h at 4 °C with gentle shaking. After centrifugation for 5 min, the supernatants were incubated with 1 μg/ml TfR1 antibody or Src antibody and were mixed with 20 μl of protein A/G-labeled agarose overnight at 4 °C. The TfR1- or Src-enriched agarose beads were washed with radioimmune precipitation assay lysis buffer six times. The samples were collected for Western blotting.
Confocal Microscopy
MDA-MB-231 cells were seeded on a coverslip in a 24-well plate for 48 h. The cells were washed with PBS and fixed by 3.7% formaldehyde for 5 min and washed three times by PBS. The cells were permeabilized with 0.1% Triton X-100 for 5 min. Nonspecific binding were blocked with normal donkey serum for 30 min. The cells were primed with mouse anti-TfR1, rabbit anti-Src antibodies, as well as normal mouse and rabbit serum as control for 2 h. After washing three times, the cells were incubated with donkey anti-rabbit labeled with Alexa Fluor 555 and donkey anti-mouse labeled with Alexa Fluor 488 antibodies. After washing, the cover glasses were taken out and mounted in-slide with mounting medium and sealed by nail oil. The samples were imaged by using a Leica TCS SP5 confocal fluorescence microscopy.
Annexin V and PI Double Apoptosis Staining
Cells were seeded into six-well plates and treated with GA. In some experiments, cells were pretreated with kinase inhibitors for 1 h and then treated with GA for various times. Both cells and cell media were collected and centrifuged at 3000 rpm for 5 min. After washing with PBS, the cells were suspended and stained with FITC-labeled annexin V and PI for 20 min. 20,000 cells were analyzed by flow cytometry.
Statistical Analyses
All values were expressed as means ± S.D. Student's t tests were used for comparison between experimental groups with a p value of < 0.05 considered statistically significant.
RESULTS
GA Induces Apoptosis in Human Breast Cancer MDA-MB-231 but Not in MCF-7 Cells
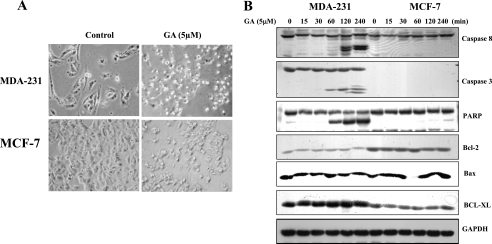
Fig. 1A shows that after treating both MDA-MB-231 and MCF-7 cells with 5 μm GA, MDA-MB-231 cells were more sensitive to GA-induced apoptosis as compared with MCF-7 cells. Caspase-3, -8, and PARP were cleaved in MDA-MB-231 cells but not in MCF-7 cells (Fig. 1B). This could be due to estrogen receptor-positive status with MCF-7 cells, which are caspase-3 deficient (22). Bcl family members such as Bcl-2, Bax, and Bcl-xL were not affected by GA in both MCF-7 and MDA-MB-231 cells (Fig. 1B). Because of the sensitivities of MDA-MB-231 cells, this cell line was further used for the present study.
FIGURE 1.
GA-induced apoptosis in breast cancer MDA-MB-231 cells but not in MCF-7 cells. A, MDA-MB-231 and MCF-7 cells were treated with 5 μm GA, and cell apoptosis was observed by microscope 4 h later. B, GA activated caspase-3 and -8 and PARP apoptotic pathways in MDA-MB-231 cells in a time-dependent manner. Caspase-3 is absent in MCF-7 cells, causing resistance to GA treatment.
GA-induced Apoptosis Is Partly TfR1-dependent
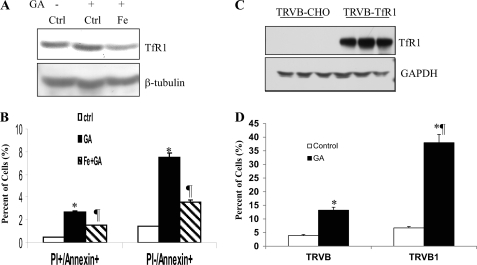
To examine whether GA-induced apoptosis is TfR1-dependent, MDA-MB-231 cells were pretreated with FeSO4 for 24 h to down-regulate levels of TfR1 (Fig. 2A) and then followed by GA treatment. Fig. 2B shows that GA significantly enhanced both apoptotic cells (PI+/annexin+) and early apoptotic cells (PI−/annexin+) as compared with the control untreated cells. Down-regulation of TfR1 by the pretreatments of FeSO4 led to a decreased level of GA-mediated apoptosis. To further relate GA-mediated apoptosis to TfR1, TfR1-deficient TRVB and TfR1-expressing TRVB1 cells were used (Fig. 2C). We found that GA induced ∼13% PI+/annexin+ apoptosis in TfR1-deficient TRVB cells but 38% apoptosis in TRVB1 cells expressing TfR1 (Fig. 2D). These results indicate that there is a TfR1-independent mechanism in GA-induced apoptosis and, thus, GA-induced apoptotic pathway is partly TfR1-dependent.
FIGURE 2.
GA-induced apoptosis is TfR1-dependent as well as TfR1-independent. A, two sets of MDA-MB-231 cells were pretreated with 5 μm FeSO4 overnight, followed by GA treatment for 4 h. First set was used for Western blot to show TfR1 down-regulation by iron. B, second set was used for flow cytometry to measure apoptosis. The apoptotic cells were counted by PI and FITC-annexin V double staining. C, confirmation of TfR1 expression by Western blots in TfR1-expressing TRVB1 cells but not in TfR1-deficient TRVB cells. Three clones were used for each of the experiments. D, after treatments of TRVB and TRVB1 cells with 5 μm GA for 4 h, PI+/annexin+ apoptotic cells were measured, indicating TfR1 expressing TRVB1 cells are more sensitive to GA-mediated apoptosis. Thus, there are TfR1-dependent and TfR1-independent mechanisms in GA-mediated apoptosis. An asterisk represents a significant difference from controls (Ctrl); ¶ represents a significant difference from GA treatment only (B) or from TRVB cells (D).
GA-mediated Cell Signaling Is Different from That of Tf
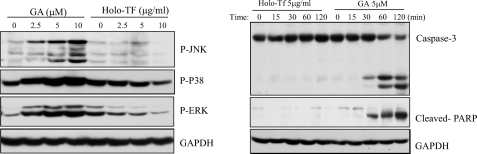
GA seems to have a different epitope from that of the well known ligand Tf (17). As shown in Fig. 3A, only GA activated phosphorylation of p38, JNK, and ERK, whereas no apparent activations were found by holo-Tf. GA, but not holo-Tf, also cleaved caspase-3 and PARP (Fig. 3B). These results further distinguished the effects of GA from those of Tf on MAPK and apoptotic signaling pathways.
FIGURE 3.
GA but not transferrin induced activation of MAPK pathway and apoptosis. A, MDA-MB-231 cells were treated with GA or holo-Tf at different doses for 60 min. Total proteins were collected, and phosphorylations of JNK, p38, and ERK were probed with antibodies against phospho-JNK, p38, and ERK. B, MDA-MB-231 cells were treated with 5 μm GA or 5 μg/ml holo-Tf for various time periods.
Src Inhibitor PP2 Promotes GA-induced Apoptosis
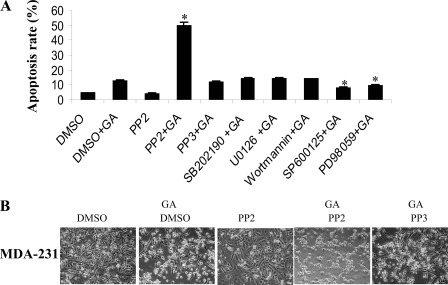
To determine which pathway(s) of the three MAPK family members is responsible for GA-induced apoptosis, MDA-MB-231 cells were pretreated with p38 inhibitor SB202190, JNK inhibitor SP600125, ERK inhibitor PD98059, and MEK1/2 inhibitor U0126, respectively, before GA treatment. Moreover, Src inhibitor PP2, its negative control PP3, and the PI3K inhibitor wortmannin were used as additional controls. Fig. 4A shows that inhibitors of the MAPK pathway SP600125 and PD98059 significantly attenuated GA-induced apoptosis, and those of PI3K pathways had no obvious effect. Interestingly, the Src inhibitor PP2 greatly enhanced GA-induced apoptosis, whereas PP2 alone or its negative control PP3 did not (Fig. 4, A and B), suggesting that Src could deter GA-induced apoptosis and serve as a negative regulator.
FIGURE 4.
Src inhibitor stimulated GA-induced apoptosis. A, MDA-MB-231 cells were pretreated with dimethyl sulfoxide (DMSO), Src inhibitor PP2, PP2-negative control PP3, p38 inhibitor SB202190, MEK1/2 inhibitor U0126, PI3K inhibitor wortmannin, JNK inhibitor SP600125, and ERK inhibitor PD98059 for overnight and then treated with 5 μm GA. The apoptosis rate was measured by PI and annexin V staining. B, representative images of morphological changes of MDA-MB-231 cells in the presence of DMSO (control), GA (treatment), PP2 alone, GA+PP2, and GA+PP3 as observed by microscope. Magnification, ×100. An asterisk indicates a significant difference from the GA treatment.
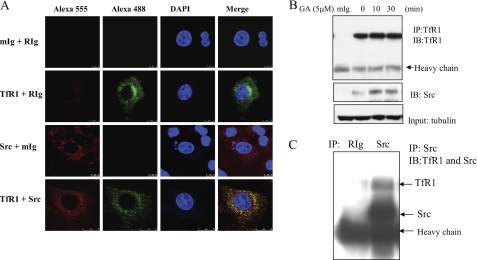
TfR1 Is Associated with Src
To elucidate the underlying molecular mechanisms by which Src inhibitor PP2 promotes GA-induced apoptosis, immunofluorescence by confocal microscopy illustrates that Src and TfR1 are co-localized in MDA-MB-231 cells (Fig. 5A). Immunoprecipitation (IP) of TfR1 followed by immunoblotting of Src or vice versa was then performed to further confirm its binding. Fig. 5B shows that Src is bound to TfR1, and GA treatments had increased the association of Src with TfR1. However, GA appeared to induce TfR1 degradation at 60 min or longer treatments (data not shown). Fig. 5C confirms that after IP of Src by its antibody, TfR1 was also pulled down.
FIGURE 5.
TfR1 is associated with Src. A, immunofluorescence and colocalization of Src and TfR1 by confocal fluorescence microscope. Src and TfR1 were stained with Alexa Fluor 555 (red) and Alexa Fluor 488 (green), respectively. Cellular nuclei were stained with DAPI (blue). Mouse (mIg) and rabbit (RIg) Ig were used as negative controls to show nonspecific bindings. B, MDA-MB-231 cells were treated with GA for different time periods, and TfR1 was immunoprecipitated by its antibody. The samples were then immunoblotted (IB) with Src antibody. C, Src was immunoprecipitated by its antibody and followed by simultaneous immunoblotting with TfR1 and Src antibodies.
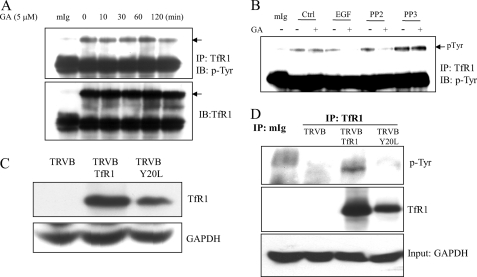
TfR1 Is Tyrosine-phosphorylated at Tyr20
We then investigated whether binding of Src leads to TfR1 tyrosine phosphorylation. Fig. 6A shows that TfR1 is tyrosine phosphorylated in MDA-MB-231 cells and treatments of GA did not affect TfR1 tyrosine phosphorylation. To confirm the role of Src in TfR1 tyrosine phosphorylation, Fig. 6B displays that Tyr20 phosphorylation in the absence of GA appeared to be Src-independent as its phosphorylation was not affected by PP2 treatment as shown in lane 6 when compared with lane 2. Co-treatments of PP2 with GA or EGF with GA abrogated TfR1 tyrosine phosphorylation but not in the control (dimethyl sulfoxide) and PP3 treatment groups.
FIGURE 6.
TfR1 is phosphorylated at Tyr20 within the cytoplasmic region of TfR1. A, MDA-MB-231 cells were treated with 5 μm GA for different time periods. The proteins were collected, and TfR1 was immunoprecipitated by TfR1 antibody and immunoblotted (IB) with antibodies against p-Tyr or TfR1. B, MDA-MB-231 cells were pretreated with the Src inhibitor PP2, PP3, and EGF for 1h, followed by GA for 4 h. The cells were lysed, and TfR1 were immunoprecipitated with antibody against TfR1. Tyr phosphorylation of TfR1 was immunoblotted with antibody against p-Tyr. C, confirmation by Western blots of TfR1 expressions in TfR1-deficient CHO TRVB cells (a negative control), TRVB1 cells expressing human TfR1, and TRVB cells transfected with Y20L point mutants. D, Y20L mutant abrogates endogenous TfR1 tyrosine phosphorylation. Proteins were collected from TRVB, TRVB1, and Y20L mutants. After IP with TfR1, they were probed with antibody against p-Tyr.
Sequence analyses reveal that there is only one tyrosine in the cytoplasmic region of TfR1, which is located at the 20YTRF23 motif. To show whether Tyr20 is the Src-mediated phosphorylation site, a Y20L point mutant was generated. After transfection, Fig. 6C shows no expression of TfR1 in TfR1-deficient TRVB cells. As expected, strong expressions of TfR1 were observed in TRVB1 cells, in which human TfR1 was stably transfected into TRVB cells (23). TfR1 was also expressed in TRVB-Y20L point mutant (Fig. 6C). As shown in Fig. 6D, Y20L mutation abrogated TfR1 tyrosine phosphorylation, indicating that Tyr20 is the Src-mediated tyrosine phosphorylation site.
Function of TfR1 Tyr20 Phosphorylation
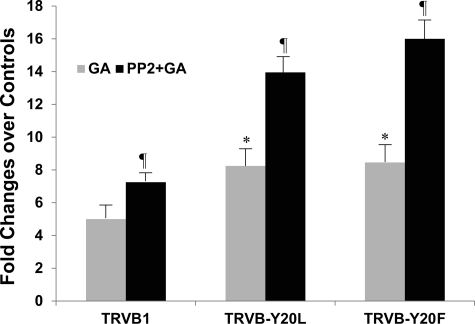
YTRF is an internalization signal and is required for Tf-induced endocytosis (24). However, Tyr20 is not necessary for TfR1 internalization. It has been shown that substitution of Tyr20 with other aromatic amino acids such as phenylalanine (Y20F) did not alter the rate constant of TfR1 internalization, whereas substitution with nonaromatic acid such as leucine (Y20L) reduced the internalization constant rate (25). Fig. 7 displays that among the TfR1-expressing cells, phosphomutants without a Tyr20 phosphorylation site were more sensitive to GA-mediated cell apoptosis, particularly in the Y20F mutants. Again, Src inhibitor PP2 increased GA-mediated apoptosis by 45% in CHO-TRVB1 cells as compared with the GA treatment only. The increases were higher by 70% in Y20L phosphomutants and by 90% in Y20F phosphomutants. PP2 had no enhancing effects on GA-mediated apoptosis in TfR1-deficient TRVB cells (data not shown). These results indicate that the role of Src in GA-mediated apoptosis is TfR1-dependent. Although phosphorylation of TfR1 at Tyr20 by Src is important, other Src-dependent TfR1 regulatory mechanisms could contribute to the apoptotic process of GA treatment.
FIGURE 7.
TfR1 Tyr20 phosphomutants were more sensitive to GA-induced apoptosis. TRVB1 cells and three TRVB-Y20L and TRVB-Y20F mutants each were pre-treated with Src inhibitor PP2 for 1 h, followed by GA treatments for 4 h. The apoptotic cells were stained with PI and FITC-annexin V and analyzed by flow cytometry. Because of variations of TfR1 levels among the different cell lines, sensitivities of the cells to GA and PP2 treatments were presented as fold change over their respective untreated controls. Asterisk indicates a significant difference from TRVB1 cells; a ¶ indicates a significant difference from their respective GA treatments only.
DISCUSSION
The findings described above provide strong evidence for TfR1 binding with Src to present survival signals in breast cancer cells. Phosphomutants of TfR1 that cannot act as a Src substrate significantly responded to GA-mediated TfR1-dependent apoptosis. This is consistent with the premise established by others that Src interacts with many membrane receptors such as EGF receptor family members and the TGF-β receptor and mediates the antiapoptosis and proliferation signal (26, 27). In the present study, we did not modify Src but modulated TfR1 by point mutations. Results show that TfR1 is a signal molecule that can be directly phosphorylated by Src.
Our study originated from the treatments of human breast cancer MDA-MB-231 and MCF-7 cells with GA (Fig. 1), a natural product from the resin of the Garcinia hanburyi tree. The resin has been used in traditional Chinese medicine (28). GA displays differential apoptosis potential in normal versus tumor cells. Here, down-regulation of TfR1 by iron treatments leads to a decrease in apoptosis with GA treatment and TRVB1 expressing human TfR1 are more sensitive to GA treatment (Fig. 2). Although TfR1 serves as an important receptor for GA, GA-mediated apoptosis appears to be only partially TfR1-dependent. These results indicate that there is a TfR1-independent mechanism since TRVB cells do not express TfR1 but these cells still underwent apoptosis in response to GA treatment (Figs. 2 and 7).
Holo-Tf and GA are known ligands of TfR1. Our study shows that GA but not holo-Tf activates three major pathways: ERK, p38, and JNK of the MAPK family (Fig. 3). In contrast, it has been shown that GA inhibits the NF-κB signaling pathway and potentiates TNF-induced apoptosis in human leukemia cancer cells (19). We thought that the GA-activated MAPK signaling mechanism might play an important role in its potent anticancer activity in MDA-MB-231 cells. Indeed, MAPK inhibitors reduced GA-mediated apoptotic effects. Our results are in agreement with the previous report showing that MAPK inhibitors could affect apoptosis, in the presence of Aplidin, a novel antitumor agent of marine origin (29). Unexpectedly, the inclusion of a Src inhibitor PP2 was found to sensitize GA-mediated apoptosis (Fig. 4), suggesting that Src may be involved in GA-mediated apoptotic process.
A function for Src tyrosine kinase in normal cell growth was first demonstrated with the binding of family member p56lck to the cytoplasmic tail of CD4 and CD8 co-receptors on T cells (30). Src plays a crucial role in the coordination and facilitation of cell signaling pathways controlling a wide range of cellular functions, including growth, survival, invasion, adhesion, and migration (31, 32). Src is highly expressed in MDA-MB-231 cells and activation of a dominant negative form of Src has been shown to decrease their tumorigenicity and their ability to induce lung and bone metastasis in animals (33). Use of Src inhibitors leads to decreased phosphorylation of multiple Src substrates such as MAPK, Akt, and subsequent inhibition of growth and migration (34). Here, our study showed that the Src inhibitor PP2 significantly enhanced GA-mediated apoptosis (Fig. 4), providing evidence of the therapeutic potential of GA and PP2 as chemo-sensitizing agents in treating cancer cells.
The pathway by which iron-bound Tf is internalized by TfR1 has been studied extensively (3, 35–37). Diferric Tf binds the receptor and both are internalized in clathrin-coated pits through receptor-mediated endocytosis. Our study is different from the one showing that Src-mediated phosphorylation of dynamin 2 and cortactin are required for regulating the constitutive endocytosis of Tf by TfR1 (13). Instead, we showed that Src is associated with TfR1, regardless of GA treatment (Fig. 5). To support our finding, Src was found to bind to an EGF receptor and Her2 and regulates their phosphorylation (27). Association of Src with Her2 results in increased Src kinase activity and plays a role in Her2-mediated invasion and metastasis (38). Phosphorylation of the EGF receptor by Src is critical for mitogenic signaling initiated by the EGF receptor itself (39). In the present study, we found that, in addition to PP2, EGF treatment abrogated Tyr20 phosphorylation (Fig. 6B). In consideration of the importance of Src in EGF-mediated EGF receptor phosphorylation (38, 39), one possible explanation is that EGF could decrease Src availability for TfR1 Tyr20 phosphorylation.
To identify which tyrosine residue within the cytoplasmic region is phosphorylated by Src, we used CHO-TRVB cell lines, which do not express TfR1 (Fig. 6C). Because there is only one tyrosine at the position 20 within the intracellular domain, point mutation assay points out that it is the Tyr20 that is phosphorylated by Src (Fig. 6D). 20YTRF23 in the cytoplasmic region of TfR1 is crucial for TfR1 internalization and endocytosis (24). The aromatic amino acid at position 20 is important for TfR1 endocytosis because replacement of Tyr20 with Phe, an aromatic amino acid, did not impair this internalization process. However, replacement Tyr with Lys, a non-aromatic amino acid, appeared to weaken it. We showed that both TRVB phosphomutants Y20L and Y20F were sensitive to GA-mediated apoptosis, but more in Y20F mutant than in Y20L mutant (Fig. 7). These results suggest that endocytosis of TfR1 with Y20F has additional effects on GA-mediated apoptosis.
If the Tyr20 is the only site responsible for the GA-mediated and TfR1-dependent apoptosis, the mutants should no longer respond to PP2 treatments. Yet, PP2 still sensitizes the phosphomutants to GA treatments (Fig. 7). These results suggest that, although this Src-mediated Tyr20 phosphorylation is an important mechanism, other Src-dependent mechanisms such as phosphorylation of dynamin 2 and cortactin and their regulation on TfR1 endocytosis could also contribute to the apoptotic process of GA treatments (13). This awaits further investigation.
In conclusion, high expression of TfR1 in breast cancer cells is to meet the high iron demand for cancer cell outgrowth. The present study provides evidence that additional functions of high expression of TfR1 exist in cancer cells. This includes, but may not be limited to, anti-apoptotic and cancer cell survival signals by Src phosphorylation of TfR1 at Tyr20. Our study is particularly significant in young breast cancer patients because of the high prevalence of iron deficiency anemia in young women (40) and poor breast cancer outcomes in this young patient population (41, 42). Future studies of this newly identified TfR1 signaling pathway are likely to provide insight into iron homeostasis in cancer cells and its relation to cancer cell survival and drug resistance.
Acknowledgments
We thank Dr. T. McGraw (Weill Cornell Medical College) for generously providing CHO-TRVB cells (a TfR1-deficient CHO cell line) and TRVB1 (CHO cells expressing human TfR1) as well as Dr. J. E. Pessin (Albert Einstein College of Medicine) for pcDNA3-TfR1 vector.
This work was supported in part by National Institutes of Health Grants ES00260, CA34588, and CA16087 and by NCI, National Institutes of Health Grant R21 CA132684 (to X. H.).
- TfR1
- transferrin receptor 1
- Tf
- transferrin
- GA
- gambogic acid
- PI
- propidium iodide.
REFERENCES
- 1. Neckers L. M., Trepel J. B. (1986) Cancer Invest. 4, 461–470 [DOI] [PubMed] [Google Scholar]
- 2. Levy J. E., Jin O., Fujiwara Y., Kuo F., Andrews N. C. (1999) Nat. Genet. 21, 396–399 [DOI] [PubMed] [Google Scholar]
- 3. Daniels T. R., Delgado T., Rodriguez J. A., Helguera G., Penichet M. L. (2006) Clin. Immunol. 121, 144–158 [DOI] [PubMed] [Google Scholar]
- 4. Collawn J. F., Stangel M., Kuhn L. A., Esekogwu V., Jing S. Q., Trowbridge I. S., Tainer J. A. (1990) Cell 63, 1061–1072 [DOI] [PubMed] [Google Scholar]
- 5. Muckenthaler M. U., Galy B., Hentze M. W. (2008) Annu. Rev. Nutr. 28, 197–213 [DOI] [PubMed] [Google Scholar]
- 6. Bianchi L., Tacchini L., Cairo G. (1999) Nucleic Acids Res. 27, 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eckard J., Dai J., Wu J., Jian J., Yang Q., Chen H., Costa M., Frenkel K., Huang X. (2010) Cancer Cell Int. 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tacchini L., Gammella E., De Ponti C., Recalcati S., Cairo G. (2008) J. Biol. Chem. 283, 20674–20686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai J., Jian J., Bosland M., Frenkel K., Bernhardt G., Huang X. (2008) Breast 17, 172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tonik S. E., Shindelman J. E., Sussman H. H. (1986) Breast Cancer Res. Treat. 7, 71–76 [DOI] [PubMed] [Google Scholar]
- 11. Walker R. A., Day S. J. (1986) J. Pathol. 148, 217–224 [DOI] [PubMed] [Google Scholar]
- 12. Habashy H. O., Powe D. G., Staka C. M., Rakha E. A., Ball G., Green A. R., Aleskandarany M., Paish E. C., Douglas Macmillan R., Nicholson R. I., Ellis I. O., Gee J. M. (2010) Breast Cancer Res. Treat. 119, 283–293 [DOI] [PubMed] [Google Scholar]
- 13. Cao H., Chen J., Krueger E. W., McNiven M. A. (2010) Mol. Cell. Biol. 30, 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng Y., Zak O., Aisen P., Harrison S. C., Walz T. (2004) Cell 116, 565–576 [DOI] [PubMed] [Google Scholar]
- 15. Li L., Fang C. J., Ryan J. C., Niemi E. C., Lebrón J. A., Björkman P. J., Arase H., Torti F. M., Torti S. V., Nakamura M. C., Seaman W. E. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3505–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Radoshitzky S. R., Abraham J., Spiropoulou C. F., Kuhn J. H., Nguyen D., Li W., Nagel J., Schmidt P. J., Nunberg J. H., Andrews N. C., Farzan M., Choe H. (2007) Nature 446, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kasibhatla S., Jessen K. A., Maliartchouk S., Wang J. Y., English N. M., Drewe J., Qiu L., Archer S. P., Ponce A. E., Sirisoma N., Jiang S., Zhang H. Z., Gehlsen K. R., Cai S. X., Green D. R., Tseng B. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12095–12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Q., Cheng H., Zhu G., Yang L., Zhou A., Wang X., Fang N., Xia L., Su J., Wang M., Peng D., Xu Q. (2010) Biol. Pharm. Bull 33, 415–420 [DOI] [PubMed] [Google Scholar]
- 19. Pandey M. K., Sung B., Ahn K. S., Kunnumakkara A. B., Chaturvedi M. M., Aggarwal B. B. (2007) Blood 110, 3517–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanzaki M., Furukawa M., Raab W., Pessin J. E. (2004) J. Biol. Chem. 279, 30622–30633 [DOI] [PubMed] [Google Scholar]
- 21. McGraw T. E., Greenfield L., Maxfield F. R. (1987) J. Cell Biol. 105, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blanc C., Deveraux Q. L., Krajewski S., Jänicke R. U., Porter A. G., Reed J. C., Jaggi R., Marti A. (2000) Cancer Res. 60, 4386–4390 [PubMed] [Google Scholar]
- 23. Carlson H., Zhang A. S., Fleming W. H., Enns C. A. (2005) Blood 105, 2564–2570 [DOI] [PubMed] [Google Scholar]
- 24. Collawn J. F., Lai A., Domingo D., Fitch M., Hatton S., Trowbridge I. S. (1993) J. Biol. Chem. 268, 21686–21692 [PubMed] [Google Scholar]
- 25. McGraw T. E., Maxfield F. R. (1990) Cell Regul. 1, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galliher A. J., Schiemann W. P. (2007) Cancer Res. 67, 3752–3758 [DOI] [PubMed] [Google Scholar]
- 27. Luttrell D. K., Lee A., Lansing T. J., Crosby R. M., Jung K. D., Willard D., Luther M., Rodriguez M., Berman J., Gilmer T. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Asano J., Chiba K., Tada M., Yoshii T. (1996) Phytochemistry 41, 815–820 [DOI] [PubMed] [Google Scholar]
- 29. Cuadrado A., Garcia-Fernandez L. F., Gonzalez L., Suarez Y., Losada A., Alcaide V., Martinez T., Fernandez-Sousa J. M., Sanchez-Puelles J. M., Munoz A. (2003) J. Biol. Chem. 278, 241–250 [DOI] [PubMed] [Google Scholar]
- 30. Rudd C. E., Trevillyan J. M., Dasgupta J. D., Wong L. L., Schlossman S. F. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5190–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Finn R. S. (2008) Ann. Oncol. 19, 1379–1386 [DOI] [PubMed] [Google Scholar]
- 32. Mayer E. L., Krop I. E. (2010) Clin. Cancer Res. 16, 3526–3532 [DOI] [PubMed] [Google Scholar]
- 33. Rucci N., Recchia I., Angelucci A., Alamanou M., Del Fattore A., Fortunati D., Susa M., Fabbro D., Bologna M., Teti A. (2006) J. Pharmacol. Exp. Ther. 318, 161–172 [DOI] [PubMed] [Google Scholar]
- 34. Jallal H., Valentino M. L., Chen G., Boschelli F., Ali S., Rabbani S. A. (2007) Cancer Res. 67, 1580–1588 [DOI] [PubMed] [Google Scholar]
- 35. Aisen P. (2004) Int. J. Biochem. Cell Biol. 36, 2137–2143 [DOI] [PubMed] [Google Scholar]
- 36. Kwok J. C., Richardson D. R. (2002) Crit. Rev. Oncol. Hematol. 42, 65–78 [DOI] [PubMed] [Google Scholar]
- 37. Pantopoulos K. (2004) Ann. N.Y. Acad. Sci. 1012, 1–13 [DOI] [PubMed] [Google Scholar]
- 38. Marcotte R., Zhou L., Kim H., Roskelly C. D., Muller W. J. (2009) Mol. Cell. Biol. 29, 5858–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Biscardi J. S., Ishizawar R. C., Silva C. M., Parsons S. J. (2000) Breast Cancer Res. 2, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zimmermann M. B., Hurrell R. F. (2007) Lancet 370, 511–520 [DOI] [PubMed] [Google Scholar]
- 41. Aebi S. (2005) Breast 14, 594–599 [DOI] [PubMed] [Google Scholar]
- 42. Gabriel C. A., Domchek S. M. (2010) Breast Cancer Res. 12, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]