Background: Long terminal repeats (LTRs) of endogenous retroviruses contain promoters active in cells.
Results: An ancient human LTR is an alternative promoter for the IL2RB gene in placental trophoblast.
Conclusion: The cytoplasmic signaling domain of IL2RB is present in placenta due to LTR activity.
Significance: These findings implicate functions for IL2RB in the trophoblast and reveal an example of LTR exaptation to drive tissue-specific expression.
Keywords: DNA Methylation, Epigenetics, Gene Regulation, Gene Transcription, Gene Transposable Elements, Immunology, Receptors, Alternative Promoters, Endogenous Retroviruses, Placenta
Abstract
The long terminal repeat (LTR) sequences of endogenous retroviruses and retroelements contain promoter elements and are known to form chimeric transcripts with nearby cellular genes. Here we show that an LTR of the THE1D retroelement family has been domesticated as an alternative promoter of human IL2RB, the gene encoding the β subunit of the IL-2 receptor. The LTR promoter confers expression specifically in the placental trophoblast as opposed to its native transcription in the hematopoietic system. Rather than sequence-specific determinants, DNA methylation was found to regulate transcription initiation and splicing efficiency in a tissue-specific manner. Furthermore, we detected the cytoplasmic signaling domain of the IL-2Rβ protein in the placenta, suggesting that IL-2Rβ undergoes preferential proteolytic cleavage in this tissue. These findings implicate novel functions for this cytokine receptor subunit in the villous trophoblast and reveal an intriguing example of ancient LTR exaptation to drive tissue-specific gene expression.
Introduction
Sequences derived from transposable elements make up nearly 50% of the human genome (1). Endogenous retrovirus-related elements (ERVs),2 one type of transposable element family, are the ancient descendants of exogenous retroviruses, whose life cycle involved a retrotransposition step and integration of the viral DNA into the host genome (2). Transcription of ERV protein-coding genes is regulated by the long terminal repeats (LTRs), regions flanking the ERV genes that contain promoter elements and polyadenylation sites, but genetic recombination events often lead to the formation of solitary LTRs (3). LTRs are sometimes exapted (domesticated) as alternative promoters of nearby cellular genes, a process that may simply serve to augment transcription from the native promoter with little functional impact, but that can have drastic effect on gene transcription patterns (4).
At least 50% of human genes employ alternative promoters, which are known to contribute to the diversity of gene expression patterns (5, 6). Regulation of transcription factor activity and/or binding site accessibility by epigenetic modifications is known to influence tissue-specific expression from alternative promoters and can result in the production of identical or different proteins depending on the position of the translation start site (7). These characteristics are also true for those LTRs that have been exapted as alternative gene promoters. Examples include the placenta-specific LTR-driven transcription of EDNRB by tissue-specific transcription factors (8) and demethylation of an LTR resulting in up-regulation of CSF1R in Hodgkin lymphoma (9).
Numerous studies have found that the placenta is generally permissive to transcription initiating from LTRs, and some ERV copies are expressed at higher levels in placenta than other tissues (4, 10, 11). In fact, ERVs may have been central in the evolution of placental mammals, and domestication of ERV proteins has led to their serving important functions in this tissue (12–14). For example, the syncytin proteins, derived from ERV envelope genes, confer the ability of placental trophoblast cells to form multinucleate syncytia through cell fusion (14–16).
IL2RB encodes the β subunit of the interleukin-2 receptor (IL-2Rβ) and is primarily expressed in the hematopoietic system, where it is involved in the activation of T and NK cell subsets (17). The IL-2Rβ chain can exist in one of two heterotrimeric complexes to form a functional cytokine receptor that responds to either interleukin-2 (IL-2) or interleukin-15 (IL-15) (17, 18). Roles for cytokine signaling at the maternal-fetal interface have been described, with IL-2 and IL-15 both required for differentiation and proliferation of decidual NK cells (19, 20). Here we demonstrate that the human IL2RB gene has an LTR alternative promoter that drives expression specifically in the placenta. We found that the tissue specificity is predominantly regulated by alterations in DNA methylation of the LTR and that splicing from the LTR into the downstream exon of IL2RB may be more efficient in placenta. We have shown transcription of IL2RB mRNA in the fetal trophoblast rather than the maternal decidua and found that the IL-2Rβ protein undergoes a post-translational cleavage. Our data outline novel transcriptional mechanisms by which this may occur.
EXPERIMENTAL PROCEDURES
Primary Tissue Samples
A panel of normal human RNAs was obtained from Ambion. Anonymous placental samples (whole chorionic villi with surface membranes removed) from normal term delivery (n = 12), first trimester terminations (n = 2), and second trimester terminations or premature births (n = 2) were obtained from the British Columbia Women's Hospital and Health Centre. Normal peripheral blood mononuclear cell (PBMC) samples were obtained from the Stem Cell Assay service at the Terry Fox Laboratory by Ficoll density separation. All samples were obtained with appropriate ethical approval from the University of British Columbia. Placenta RNA samples were obtained from sampling the villous tree in several areas and were stored in RNAlater (Ambion) until required. Decidua was dissected by manual visualization, and trophoblast and mesenchyme were separated by an enzymatic digestion method as described previously (21). Trophoblast and mesenchyme purity was typically found to be 85–90% as determined using a pyrosequencing assay for CpG methylation of genes that are known to be specifically methylated or unmethylated in each tissue (data not shown).
RNA Extraction, Reverse Transcription, and Quantitative PCR
Expression levels of IL2RB were measured by qRT-PCR. RNA extraction was performed with the RNeasy kit (Qiagen) and reversed-transcribed with SuperScript III (Invitrogen). Quantitative PCR was performed with the Applied Biosystems Fast SYBR master mix on an ABI7500 Fast system with normal cycling parameters. Primer sequences are given in supplemental Table S1, and relative positions are indicated in Fig. 1A. The two splice donor sites within the LTR were used equally (data not shown) so that the transcript level from each is summed and presented as total LTR expression. When primers were used to detect unspliced mRNA, a no RT control was performed to ensure measurement of mRNA and not genomic DNA. Expression levels were normalized to a control gene, succinate dehydrogenase complex subunit A (SDHA), by the ΔΔCT method. Promoter use was determined as a percentage of total IL2RB expression.
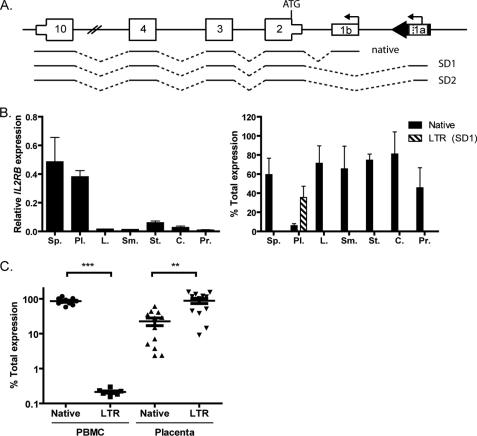
FIGURE 1.
Expression and promoter usage of IL2RB. A, schematic representation (not to scale) of human IL2RB, with the THE1D LTR ∼25 kb upstream (chr22:37570916–37571292, human reference sequence GRCh37/hg19). Three predicted mRNAs are shown, one from the native promoter and two from the LTR (SD1 and SD2) that utilize different splice donor sites. Bent arrows indicate transcription start sites, numbered boxes are exons, thick black arrow is the LTR, and dotted lines represent splicing events. B, total IL2RB expression (left panel) and promoter use (right panel) determined by qRT-PCR in a panel of normal tissues: Sp., spleen; Pl., placenta; L., liver; Sm., small intestine; St., stomach; C., colon; Pr., prostate. Error bars represent the S.E. of three independent experiments. Black bars represent native promoter use; hatched bars indicate LTR promoter use. Only transcription of the first LTR splice form (SD1) is shown; hence, the total expression in placenta is not 100%. C, IL2RB promoter use in normal PBMC (n = 10) and placenta (n = 12) samples. Each dot represents an individual biological replicate, and error bars represent the S.E. Statistical significance determined by Student's t test is indicated as follows: **, p < 0.01; ***, p < 0.001.
Bisulfite Sequencing Analysis
DNA extraction was performed using DNAzol (Invitrogen). Bisulfite conversion was performed as described previously (22). Converted DNA was used as a template for 40 cycles of PCR with AmpliTaq Gold DNA polymerase (Applied Biosystems) using the primers given in supplemental Table S1. Two independent PCRs were performed for each primer pair to eliminate amplification bias of methylated or unmethylated sequences. PCR products were gel-purified (Invitrogen PureLink kit) and cloned using the TOPO TA cloning vector (Invitrogen). Plasmid preparation and DNA sequencing were performed by McGill University and the Genome Québec Innovation Centre Sequencing platform. At least eight independent clones were obtained for each region of interest. Data analysis was performed using the QUMA analysis program from RIKEN.
Cell Culture, Reporter Assays, and Drug Treatments
JEG-3 (human choriocarcinoma, placental epithelial origin) and U87-MG (human glioblastoma-astrocytoma, brain epithelial origin) cell lines were cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 units/ml streptomycin. Promoter regions of interest were PCR-amplified with primers given in supplemental Table S1, some of which inserted restriction sites. PCR products were cloned into the TOPO TA cloning vector (Invitrogen), and reporter constructs were generated by subcloning into the pGL4-1.0 vector (Promega). pGL4-spl1-AdSA was generated by additionally subcloning a region encoding the adenovirus splice acceptor site (AdSA) between the promoter sequence and luciferase gene. Point mutations were generated using the QuikChange II site-directed mutagenesis kit (Stratagene). In vitro methylation (or mock control) was performed with SssI methylase (Invitrogen), after which the LTR region was excised by digestion with NsiI/BglII, gel-purified, and religated into an unmethylated vector prior to transfection.
JEG-3 and U87-MG cells at 50,000 cells/well in 24-well plates were co-transfected with 1 μg of pGL4 construct and 100 ng of pRL-TK (Promega) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 24 h, luciferase activity was measured using the Dual-Luciferase assay reporter system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized relative to Renilla luciferase activity for each transfection and calculated as -fold increase over a promoterless control vector, pGL4.10-BASIC (pGL4B), or a stuffer construct with a random sequence of the appropriate length.
JEG-3 and U87-MG cells were treated with 5-azacytidine and trichostatin A (TSA), inhibitors of DNA methylation and histone deacetylase activity, respectively. A wide range of drug concentrations was tested to establish a range in which cytotoxicity was not too great but a biological effect was seen. Cells were plated at 105 cells/well in 6-well plates overnight. 5-Azacytidine was added at 24, 48, and 72 h, and cells were assayed at 96 h. TSA was added after 24 h, and cells were assayed at 96 h. Expression from the LTR was determined by qRT-PCR as above.
Western Blotting
Cell lysates were made in radioimmunoprecipitation buffer with protease inhibitors. The positive control used in this assay is a human IL2RB-transfected 293T lysate from Santa Cruz Biotechnology (sc-114166). Placental villi were separated by dissection and washed to remove contaminating blood cells prior to protein extraction, and these samples were homogenized in a Dounce homogenizer to achieve cell separation. Protein concentration was determined with a Qubit fluorometer (Qiagen). Proteins were separated by denaturing SDS-PAGE and transferred to PVDF membrane in 25 mm Tris-HCl, pH 8, 192 mm glycine. Primary antibodies used were anti-IL2RB (intracellular domain) ab61195 (Abcam) with sc-672 (Santa Cruz Biotechnology), anti-GRB2 (Santa Cruz Biotechnology), or anti-actin (Sigma A5441), as loading controls. Secondary antibodies were HRP-conjugated anti-rabbit IgG (Sigma) and HRP-conjugated anti-mouse IgG (Sigma). Proteins were detected with ECL detection reagent (GE Healthcare) on x-ray film. Supplemental Fig. S5 shows three different and independent Western blots, with the 293T-positive control protein extract blotted with both anti-IL2RB antibodies (supplemental Fig. S5, A and B) and also the placenta samples blotted with the Abcam antibody (supplemental Fig. S5C). To verify the specificity of the observed truncated IL2RB in the placentas, we performed a competition assay using the anti-IL2RB from Santa Cruz Biotechnology and the blocking peptide from the same manufacturer (sc-672 P) (see Fig. 5E). The competition assay and Western blot were done in parallel. Two gels were loaded equally with the placenta protein extracts and the positive control. After transfer, one membrane was blotted with a preincubated anti-IL2RB antibody with peptide, and the other was blotted with anti-IL2RB only. The bands present in both membranes are nonspecific, whereas bands present solely in the IL2RB-only blot are specific. Note that the amount of 293T protein extract loaded into the competition assay gels was very low to avoid well-to-well competition. For this reason, no actin is observed in the competition blots, but supplemental Fig. S5 shows a higher concentration of the 293T protein extract and the presence of the actin and IL2RB proteins.
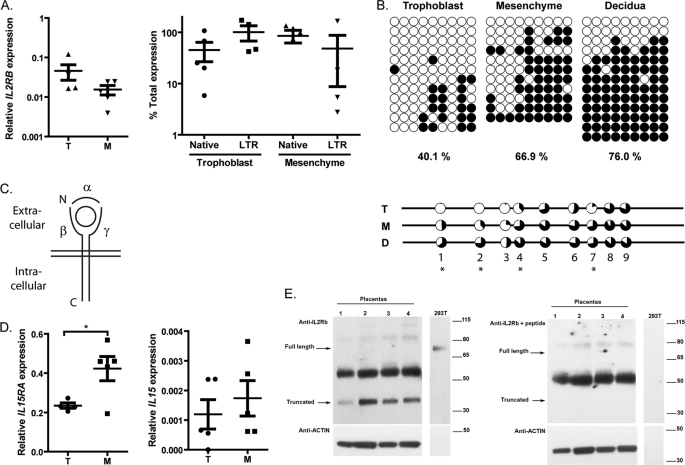
FIGURE 5.
Regulation of IL2RB and associated proteins in placental tissues. A, total IL2RB expression (left) and promoter use (right) in paired trophoblast (T) and mesenchyme (M) samples (n = 5). Each dot represents an individual biological replicate, and error bars represent the S.E. GAPDH was used as a control. B, DNA methylation of the IL2RB LTR in trophoblast, mesenchyme, and decidua from one individual (upper) and average of multiple samples (n = 3) (lower) determined by DNA methylation as represented in Fig. 3. Percentages shown are the average methylation of all samples. C, schematic representation of the IL-2 or IL-15 receptor with α, β, and γ chains represented by solid lines and the bound cytokine represented as a circle. The plasma membrane is indicated by the parallel lines. D, expression of IL15RA (left) and IL15 (right) in trophoblast (T) and mesenchyme (M) samples (n = 5). Each dot represents an individual biological replicate, and error bars represent the S.E. Asterisks indicate statistical significance (*, p < 0.05) as determined by Student's t test. E, Western blot on protein extracts from four placental samples and a positive control (293T transfected cells) detected with anti-IL2RB (left panel) and anti-IL2RB preincubated with a competing peptide (right panel) and anti-actin loading control (lower panels). Bands present only in the left panel are specific IL2RB proteins, whereas the strong ∼52-KDa band in placenta is nonspecific. Please note that the amount of 293T protein extract is low, and therefore, no actin is observed. For a Western blot containing more protein extract, see supplemental Fig. S5A. The same results were obtained with another anti-IL2RB antibody and can be seen on supplemental Fig. S5, B and C. Placentas 1, 2, 3, and 4 correspond to samples 114, 144, 133, and 132.
Determination of CpG Content in THE1D LTRs
To obtain the genomic distribution of the number of CpGs within each THE1D LTR, the DNA sequences of all THE1D LTRs annotated by RepeatMasker in the human genome were downloaded from the University of California, Santa Cruz (UCSC) Genome Browser. The total number of CpG dinucleotides present in each THE1D fragment with a size ranging from 320 to 420 bp was counted. A histogram distribution of the CpG count is shown in supplemental Fig. S1.
Statistical Analysis
Gene expression levels, promoter activities, and DNA methylation were analyzed by one-way analysis of variance or Student's t test using GraphPad Prism version 5 as indicated in the figure legends.
RESULTS
Identification of an LTR Element Upstream of IL2RB with Potential Promoter Activity
LTR elements contain promoter sequences and thus have the capability to become alternative promoters of nearby cellular genes (4). We and others have previously identified such promoters by computational methods to search for LTR sequences that overlap with the 5′ end of RefSeq genes or expressed sequence tags (ESTs) (4, 23, 24). The case of IL2RB was particularly striking because over 50 EST sequences appear to initiate within a THE1D LTR element ∼25 kb upstream of the native promoter and splice into a common downstream exon of the gene (Fig. 1A). IL2RB ESTs initiating from within this LTR utilize one of two splice donor (SD) sites within the LTR, and the resulting fusion transcript is predicted to produce the same protein as that from the native promoter because the ATG is in a common downstream exon (Fig. 1A, exon 2). Notably, all of the ESTs initiating within the LTR are from placenta, suggesting that activity of the LTR promoter may be placenta-specific. THE1D elements were actively retrotransposing 40–50 million years ago and comprise one subfamily of the larger THE1 retroelement family (25). This particular LTR (hereafter designated the IL2RB LTR) became fixed before the New World-Old World primate divergence because it is present in the genomes of Old World primates and the New World monkey marmoset as evident from genomic sequence alignments (data not shown).
The LTR Is a Placenta-specific Alternative Promoter of IL2RB
To validate the bioinformatics evidence that the IL2RB LTR confers placenta-specific gene expression, we performed qRT-PCR with promoter-specific primer pairs on a panel of commercial normal human RNA samples. Spleen, a positive control rich in hematopoietic cells, and placenta both expressed high levels of IL2RB RNA when compared with other tissues, and the LTR promoter was indeed only active in placenta (Fig. 1B). We corroborated these findings by testing IL2RB expression in several primary human PBMC and term placenta samples (Fig. 1C). Transcription in PBMC arises predominantly from the native promoter (85% of transcripts), whereas in the placenta samples, the LTR promoter was more active (88% of transcripts). There was a much higher variability in the levels of IL2RB transcripts in the placenta than in the PBMC (Fig. 1C), which is consistent with previous studies that have identified widely variable expression and DNA methylation levels as mechanisms that enable the placenta to adapt to its surroundings (26). From these results, it appears that the IL2RB LTR does indeed confer placenta-specific transcription.
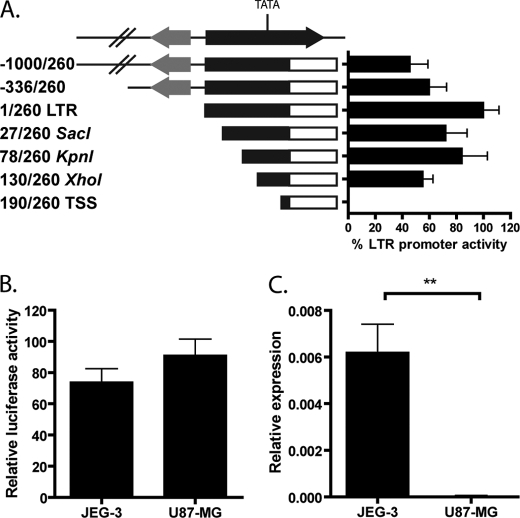
Promoter Activity in Cell Lines Does Not Reflect Actual Expression Levels
To test the IL2RB LTR promoter activity and tissue specificity in vitro, we generated a reporter construct in which the sequence encoding the 260 bp of the LTR 5′ to the first splice donor site was inserted upstream of a luciferase gene (pGL4-LTR). The LTR promoter did exhibit considerable promoter activity in JEG-3 cells (Fig. 2A), a choriocarcinoma cell line commonly used as a placenta model (27). To facilitate our search for potential DNA-binding motifs that may confer the tissue specificity of the LTR promoter observed in placenta, we generated a series of deletion constructs from 1 kb upstream of the promoter to the transcription start site (Fig. 2A). Significant loss of promoter activity occurred only upon deletion of a TATA box located at positions 170–174 in the LTR and 24 bp 5′ to the probable major transcriptional start site as judged by the number of ESTs initiating at this site. A survey of this core regulatory region using transcription factor databases failed to identify any placenta-specific transcription factor-binding sites (data not shown). Not surprisingly, given this information, the full-length construct as well as the deletion series exhibited comparable activity in U87-MG cells (Fig. 2B and data not shown), a glioblastoma-astrocytoma cell line that does not express IL2RB (Fig. 2C). These findings indicate that there is no sequence within the LTR or 1 kb upstream that is directly responsible for the tissue specificity of the promoter and point to some other mechanism of regulating the IL2RB LTR.
FIGURE 2.
LTR expression and promoter activity in cell lines. A, promoter activity of pGL4-LTR and derivative constructs in JEG-3 cells measured by luciferase reporter assay. The construct name indicates the number of bases and restriction site used for cloning upstream of the luciferase gene. The black arrow is the THE1D LTR, and the gray arrow is a nearby MLT1L LTR. Luciferase activity was determined when compared with a promoterless vector, and error bars represent the S.E. of at least three independent experiments. B, promoter activity of pGL4-LTR in JEG-3 and U87-MG cells measured as in A. C, total IL2RB expression in JEG-3 and U87-MG cell lines measured by qRT-PCR as in Fig. 1. **, p < 0.01.
DNA Methylation Determines Tissue Specificity of the LTR Promoter
Having found that there were no placenta-specific enhancer sequences within or upstream of the IL2RB LTR, we decided to probe the chromatin surrounding the LTR in placenta and PBMC for epigenetic modifications. Most transposable elements are thought to be silenced in differentiated tissues by DNA methylation at CpG dinucleotides, most likely a host defense mechanism to curb transpositional activity (28). Specifically, the THE family of LTR elements is generally heavily methylated in normal human somatic tissues (29). Methylated cytosines are preferentially mutated to thymidine, leading to an increased evolutionary rate of CpG to TpG mutation within transposable elements (30). Therefore, it was surprising to find that nine CpG sites have been retained within the LTR upstream of IL2RB, whereas other THE1D LTRs contain an average of only two CpGs (supplemental Fig. S1). Accordingly, we concluded that the IL2RB LTR is likely to have been relatively protected from methylation during evolution and postulated a role of DNA methylation in regulating its promoter activity.
We determined the DNA methylation status of nine CpG sites within the LTR and five CpG sites in the 320 bp immediately downstream of the LTR in four PBMC samples and six placenta samples by bisulfite sequencing (Fig. 3A). Overall levels of methylation within the LTR were significantly lower in the placenta (55%) than in the PBMC (88%) (Fig. 3A). When individual CpG residues were considered, seven sites had significantly lower methylation in the placenta, including one CpG (site 2) that overlies one of the splice donor sites. There was a significant inverse correlation between the CpG methylation of individual residues and the log expression level at five sites (supplemental Fig. S2). Both sample types displayed a heterogeneous pattern of methylation, which could be an inherent stochastic variability or due to the mix of cell types within the samples (26). Taken together, these data demonstrate that DNA methylation of the LTR is significantly different between the placenta and PBMC and indicate a probable regulatory role for DNA methylation in conferring tissue specificity of the IL2RB LTR promoter.
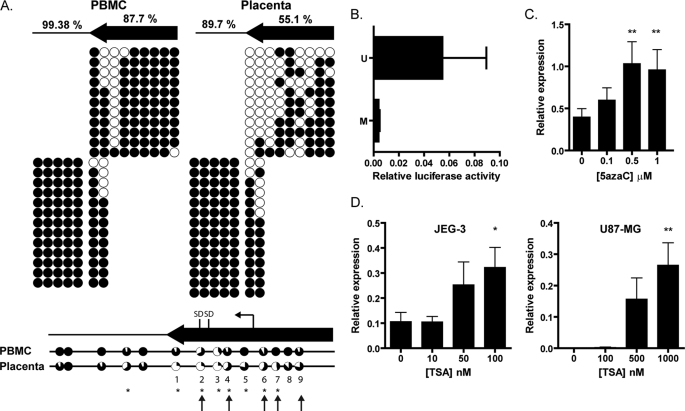
FIGURE 3.
Effect of chromatin context on tissue specificity of expression. A, DNA methylation of the LTR promoter and region immediately downstream in PBMC and placenta samples as determined by bisulfite sequencing. Upper panel, black circles show methylated CpGs, and white circles show unmethylated CpGs. Columns represent individual CpG sites, and rows represent individual sequence clones. Results from individual PBMC (left) and placenta (right) samples are shown, with the percentage of methylation in the LTR (arrow) or downstream region (line) indicated for the average of four PBMC and six placenta samples. Two PCRs were required to amplify the whole region, as represented by the individual blocks. Lower panel, combined results from PBMC (n = 4) and placenta (n = 6) samples. The portion of black for each circle represents the percentage of methylation at that site for all samples combined. Asterisks indicate statistically significant differences in DNA methylation between the two groups at individual CpGs (Fisher's exact test). Arrows indicate CpG residues for which the DNA methylation correlated significantly with the expression level (supplemental Fig. S2). CpG sites within the LTR are numbered 1–9. B, luciferase reporter activity of pGL4-LTR with the LTR sequence either in vitro methylated (M) or in vitro unmethylated (U). Error bars represent the S.E. from four independent experiments. There was no significant difference between activities of the methylated and unmethylated promoters due to the high variability of the assay. C, expression from the IL2RB LTR promoter in JEG-3 cells determined as in Fig. 1 following 5-azacytidine ([5azaC]) treatment. D, expression from the IL2RB LTR promoter in JEG-3 and U87-MG cells determined as in Fig. 1 following TSA treatment. Statistical significance determined by one-way analysis of variance is indicated as follows: *, p < 0.05; **, p < 0.01.
To investigate whether DNA methylation influences LTR promoter activity, we performed in vitro methylation of the IL2RB LTR in the pGL4-LTR construct used above. We found that DNA methylation inhibits the LTR promoter activity, indicating that this modification is sufficient to repress transcription initiation (Fig. 3B). Additionally, we treated JEG-3 cells with 5-azacytidine, an inhibitor of DNA methylation, and then measured transcription from the LTR promoter. There was a modest dose-dependent increase in expression from the LTR up to 1 μm 5-azacytidine, but higher concentrations were toxic to the treated cells (Fig. 3C). The increased expression is probably a direct effect of the drug treatment because the LTR is indeed demethylated after 5-azacytidine exposure (supplemental Fig. S3). We also detected an increase in expression when U87-MG cells were treated with 5-azacytidine, but this was probably an indirect effect because the LTR was mostly unmethylated in this cell line, a finding that suggests that other epigenetic factors play a role in transcriptional regulation of the IL2RB LTR promoter in these cells (supplemental Fig. S3). Nevertheless, these experiments showed that DNA methylation is a key regulatory mechanism by which placenta specificity of IL2RB expression is achieved.
Histone H3 Acetylation Is No Different between Placenta and Brain Cell Lines
Histone modifications are another class of epigenetic mechanisms that regulate transcription and determine tissue specificity (31). Acetylation of histone H3 is associated with actively transcribing genes and can be increased by treating cells with TSA, an inhibitor of histone deacetylases (32). TSA treatment modestly increased IL2RB transcription from the LTR promoter in JEG-3 cells, but a more striking effect was seen in U87-MG cells, where levels of transcription were up-regulated 400-fold, comparable with expression levels in JEG-3 cells (Fig. 3D). This led us to postulate that the IL2RB LTR may already be marked by H3 acetylation in JEG-3 cells, but induction of this mark in the U87-MG cells was sufficient to promote transcription from the IL2RB LTR. We therefore measured the level of histone acetylation in JEG-3 and U87-MG cells using chromatin immunoprecipitation but found no enrichment of H3Ac at the LTR in either cell line (data not shown). Therefore, the increased expression of IL2RB in U87-MG cells upon TSA treatment is an indirect effect, possibly due to activation of a secondary transcription factor or to the activation of a nearby cis-regulatory element.
LTR-initiating Transcripts Are Spliced More Efficiently in a Placenta Cell Line
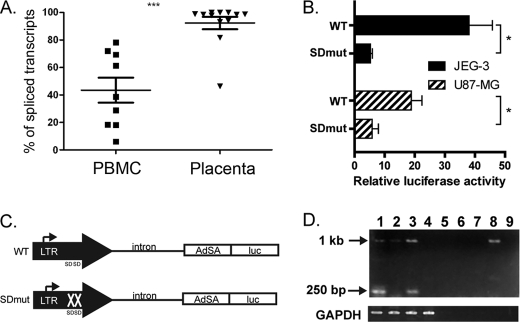
In integrated retroviruses, transcription initiates within the LTR in a similar manner to that seen for the IL2RB LTR. The retroviral splice donor site for the splicing event required to produce functional env transcripts may occur within the LTR itself or downstream, between the LTR and the gag ORF (33, 34). We noticed that the two splice donor sites in the IL2RB LTR are conserved in the THE1D LTR consensus sequence from Repbase (ATT|GTAAG and ACT|GTGAG, where | indicates the splicing event) and wondered whether this characteristic might contribute to the tissue specificity of the LTR promoter. Primer sequences located in the LTR and immediately downstream in the genomic DNA were used to amplify a putative unspliced mRNA initiating within the LTR and proceeding into downstream flanking sequences. The levels of unspliced transcripts were compared with the total levels of transcripts initiating in the LTR. For PBMC samples, not only was the overall level of LTR-initiated transcripts much lower than in placenta, but a much higher fraction of these transcripts was unspliced and presumably non-functional (Fig. 4A). In contrast, except for one sample, nearly all the LTR-initiated transcripts in placenta were spliced.
FIGURE 4.
Determination of splicing efficiency between the LTR and IL2RB exon 2. A, fractions of unspliced and spliced mRNA from the LTR promoter measured by qRT-PCR expressed as a percentage of total transcription initiation from the LTR (using data from Fig. 1). Each dot represents an individual biological replicate, and error bars represent the S.E. ***, p < 0.001. B, luciferase reporter activity of pGL4-spl1-AdSA (WT) and pGL4-spl1-AdSA-SDmut (SDmut) constructs in JEG-3 and U87-MG cells. Asterisks indicate statistical significance (*, p < 0.05) as determined by Student's t test. C, schematic of pGL4-spl1-AdSA and pGL4-spl1-AdSA-SDmut constructs (not to scale). Bent arrows indicate the transcription start site, AdSA is the adenovirus splice acceptor site, and luc is the luciferase gene. D, RT-PCR to detect splicing of the mRNA from pGL4-spl1-AdSA (lanes 1 and 3) and pGL4-spl1-AdSA-SDmut (lanes 2 and 4) in JEG-3 cells (lanes 1 and 2) and U87-MG cells (lanes 3 and 4). Lanes 5–8 are as lanes 1–4 but with no enzyme in the RT reaction, and lane 9 is a no template control. The 1-kb and 250-bp bands corresponding to the unspliced and spliced forms are indicated. GAPDH was used as a control.
To investigate this phenomenon further, a splicing reporter construct was generated (pGL4-spl1-AdSA), consisting of the entire IL2RB LTR, 1 kb of the downstream intron, and a known adenovirus splice acceptor site proximal to the luciferase gene (Fig. 4C). When transfected into the JEG-3 and U87-MG cell lines, the wild-type (WT) construct showed considerable reporter activity when compared with a promoterless vector, but a construct bearing mutations in the splice donor sites (pGL4-spl1-AdSA-SDmut) showed a significant reduction in promoter activity (Fig. 4B). This result suggests that a splicing defect may be responsible for the inability to produce correctly processed transcripts in non-placental tissue. To clarify this issue, we performed RT-PCR on mRNA extracted from the transfected cells (Fig. 4D). A 250-bp band corresponding to the spliced form was seen in both JEG-3 and U87-MG cells transfected with the WT but not the mutant construct, confirming that mutation of the SD site inhibits splicing. (The full-length band of 1 kb was observed in most lanes due to plasmid contamination.) These results are consistent with our finding that tissue specificity of the LTR promoter activity is lost when the sequence is ectopically introduced in a reporter construct. Therefore, although we do see differences in splicing efficiency between the PBMC and placenta samples, the LTR sequence alone does not contain all the regulatory components necessary to confer this specificity. It is possible that epigenetic factors such as DNA methylation affect efficient splicing and formation of the LTR-IL2RB fusion transcript.
IL2RB Is Predominantly Expressed in Placental Trophoblast
Because IL2RB is normally expressed in the hematopoietic system, we sought to determine its functional role in the placenta. We first investigated which placental cell type expresses IL2RB given that the placenta is a heterogeneous organ. Placental decidua and villi were dissected, and populations of trophoblast and mesenchyme were obtained by enzymatic digestion of the villi (see “Experimental Procedures”). The overall level of IL2RB expression was slightly higher in trophoblast than in mesenchyme samples, and the LTR promoter was predominantly used in the trophoblast (Fig. 5A). Nevertheless, these differences were not statistically significant and were likely due to variability between the biological samples tested (compare also Fig. 1C). However, average DNA methylation was significantly different between three trophoblast samples and three mesenchyme samples, 40.1% versus 66.9%, respectively (Fig. 5B). Furthermore, DNA methylation levels were inversely correlated with expression level, leading us to conclude that the trophoblast is the predominant tissue in which IL2RB is expressed (Fig. 5B). The trend of lower DNA methylation in trophoblast was also true for first and second trimester samples (supplemental Fig. S4). A statistical analysis showed that differential methylation occurs specifically at CpG residues 1, 2, 4, and 7 (Fig. 5B). Although no RNA samples were available for the maternal decidua, this tissue was almost completely methylated and is therefore unlikely to be a significant source of IL2RB expression (Fig. 5B). Taken together, these results demonstrate that IL2RB is expressed primarily in placental trophoblast cells from the LTR promoter and that its expression is regulated by DNA methylation.
Transcription of Other Members of the IL-2 and IL-15 Receptor Complexes in Placenta
In the hematopoietic system, the IL-2 receptor β subunit forms a functional heterotrimeric receptor in complex with the common γ chain (encoded by IL2RG) and either the IL-2 receptor α chain or the IL-15 receptor α chain (encoded by IL2RA and IL15RA, respectively) as shown schematically in Fig. 5C (17). We analyzed the transcript levels of these other receptor subunits in the placenta to determine whether or not a functional receptor could be formed in this tissue. Among IL15RA, IL2RA, and IL2RG, transcripts were detectable for all three genes in placenta, with IL15RA being significantly higher than the other two (data not shown). We subsequently tested the expression of IL15RA and IL15 in the purified trophoblast and mesenchyme. Interestingly, the IL15RA subunit was expressed more highly in the mesenchyme cells (Fig. 5D), perhaps indicating a mode of trans-presentation with the α subunit present on the mesenchyme and the β subunit present on the trophoblast. IL15 mRNA but not IL2 mRNA was detectable in both trophoblast and mesenchyme (Fig. 5D and data not shown). These data indicate that the IL-15 signaling pathway is more likely to be active in the placental villi.
Expression and Efficient Cleavage of the IL-2Rβ Protein in Placenta
We next analyzed the protein product of the LTR-driven IL2RB mRNA by Western blot on protein extracts from four placental villi (Fig. 5E and supplemental Fig. S5). The membrane was probed with an antibody directed toward the C-terminal signaling domain of IL-2Rβ, and a very strong band at 37 kDa was seen in the placenta samples, not the expected 75-kDa full-length form observed in our positive control. A competition assay using the same antibody preincubated with a peptide shows the 37-kDa protein to be specific (Fig. 5E). Furthermore, we have tested another C-terminal specific antibody and have obtained similar results (supplemental Fig. S5). IL-2Rβ has been shown to undergo cleavage by a matrix metalloprotease (MMP) in leukemic cell lines, resulting in the formation of a 50-kDa fragment corresponding to the extracellular domain and a 37-kDa fragment encompassing the C-terminal domain (35, 36). Thus, although we cannot rule out that the isoform detected in placenta is produced by alternative splicing, the band size corresponds precisely to the intracellular domain that would result from proteolytic cleavage of IL-2Rβ. Moreover, matrix metalloproteases are known to be expressed in the villous trophoblast (37). The presence of the 37-kDa isoform and a demonstrable absence of the full-length protein indicate that IL-2RB is preferentially cleaved in placenta.
DISCUSSION
Here we have described a new instance of an LTR promoter that confers tissue-specific expression of IL2RB in the placenta. When introduced into a reporter construct, the LTR demonstrated nonspecific promoter activity, indicating that chromatin context regulates its tissue specificity. Indeed, we found that DNA methylation is a key determinant of the expression pattern, but histone acetylation near the LTR itself did not seem to play a major role. The fusion transcript between the LTR and downstream IL2RB exons requires efficient splicing from the LTR-derived SD sites, which occurs preferentially in the placenta and also appears to be regulated by epigenetic factors. We identified the trophoblast as the subpopulation of placental cells in which the majority of LTR-derived IL2RB transcription occurs and found that the IL-2Rβ protein undergoes post-translational cleavage to produce a membrane-bound signaling domain.
Several other examples are known in which an LTR confers placenta-specific gene expression, although in at least some of these cases, there are sequence determinants within the LTR itself that confer the tissue-specific effects (8, 38). For example, the LTR upstream of EDNRB was shown to have promoter activity in a reporter assay specifically in JEG-3 cells, quite in contrast to the findings here, and serial deletions identified two enhancers within the LTR that probably bind placenta-specific transcription factors (8). The LTR promoter of syncytin, however, has core promoter activity, but the placental specificity of transcription is conferred by binding of Gcm1, a placenta-specific transcription factor, to DNA upstream of the LTR (39, 40). In this case, the genomic position of the LTR insertion has serendipitously resulted in tissue-specific expression and the subsequent domestication of the env ORF.
DNA methylation is an epigenetic modification normally associated with gene silencing, which may have evolved as a host-defense mechanism to prevent potentially detrimental transposition events of transposable elements (28, 41). Most CpG dinucleotides in the genome are normally methylated in somatic tissues, with the exception of those located within CpG islands, regions of high CpG density that escape the global DNA methylation that occurs during early development (41). Therefore, LTR elements that are unmethylated can be considered to be specifically derepressed. Some studies have shown that the placenta is globally hypomethylated when compared with other somatic tissues (42), and it is possible that this hypomethylation is at least partly responsible for the high expression of ERVs in the placenta. Several examples of LTR elements that act as alternative promoters in the placenta have been shown to be unmethylated (22, 43), but this effect may be secondary to the LTR or closely linked sequence determinants in terms of causing tissue specificity. In contrast, we have shown here that demethylation of the IL2RB LTR promoter is a primary mechanism by which tissue-specific transcription is achieved. These findings support previous work from our laboratory showing that demethylation of LTR promoters in placenta is specific and not merely a consequence of global placental hypomethylation (22).
The ability of a solitary LTR to exhibit promoter activity and form a productive fusion transcript with a downstream gene depends upon the presence of a splice donor site in the genomic DNA flanking the LTR (44) or within the LTR itself (45), as is the case here. The two splice donor sites used in the IL2RB LTR are present in the consensus sequence for the THE1D family of ERVs. This might result in THE1D LTRs more frequently being exapted as LTR promoters as they are not dependent on nearby splice donor sites to form fusion transcripts with nearby exons. Although no such genome-wide study has been performed, another intriguing example of a THE1 LTR promoter was recently described (9). These authors found that a THE1B LTR was exapted as an alternative promoter of CSF1R in Hodgkin lymphoma and that this gene expression contributed to tumor cell survival. Using degenerate PCR, they demonstrated that other THE1B copies are expressed in these cells and probably do form fusion transcripts with cellular genes. The CSF1R LTR promoter was found to be derepressed by DNA demethylation in a number of lymphoma cell lines when compared with normal PBMC. Therefore, it is not unreasonable to speculate that other members of the THE1 family of LTRs may act as tissue-specific promoters in the placenta. Our data show that the splicing is more efficient in the placenta and is probably also dependent on the chromatin context because the tissue specificity of splicing efficiency is lost in a transfected splicing-dependent reporter construct.
The divergence of placentation in mammals has likely been impacted by the differences in transposable elements between species (13). The maternal decidua is created by transformation of the uterine mucosa and is characterized by the presence of uterine natural killer (uNK cells) and a number of other immune cell types including macrophages and T lymphocytes (46). Successful human pregnancy can be viewed as a delicate balance in which the extent of trophoblast invasion required for sufficient fetal nutrition is offset by potentially detrimental disruption to the uterine lining (46). This stabilization is achieved in part by inflammatory processes, including cytokine signaling, that modulate the activities of decidual leukocytes (46, 47). IL-15 is known to increase the differentiation, proliferation, and cytotoxic capability of uNK cells (19). IL-2 can also activate regulatory T helper cells, whose presence dampens the maternal immune response to the fetus (48). Nevertheless, these cell types are derived from peripheral blood precursors so that their expression of IL2RB is mostly likely from the native promoter rather than the LTR. In contrast, little is known about the effects of IL-2 or IL-15 on the fetal villi, although the proliferation of JEG-3 cells is unaffected by IL-2 (Ref. 51 and data not shown). The villi consist of a mesenchymal core, containing the fetal blood vessels, surrounded by the trophoblast cell layer (46). The trophoblast represents a heterogeneous mix of cytotrophoblast cells at various stages of differentiation and syncytiotrophoblast cells formed by fusion events that secrete hormones and are involved in decidual invasion. Scant evidence suggests that IL-2 may be expressed in the syncytiotrophoblast (49) and may be secreted from cultured trophoblast cells (50), although we did not detect IL-2 mRNA in our purified trophoblast and mesenchyme samples. Our data confirmed previous results that IL-15 is expressed by the syncytiotrophoblast and villi mesenchyme (19, 52), and IL-15 has been shown to increase the invasion of JEG-3 cells in a model system (53). We also found that the trophoblast and mesenchyme express the other subunits required to form functional cytokine receptors. Taken together, our findings support the possibility that IL-2 and/or IL-15 may be able to have direct effects on the trophoblast as well as on the decidual leukocytes.
Several cytokine receptors are known to undergo proteolytic cleavage or alternative splicing events that lead to the formation of soluble extracellular domains (54). These domains have the potential to regulate cytokine activity in a number of ways, including inhibition by direct competition for cytokine binding or activation by increasing cytokine stability or the affinity of cytokine-receptor interactions (55). Indeed, the IL-2 and IL-15 receptor α chains are both known to undergo such ectodomain shedding, and the presence of soluble IL-2Rα is a clinical indicator for disease progression in a number of infectious and autoimmune diseases, hematologic malignancies, and solid tumors (56, 57). Two studies have shown that IL-2Rβ undergoes proteolytic cleavage by a matrix metalloprotease to form a 50-kDa soluble form consisting of the extracellular domain and a 37-kDa cytoplasmic domain that is capable of cytokine signaling (35, 36). The cleaved forms were detectable at low levels in activated PBMC but were more frequent in leukemic and lymphoid cell lines. Here we have shown for the first time that IL-2Rβ appears to be constitutively cleaved in the trophoblast of the placenta. The bare C-terminal domain may confer proliferative capacity to the trophoblast cells independently of IL-2 or IL-15 because this subunit was phosphorylated in Baf-β cells undergoing cytokine starvation (36). Therefore, the soluble extracellular domain of IL-2Rβ could have either positive or negative effects on the trophoblast or on juxtaposed maternal peripheral blood leukocytes in the interstitial space.
The above speculations all assume that the cleaved form is beneficial to the development of the placenta, and to fetal growth. Without knowing more about the role of this protein in normal pregnancy or in disorders such as intrauterine growth restriction or pre-eclampsia, we cannot rule out that the expression of IL2RB from the LTR promoter plays no functional role or is in fact detrimental. In the latter case, perhaps proteolytic cleavage of the protein product is a host defense mechanism required to counter the effects of LTR promoter activity that have escaped normal repression by DNA methylation. Nevertheless, this scenario is less likely because detrimental transposable element insertions are usually selected against in evolution.
In summary, we have described the expression of IL2RB from an LTR promoter. The co-option of this LTR as a promoter contributes novel tissue specificity of IL2RB expression that may have a functional impact in placenta biology. Further studies will be necessary to elucidate the potential functions of both the extracellular and the intracellular subunits of IL-2Rβ in placentation.
Supplementary Material
Acknowledgments
We thank Ruby Jiang, Maria Peñaherrera, and Dan Diego-Alvarez for help with placenta samples and expertise in trophoblast separation and C. Benjamin Lai and Liane Gagnier for technical assistance. We also thank Mark Romanish for comments on the manuscript.
This work was supported by a grant from the Canadian Institute for Health Research (to D. L. M.) with core support provided by the BC Cancer Agency.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- ERV
- endogenous retrovirus
- IL2RB
- interleukin-2 receptor β subunit
- EST
- expressed sequence tag
- PBMC
- peripheral blood mononuclear cell(s)
- AdSA
- adenovirus splice acceptor site
- TSA
- trichostatin A
- qRT-PCR
- quantitative reverse transcription
- SD
- site donor.
REFERENCES
- 1. International Human Genome Sequencing Consortium (2001) Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 2. Jern P., Coffin J. M. (2008) Annu. Rev. Genet. 42, 709–732 [DOI] [PubMed] [Google Scholar]
- 3. Belshaw R., Watson J., Katzourakis A., Howe A., Woolven-Allen J., Burt A., Tristem M. (2007) J. Virol. 81, 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen C. J., Lock W. M., Mager D. L. (2009) Gene 448, 105–114 [DOI] [PubMed] [Google Scholar]
- 5. Landry J. R., Mager D. L., Wilhelm B. T. (2003) Trends Genet. 19, 640–648 [DOI] [PubMed] [Google Scholar]
- 6. Baek D., Davis C., Ewing B., Gordon D., Green P. (2007) Genome Res. 17, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davuluri R. V., Suzuki Y., Sugano S., Plass C., Huang T. H. (2008) Trends Genet. 24, 167–177 [DOI] [PubMed] [Google Scholar]
- 8. Landry J. R., Mager D. L. (2003) J. Virol. 77, 7459–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lamprecht B., Walter K., Kreher S., Kumar R., Hummel M., Lenze D., Köchert K., Bouhlel M. A., Richter J., Soler E., Stadhouders R., Jöhrens K., Wurster K. D., Callen D. F., Harte M. F., Giefing M., Barlow R., Stein H., Anagnostopoulos I., Janz M., Cockerill P. N., Siebert R., Dörken B., Bonifer C., Mathas S. (2010) Nat. Med. 16, 571–579 [DOI] [PubMed] [Google Scholar]
- 10. Okahara G., Matsubara S., Oda T., Sugimoto J., Jinno Y., Kanaya F. (2004) Genomics 84, 982–990 [DOI] [PubMed] [Google Scholar]
- 11. Blikstad V., Benachenhou F., Sperber G. O., Blomberg J. (2008) Cell. Mol. Life Sci. 65, 3348–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Varela M., Spencer T. E., Palmarini M., Arnaud F. (2009) Ann. N.Y. Acad. Sci. 1178, 157–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rawn S. M., Cross J. C. (2008) Annu. Rev. Cell Dev. Biol. 24, 159–181 [DOI] [PubMed] [Google Scholar]
- 14. Sugimoto J., Schust D. J. (2009) Reprod. Sci. 16, 1023–1033 [DOI] [PubMed] [Google Scholar]
- 15. Mi S., Lee X., Li X., Veldman G. M., Finnerty H., Racie L., LaVallie E., Tang X. Y., Edouard P., Howes S., Keith J. C., Jr., McCoy J. M. (2000) Nature 403, 785–789 [DOI] [PubMed] [Google Scholar]
- 16. Frendo J. L., Olivier D., Cheynet V., Blond J. L., Bouton O., Vidaud M., Rabreau M., Evain-Brion D., Mallet F. (2003) Mol. Cell. Biol. 23, 3566–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma A., Koka R., Burkett P. (2006) Annu. Rev. Immunol. 24, 657–679 [DOI] [PubMed] [Google Scholar]
- 18. Lan R. Y., Selmi C., Gershwin M. E. (2008) J. Autoimmun 31, 7–12 [DOI] [PubMed] [Google Scholar]
- 19. Verma S., Hiby S. E., Loke Y. W., King A. (2000) Biol. Reprod 62, 959–968 [DOI] [PubMed] [Google Scholar]
- 20. Moffett-King A. (2002) Nat. Rev. Immunol. 2, 656–663 [DOI] [PubMed] [Google Scholar]
- 21. Henderson K. G., Shaw T. E., Barrett I. J., Telenius A. H., Wilson R. D., Kalousek D. K. (1996) Hum. Genet. 97, 650–654 [DOI] [PubMed] [Google Scholar]
- 22. Reiss D., Zhang Y., Mager D. L. (2007) Nucleic Acids Res. 35, 4743–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van de Lagemaat L. N., Landry J. R., Mager D. L., Medstrand P. (2003) Trends Genet. 19, 530–536 [DOI] [PubMed] [Google Scholar]
- 24. Conley A. B., Piriyapongsa J., Jordan I. K. (2008) Bioinformatics 24, 1563–1567 [DOI] [PubMed] [Google Scholar]
- 25. Smit A. F. (1993) Nucleic Acids Res. 21, 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Avila L., Yuen R. K., Diego-Alvarez D., Peñaherrera M. S., Jiang R., Robinson W. P. (2010) Placenta 31, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 27. Kilic M., Flossmann E., Flossmann O., Vogelsang H., Junker U., Chaouat G., Markert U. R. (1999) Am. J. Reprod Immunol. 41, 61–69 [DOI] [PubMed] [Google Scholar]
- 28. Yoder J. A., Walsh C. P., Bestor T. H. (1997) Trends Genet. 13, 335–340 [DOI] [PubMed] [Google Scholar]
- 29. Szpakowski S., Sun X., Lage J. M., Dyer A., Rubinstein J., Kowalski D., Sasaki C., Costa J., Lizardi P. M. (2009) Gene 448, 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walser J. C., Ponger L., Furano A. V. (2008) Genome Res. 18, 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 32. Yoshida M., Horinouchi S. (1999) Ann. N.Y. Acad. Sci. 886, 23–36 [DOI] [PubMed] [Google Scholar]
- 33. Majors J. (1990) Curr. Top. Microbiol Immunol. 157, 49–92 [DOI] [PubMed] [Google Scholar]
- 34. Muranyi W., Flügel R. M. (1991) J. Virol. 65, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Honda M., Kitamura K., Takeshita T., Sugamura K., Tokunaga T. (1990) J. Immunol. 145, 4131–4135 [PubMed] [Google Scholar]
- 36. Montes de Oca P., Malardé V., Proust R., Dautry-Varsat A., Gesbert F. (2010) J. Biol. Chem. 285, 22050–22058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cohen M., Meisser A., Bischof P. (2006) Placenta 27, 783–793 [DOI] [PubMed] [Google Scholar]
- 38. Landry J. R., Rouhi A., Medstrand P., Mager D. L. (2002) Mol. Biol. Evol. 19, 1934–1942 [DOI] [PubMed] [Google Scholar]
- 39. Yu C., Shen K., Lin M., Chen P., Lin C., Chang G. D., Chen H. (2002) J. Biol. Chem. 277, 50062–50068 [DOI] [PubMed] [Google Scholar]
- 40. Prudhomme S., Oriol G., Mallet F. (2004) J. Virol. 78, 12157–12168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki M. M., Bird A. (2008) Nat. Rev. Genet. 9, 465–476 [DOI] [PubMed] [Google Scholar]
- 42. Fuke C., Shimabukuro M., Petronis A., Sugimoto J., Oda T., Miura K., Miyazaki T., Ogura C., Okazaki Y., Jinno Y. (2004) Ann. Hum. Genet. 68, 196–204 [DOI] [PubMed] [Google Scholar]
- 43. Matousková M., Blazková J., Pajer P., Pavlícek A., Hejnar J. (2006) Exp. Cell Res. 312, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 44. Medstrand P., Landry J. R., Mager D. L. (2001) J. Biol. Chem. 276, 1896–1903 [DOI] [PubMed] [Google Scholar]
- 45. Dunn C. A., Medstrand P., Mager D. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12841–12846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moffett A., Loke C. (2006) Nat. Rev. Immunol. 6, 584–594 [DOI] [PubMed] [Google Scholar]
- 47. Bulmer J. N., Williams P. J., Lash G. E. (2010) Int. J. Dev. Biol. 54, 281–294 [DOI] [PubMed] [Google Scholar]
- 48. Aluvihare V. R., Kallikourdis M., Betz A. G. (2004) Nat. Immunol. 5, 266–271 [DOI] [PubMed] [Google Scholar]
- 49. Boehm K. D., Kelley M. F., Ilan J., Ilan J. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rein D. T., Breidenbach M., Hönscheid B., Friebe-Hoffmann U., Engel H., Göhring U. J., Uekermann L., Kurbacher C. M., Schöndorf T. (2003) Cytokine 23, 119–125 [DOI] [PubMed] [Google Scholar]
- 51. Hamai Y., Fujii T., Yamashita T., Miki A., Hyodo H., Kozuma S., Geraghty D. E., Taketani Y. (1999) Am. J. Reprod Immunol. 41, 153–158 [DOI] [PubMed] [Google Scholar]
- 52. Amash A., Huleihel M., Eyal S., Maor E., Myatt L., Holcberg G. (2007) Eur. Cytokine Netw 18, 188–194 [DOI] [PubMed] [Google Scholar]
- 53. Zygmunt M., Hahn D., Kiesenbauer N., Münstedt K., Lang U. (1998) Am. J. Reprod Immunol. 40, 326–331 [DOI] [PubMed] [Google Scholar]
- 54. Levine S. J. (2008) J. Biol. Chem. 283, 14177–14181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fernandez-Botran R. (1999) Crit. Rev. Clin. Lab. Sci. 36, 165–224 [DOI] [PubMed] [Google Scholar]
- 56. Murakami S. (2004) Front. Biosci. 9, 3085–3090 [DOI] [PubMed] [Google Scholar]
- 57. Witkowska A. M. (2005) Mediators Inflamm. 2005, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.