Background: The mitochondrial ATP synthase adopts oligomeric structures.
Results: Cross-linking between two subunits 6 (a) indicates their proximity in the ATP synthase dimer.
Conclusion: Subunit 6 participates in the ATP synthase monomer-monomer interface.
Significance: Learning how the ATP synthases assemble is essential for understanding their involvement in the control of the biogenesis of the inner mitochondrial membrane.
Keywords: F1F0-ATPase, Mitochondria, Protein Cross-linking, Proteomics, Yeast
Abstract
The involvement of subunit 6 (a) in the interface between yeast ATP synthase monomers has been highlighted. Based on the formation of a disulfide bond and using the unique cysteine 23 as target, we show that two subunits 6 are close in the inner mitochondrial membrane and in the solubilized supramolecular forms of the yeast ATP synthase. In a null mutant devoid of supernumerary subunits e and g that are involved in the stabilization of ATP synthase dimers, ATP synthase monomers are close enough in the inner mitochondrial membrane to make a disulfide bridge between their subunits 6, and this proximity is maintained in detergent extract containing this enzyme. The cross-linking of cysteine 23 located in the N-terminal part of the first transmembrane helix of subunit 6 suggests that this membrane-spanning segment is in contact with its counterpart belonging to the ATP synthase monomer that faces it and participates in the monomer-monomer interface.
Introduction
F1F0-ATP synthase is a molecular rotary motor that is responsible for aerobic synthesis of ATP. It exhibits a headpiece (catalytic sector), a base piece (membrane sector), and two connecting stalks. The F1 sector is a water-soluble unit retaining the ability to hydrolyze ATP when in soluble form. F0 is embedded in the membrane and is mainly composed of hydrophobic subunits forming a specific proton-conducting pathway. When the F1 and F0 sectors are coupled, the enzyme functions as a reversible H+-transporting ATPase or ATP synthase (1) for review.
Crystallization experiments led to a model at 3.9 Å resolution of a Saccharomyces cerevisiae F1c10-ATP synthase subcomplex (2). More recently, the model was refined at 3.4 Å resolution (3). So far, the peripheral stalk and the membranous part of the stator are lacking in crystals for either yeast or bovine enzyme (4). At a lower resolution, only cryo-electron microscopy has provided a complete view of the yeast ATP synthase to date (5).
Mitochondrial F1F0-ATP synthases adopt oligomeric structures. This has been shown in native polyacrylamide gels of detergent extracts from a wide range of species including yeast, mammals, and plants (6–8). Moreover, low resolution structural data obtained by single particle electron microscopy analyses are available for dimeric forms in detergent extracts (9–14). In all cases, the ATP synthase dimers were found associated by their F0 domains. Previously, the association of ATP synthases in the mitochondrial membrane was evidenced from chemical cross-linking (15, 16) and in vivo cross-linking experiments (17). More recently, atomic force microscopy and electron microscopy experiments revealed the oligomeric state of the enzyme in the inner mitochondrial membrane of yeast (12, 18) and the presence of dimer ribbons of ATP synthase in mammalian mitochondria (19) and Polytomella mitochondria (20). Rows of F1 were also observed by cryo-electron tomography of Neurospora crassa mitochondria (21).
Several components that are involved in the association of ATP synthases in yeast have been proposed. The dimer-specific subunits e and g were identified on the basis that the deletion of their genes leads to ATP synthase monomers in native polyacrylamide gels (6). However, these subunits stabilize the dimeric structures of ATP synthases but are not essential for their formation (22, 23). On the basis of cross-linking experiments, we found that subunits e and g participate most probably in the dimer-dimer interface to constitute ATP synthase oligomers (24, 25).
Recently, it was proposed that the highly hydrophobic mitochondrially encoded subunit a (subunit 6 in yeast) is essential for dimerization because the dimeric a-subunit associated with two c-rings (c10a2c10) was identified in SDS gels, thus suggesting that the a-subunit may constitute an important part of the monomer-monomer interface of ATP synthases (26). Mature subunit 6 is a hydrophobic protein with a molecular mass of 27,958 Da and a stoichiometry of 1. It belongs to the F0 domain and constitutes the proton channel together with the subunit c ring. Subunit 6 is homologous to the Escherichia coli subunit a. From insertion of cysteine residues, a topological model with five TMHs3 was proposed for the prokaryotic protein subunit a, with the NH2 terminus in the periplasm and the COOH terminus in the cytoplasm (27). Little is known about the structure of the eukaryotic subunit a. Although the protein sequences of eukaryotic and prokaryotic proteins are only weakly related, their hydrophobic profiles suggest that they have common secondary structures (28). Thus, the N terminus of the yeast subunit 6 was found located in the mitochondrial intermembrane space (29), showing that the two proteins display some similarities.
The present work investigates the involvement of the S. cerevisiae subunit 6 in the interface between ATP synthase monomers. Based on cross-linking experiments and using the unique cysteine 23 as target, we show that two subunits 6 are close in the inner mitochondrial membrane and also in the solubilized supramolecular forms of the yeast ATP synthase.
EXPERIMENTAL PROCEDURES
Materials
Digitonin was from Sigma. All other reagents were of reagent grade quality.
Strains
The wild-type yeast strain MR6 (Mata ade2-1 his3-11,15 trp1-1 leu2-3,112 ura3-1 CAN1 arg8::HIS3) was kindly provided by Dr. Jean Paul di Rago (Institut de Biochimie et Génétique Cellulaires (IBGC), Bordeaux, France). This strain, which has the nuclear genotype of the strain W303-1B, was built to introduce mutations in the mitochondrially encoded ATP synthase subunits (30). The null mutant ΔTIM11 was constructed by PCR-based mutagenesis (31). The latter strain is devoid not only of subunit e but also of subunit g, which is the consequence of the absence of subunit e (6).
Biochemical Procedures
Cells were grown aerobically at 28 °C in a complete liquid medium containing 2% lactate as carbon source (32). Mitochondria were prepared from protoplasts as described previously (33). Protein amounts were determined according to Ref. 34 in the presence of 5% SDS using bovine serum albumin as standard. The ATPase activity with and without oligomycin was measured at pH 8.4 in the presence of 0.375% Triton X-100 to remove the endogenous IF1 inhibitor (35). Cross-linking experiments were performed as described in Ref. 24 at either 4 °C or 30 °C with 2 mm CuCl2 as oxidative agent. Triton X-100 extracts were performed as follows. 200 μl of 2% Triton X-100 (w/v) were added to the same volume of mitochondrial suspension (10 mg of protein/ml) in 0.1 m mannitol, 50 mm HEPES, pH 7.4. Incubation was performed at 4 °C for 30 min, and the extract was centrifuged at 30,000 × g for 15 min. The supernatant was then incubated with CuCl2. Organic solvent extracts containing subunit 6 and the 47-kDa adduct were obtained from small scale experiments as described in Ref. 36.
Electrophoretic and Western Blot Analyses
SDS-gel electrophoreses were done as described (37). Western blot analyses were conducted using nitrocellulose membranes (Whatman PROTEANR BA83, 0.2 μm), and polyclonal antibodies against subunit 6 were used as described previously (38). Peroxidase activity of secondary antibodies was visualized on Amersham Biosciences HyperfilmTM MP (GE Healthcare) after incubating the nitrocellulose membrane for 1 min in 0.4 mm coumaric acid, 2.5 mm luminol, 0.0165% (v/v) H2O2, 0.1 m Tris, pH 8.5. The amount of subunit 6 and 47-kDa cross-linked product was determined by using the G:BOX Chemi IR6 (Syngene) and Western Lighting Plus-ECL (PerkinElmer Life Sciences) as enhanced chemiluminescence substrate.
CN-PAGE experiments were done as described in Ref. 39. After electrophoresis, the gel was incubated in 5 mm ATP, 5 mm MgCl2, 0.05% lead acetate, 50 mm glycine-NaOH, pH 8.4, to identify the bands showing an ATPase activity (40, 41). The bands were revealed by the white precipitate resulting from ATP hydrolysis. They were cut, and fragments were submitted to an SDS-PAGE gel that was transferred to a nitrocellulose membrane. Subunit 6 and cross-links were revealed by Western blotting. Silver staining of SDS-polyacrylamide gels has been described previously (42). Selected bands were excised from gels and digested with trypsin using standard protocols, the resulting peptide mixture being analyzed by MALDI-MS, MALDI-MS/MS, and/or LC-MS/MS. MALDI-MS and MALDI-MS/MS analyses were performed on a MALDI-TOF/TOF Ultraflex III mass spectrometer (Bruker) with α-cyano-4-hydroxy-cinnamic acid (Sigma-Aldrich) used as a matrix. For LC-MS/MS analysis, the peptide mixture was analyzed on a Dionex U-3000 Ultimate nano LC system coupled to a nanospray LTQ-Orbitrap XL mass spectrometer (Thermo-Finnigan). Data were searched by SEQUEST through the Bioworks 3.3.1 interface (Thermo-Finnigan) against a Swiss-Prot database restricted to S. cerevisiae species (7245 entries, version 2010.12) supplemented by mature ATP6 sequence.
RESULTS
In an Oxidative Environment, the Yeast ATP Synthase Subunit 6 Cross-links to Produce a 47-kDa Adduct
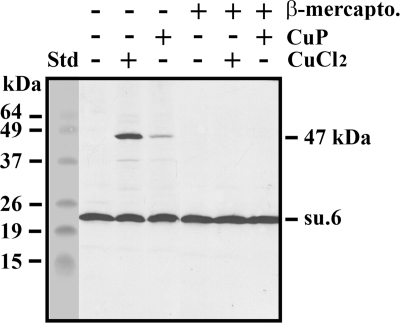
The mature yeast subunit 6 displays a unique cysteine residue in its sequence at position 23. This residue, which is non-conserved, was susceptible to cross-linking from the intermembrane space with three components of the peripheral stalk (subunits 4, 8, and i) that are located in its neighborhood (29, 43, 44). In this work, we found that a cross-linking product involving subunit 6 and showing a relative mass of 47,000 was obtained upon incubation of wild-type mitochondria with CuCl2 (Fig. 1). This adduct disappeared upon the addition of β-mercaptoethanol, thus indicating the possibility that subunit 6 can establish a disulfide bridge with another protein bearing a cysteine residue. Surprisingly, the intensity of the 47-kDa band was highly decreased in the presence of Cu(1,10-phenanthroline)2SO4, a reagent widely used to promote disulfide bond formation. Among the various parameters influencing the cross-linking, temperature was critical because the formation of the disulfide bridge was prevented at 4 °C (Fig. 2). Furthermore, as shown on the figure, a similar amount of 47-kDa adduct was obtained after 15 and 30 min of incubation with CuCl2. In fact, a kinetic analysis from 2 to 15 min displayed similar amounts of 47-kDa product (not shown). Careful quantification (see “Experimental Procedures”) of several Western blots showed that 38% ± 2% of subunit 6 was present in the 47-kDa cross-linked product. The effect of such a cross-linking on ATPase activity was an important issue. In fact, the ATPase activity of CuCl2-treated mitochondria was not modified. In addition, oligomycin sensitivity was not affected, indicating that physical constraints resulting from the formation of the disulfide bond do not have any effect on proton channel activity (Table 1).
FIGURE 1.
Oxidation of wild-type yeast mitochondria leads to a 47-kDa adduct involving subunit 6 (su.6). Mitochondria (250 μg of protein) in 50 μl of 0.1 m mannitol, 50 mm HEPES, pH 7.4, were incubated with or without 2 mm CuCl2 or Cu(1,10-phenanthroline)2SO4 (CuP) for 15 min at 30 °C. The reactions were stopped upon the addition of 10 mm EDTA and 10 mm NEM. 2% (v/v) β-mercaptoethanol (β-mercapto.) was added to samples to reduce the disulfide bridge. Samples were dissociated, and aliquots (30 μg of protein) were analyzed by SDS-PAGE on a 16% polyacrylamide gel. The gel was transferred to a nitrocellulose membrane that was revealed with a polyclonal antibody raised against the yeast subunit 6. The BenchMarkTM prestained protein ladder was the standard (Std).
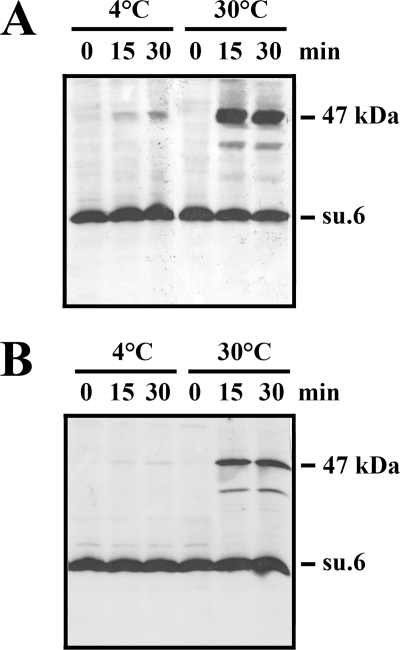
FIGURE 2.
The 47-kDa adduct involving subunit 6 (su.6) is obtained at 30 °C upon oxidation of either mitochondria or a 1% Triton X-100 extract. A and B, mitochondria (A) and a 1% (w/v) Triton X-100 extract (B) were incubated with 2 mm CuCl2 at either 4 °C or 30 °C for 0, 15, and 30 min. The reactions were stopped upon the addition of 10 mm EDTA and 10 mm NEM, and the samples were analyzed as in Fig. 1.
TABLE 1.
ATPase activity
The control strain was the MR6 strain (see “Experimental Procedures”). Mitochondria were prepared from protoplasts and incubated or not with 2 mm CuCl2 for 15 min at 30 °C. The ATPase activities and sensitivity to the F0 inhibitor oligomycin (6 μg/ml) were measured at pH 8.4 in the presence of 0.375% Triton X-100 (w/v) to remove the IF1 inhibitor.
| Samples | Control | +Oligomycin | Oligomycin sensitivity |
|---|---|---|---|
| μmol of Pi/min/mg of protein | % | ||
| MR6 mitochondria | 6.47 ± 0.33 | 0.67 ± 0.06 | 89.6 |
| CuCl2-treated | 6.39 ± 0.15 | 0.59 ± 0.05 | 90.8 |
| MR6 mitochondria | |||
The Cross-linked Product Involving Subunit 6 Was Obtained in Detergent Extracts Containing the Mitochondrial ATP Synthase
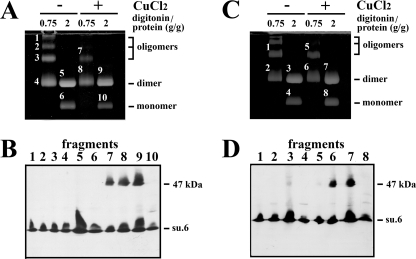
The next goal was to identify the target that was cross-linked with subunit 6. In the next experiment, Triton X-100, a non-ionic detergent that does not destroy ATP synthase interactions at low concentrations, was used to extract the mitochondrial ATP synthase. The rationale of the experiment was to determine whether it is possible to cross-link subunit 6 in Triton X-100 extracts containing the solubilized monomeric and dimeric forms of the mitochondrial ATP synthase (6). Fig. 2B shows the formation of a 47-kDa adduct in 1% Triton X-100 extract that was revealed by an antibody raised against subunit 6. As above, the cross-linked product was obtained at 30 °C and not at 4 °C. Digitonin is another mild detergent that is more efficient than Triton X-100 for isolating the supramolecular associations of multiprotein complexes in combination with native electrophoresis (6, 45). Thus, in the following experiments, mitochondria and digitonin extracts were submitted to oxidation. Then the oligomeric forms of ATP synthases were separated by CN-PAGE and revealed by the in-gel lead phosphate precipitation resulting from the ATPase activity (Fig. 3, A and C). As described previously, the detergent extract obtained with a digitonin-to-protein ratio of 0.75 g/g mainly contained ATP synthase dimer and oligomers, whereas the digitonin extract obtained with a digitonin-to-protein ratio of 2 g/g, which destabilizes oligomers, mainly contained the dimeric and monomeric forms of ATP synthase (6, 45). Bands corresponding to the different forms of ATP synthase were excised, submitted to SDS-PAGE, and analyzed by Western blotting. Fig. 3, B and D, show that the 47-kDa adduct was present only in the supramolecular forms of the ATP synthase after oxidation of both mitochondria and digitonin extracts but was absent in the ATP synthase monomer. From these data, it appears that the protein, which was cross-linked with subunit 6, could be (i) a soluble cysteine-containing protein located in the intermembrane space, (ii) a membranous protein having a cysteine residue in the intermembrane space and extractable by digitonin concentrations that solubilize the ATP synthase, or (iii) a component of the ATP synthase. In the latter case, a good candidate could be subunit 6 itself. Indeed, the relative molecular mass of this hydrophobic protein in 16% polyacrylamide gel is close to 22,000 (Fig. 1). Thus, the mass of a subunit 6 dimer could be near to that of the 47-kDa adduct. If so, then the dimerization of subunit 6 indicates the proximity in the inner mitochondrial membrane of two subunits 6, each belonging to an ATP synthase monomer.
FIGURE 3.
The 47-kDa adduct involving subunit 6 is associated to the oligomeric forms of the ATP synthase upon oxidation of either mitochondria or digitonin extracts. A and C, mitochondria (A) and digitonin extracts (C) were incubated with or without 2 mm CuCl2 for 15 min at 30 °C. The reactions were stopped upon the addition of 10 mm EDTA and 10 mm NEM. Mitochondria were then extracted with digitonin at the indicated digitonin-to-protein ratios in the presence of 10 mm NEM. Digitonin extracts were submitted to CN-PAGE. The slab gels were revealed by the in-gel ATPase activity procedure. Fragments of gels corresponding to monomeric and supramolecular forms of ATP synthase were cut, and proteins were separated by SDS-PAGE on a 16% polyacrylamide gel. The slab gel was analyzed by Western blot. B, analysis of fragments from A. su.6., subunit 6. D, analysis of fragments from C. The blots were revealed with a polyclonal antibody raised against the yeast subunit 6.
Evidence for the Formation in the Yeast Inner Mitochondrial Membrane of a Subunit 6 Dimer Mediated by Oxidation
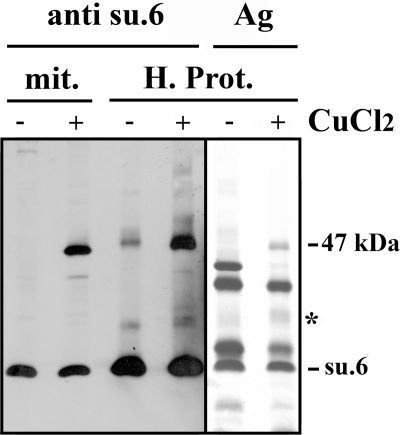
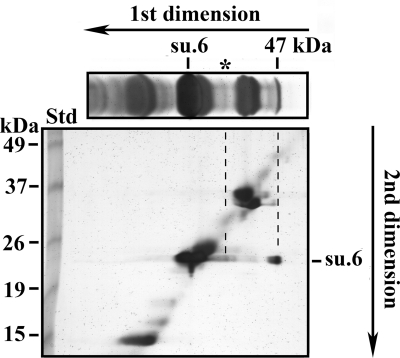
Subunit 6 is a hydrophobic protein that is soluble in organic solvents such as a mixture of chloroform/methanol (1/1). If oxidation leads to the dimerization of subunit 6, then it is possible to purify partially the 47-kDa cross-linked product by using methods previously developed for the purification of proteolipids (36). The proteins present in the organic solvent extract were separated by SDS-PAGE. A part of the gel was transferred to a nitrocellulose membrane, which was revealed by anti-subunit 6 antibodies. The other part, which was silver-stained (Fig. 4), shows that in addition to subunit 6, several proteins were also extracted by the chloroform/methanol mixture. However, subunit 6 and the 47-kDa cross-linked product were clearly identified by Western blot analysis. The data highlight the solubility of the 47-kDa adduct in organic solvents, thus underscoring the high hydrophobic nature of the partner protein cross-linked with subunit 6. The identification of proteins present in the 47-kDa band was undertaken. A slice of the polyacrylamide gel was incubated with β-mercaptoethanol to reduce the disulfide bridge between the two partners and was submitted to a second SDS-PAGE (Fig. 5). Silver staining of the gel shows (i) the migration of non-cross-linked proteins along a diagonal, (ii) the disappearance of the 47-kDa band, and (iii) the presence of two spots at the migration level of the non-cross-linked subunit 6. The spot showing the lowest intensity (indicated by the asterisk) originates from the dissociation of subunit 6 and a low molecular mass protein, whose identification was not performed. This faint intensity adduct was observed in the silver-stained SDS-PAGE gel of organic solvent extract as a diffuse band (Fig. 4 and top part of Fig. 5). It was observed only in organic solvent extracts and resulted from the enrichment procedure by the organic solvent extraction. The second spot corresponds to subunit 6, which migrated at 47 kDa in the first dimension before the disulfide bridge reduction. After reduction with β-mercaptoethanol, the proteins present in the 47-kDa band migrated as a unique band with an apparent molecular mass corresponding to subunit 6. Thus, the 47-kDa band should correspond to a subunit 6 dimer. The presence of subunit 6 in the 47-kDa band was confirmed by mass spectrometry analyses. Pieces of the subunit 6 band and the 47-kDa band were incubated with trypsin. The presence of subunit 6 in the 47-kDa band was confirmed by the identification of only two peptides (Table 2), whereas 10 tryptic peptides could potentially be produced. Until this point, the peptides linked by the two cysteines had not been found, nor had the peptide 10-52 bearing the Cys23 of the uncross-linked subunit 6. The low number of tryptic peptides that were identified could result from the inaccessibility of trypsin to its targets inside this hydrophobic protein that is subunit 6, but also from the poor recovery of hydrophobic peptides. Mass spectrometric analyses of tryptic digests by LC-MS/MS on an LTQ-Orbitrap indicated the presence of faint amounts of several proteins at the migration level of the 47-kDa band. However, the molecular masses of these contaminants were not compatible with a cross-linking involving subunit 6 because their masses were ∼50 kDa (not shown). From the above data, we conclude that the 47-kDa adduct corresponds to a dimer involving two subunits 6 linked by their Cys23. This indicates the proximity of two subunits 6 belonging to monomers that are close either in the membrane or in solubilized oligomeric forms of ATP synthase.
FIGURE 4.
The 47-kDa adduct displays a proteolipid behavior. Wild-type mitochondria were incubated with or without 2 mm CuCl2 for 15 min at 30 °C. The reaction was stopped upon the addition of 10 mm EDTA and 10 mm NEM. Mitochondria were then extracted with a mixture of CHCl3/CH3OH (1/1). The crude organic extract was washed with water, and the organic phase was dried. The hydrophobic proteins (H. Prot.) solubilized in CHCl3/CH3OH (2/1) were precipitated by the addition of diethyl ether. The pellet recovered after centrifugation was dried and solubilized in SDS, and proteins were separated by SDS-PAGE on a 10% polyacrylamide gel. A part of the gel was silver-stained (Ag). The other part was analyzed by Western blot. The blot was revealed with a polyclonal antibody raised against the yeast subunit 6 (su.6). mit., mitochondria before extraction by the organic solvent mixture and solubilized by SDS. The asterisk indicates a faint intensity adduct.
FIGURE 5.
Evidence of dimerization of yeast subunit 6 (su.6) upon oxidation of wild-type mitochondria. Wild-type mitochondria were incubated with 2 mm CuCl2 for 15 min at 30 °C. The reaction was stopped upon the addition of 10 mm EDTA and 10 mm NEM. Organic extracts were prepared as in the legend for Fig. 4 and analyzed by SDS-PAGE on a 12% polyacrylamide slab gel. A slice was cut and silver-stained (top part of the figure). Another slice was incubated at room temperature for 1 h with 1% SDS, 1% β-mercaptoethanol and analyzed by SDS-PAGE on a 12% polyacrylamide slab gel for a second dimension. After migration the gel was silver-stained. The BenchMarkTM prestained protein ladder was the standard (Std). The asterisk indicates a faint intensity adduct.
TABLE 2.
Mass spectrometry analysis of tryptic peptides of subunit 6 and 47-kDa bands
The samples were prepared as described in the legend for Fig. 4. The gel was stained with the ProteoSilverTM silver staining kit (Sigma). The revealed bands were excised and submitted to trypsin action in the gel. Tryptic peptides were extracted and analyzed by MALDI-TOF-TOF and by LC-MS/MS. The asterisks indicate that the methionine residues were oxidized.
| Sample | [M + H]+m/z |
Residues | Peptide sequence | |
|---|---|---|---|---|
| Theoretical mass | Measured | |||
| Subunit 6 band | 1104.568 | 1104. 568 | 1–9 | SPLDQFEIR |
| 729.4617 | 729.4617 | 170–176 | AISLGLR | |
| 47-kDa band | 1104.568 | 1104.568 | 1–9 | SPLDQFEIR |
| 2057.0082 | 2088.998 | 58–74 | WLISQEAIYDTIM*NM*TK | |
The Dimerization of Subunit 6 Occurs in the Absence of the So-called Dimer-specific Subunits
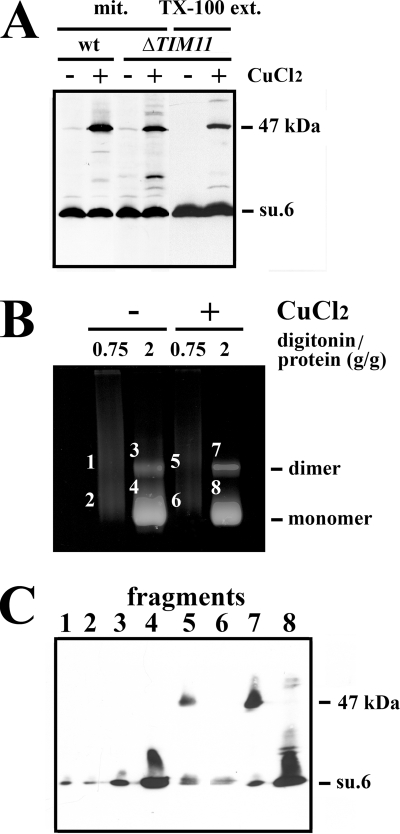
Another issue is the formation or not of the subunit 6 dimer in mitochondria devoid of subunit e because it has been reported that this supernumerary subunit is involved in the dimerization/oligomerization of ATP synthase (6, 24). To do this, ΔTIM11 mitochondria and Triton X-100 ΔTIM11 mitochondrial extract were incubated with CuCl2. As shown in Fig. 6A, the 47-kDa band was present, although to a lesser extent than for wild-type mitochondria. Quantification indicated that 18% ± 4% of subunit 6 was detected in the 47-kDa adduct resulting from oxidation of ΔTIM11 mitochondria. As described previously (6), CN-PAGE analysis of ΔTIM11 digitonin extracts shows the destabilization of supramolecular species of ATP synthase to the monomeric form (compare Figs. 3A and 6B). However, CuCl2-treated mitochondria displayed a slight increase in the amount of dimeric form at a digitonin-to-protein ratio of 2 g/g, thus suggesting a reinforcement of the link between ATP synthases mediated by the formation of the disulfide bridge between subunits 6 (Fig. 6B). This was confirmed by the presence of the 47-kDa band only in the ATP synthase dimer (Fig. 6C, fragments 5 and 7).
FIGURE 6.
The 47-kDa adduct is formed in the absence of subunit e. A, wild-type mitochondria (wt), ΔTIM11 mitochondria (mit.), and 1% (w/v) Triton X-100 ΔTIM11 mitochondrial extract (TX-100 ext.) were incubated with or without 2 mm CuCl2 for 15 min at 30 °C. The reactions were stopped upon the addition of 10 mm EDTA and 10 mm NEM. The samples were solubilized in SDS, and aliquots (30 μg of protein) were analyzed by Western blot. B, CuCl2-treated ΔTIM11 mitochondria were extracted with digitonin at the indicated digitonin-to-protein ratios in the presence of 10 mm NEM. Digitonin extracts were submitted to CN-PAGE. The slab gels were revealed by the in-gel ATPase activity procedure. C, fragment of gels corresponding to dimeric and monomeric forms of ATP synthase were cut and analyzed by Western blot as in the legend for Fig. 3. Blots were revealed with a polyclonal antibody raised against the yeast subunit 6 (su.6).
DISCUSSION
The eukaryotic ATP synthase is a membranous hetero-oligomeric complex with a mass of 0.6 MDa. In addition, it adopts supramolecular structures in the inner mitochondrial membrane that can be observed by electron microscopy and by native gel electrophoresis when using mild detergents for their extraction. The dimeric form of the enzyme appears to be the major form when mitochondria are solubilized by digitonin. In all cases, the dimers observed by electron microscopy are associated by their F0 domains (9–14). However, higher forms larger than the ATP synthase dimer such as tetrameric and hexameric forms have been reported when decreasing the digitonin concentration. One of the main challenges is to identify the components that are involved not only at the interfaces between ATP synthase monomers but also between ATP synthase dimers. Previous data have shown that disulfide bridge formation between either subunit e or subunit g in the membrane increases the stability of an oligomeric structure in detergent extracts whose mass is higher than that of the dimer (24, 25). This shows that although they are important for the stabilization of ATP synthase dimer, these two supernumerary subunits are also involved in the oligomerization of the yeast enzyme. Other candidates for the dimer-dimer interface in bovine mitochondria are thought to be carriers for inorganic phosphate and ADP/ATP (46). Disulfide bridge formation between two subunits i, two subunits h, and two subunits 4 (b), which are unique in the ATP synthase monomer, suggests that components of the second stalk are good candidates for participating at the interfaces between ATP synthase monomers (23). In fact, deletion of the ATP18 gene encoding the supernumerary subunit i only slightly alters the dimerization/oligomerization (45). Other candidates such as subunits h and 4 are structural components of the ATP synthase, and deletion of their structural genes makes it difficult to analyze their involvement at the monomer-monomer interface. However, either deletion of the first membrane-spanning segment or alteration of the intermembrane space loop of subunit 4 leads to considerable instability of the supramolecular species of ATP synthase (47, 48).
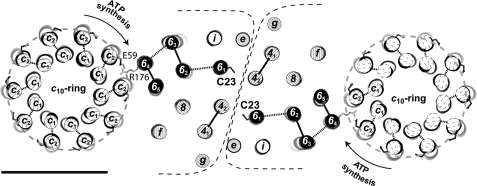
Recently, the observation of a dimeric a-subunit associated with two c-rings (c10a2c10) in yeast led to the proposal that the a-subunit may be an important part of the monomer-monomer interface (26). Subunit 6 (a) is a highly hydrophobic protein because it was extracted and purified by organic solvent mixtures (49, 50). As with very hydrophobic proteins, migration anomalies are observed in SDS-PAGE experiments with a decrease in the relative mass that parallels a decrease in the acrylamide concentration (51). For example, at a 16% polyacrylamide concentration, subunit 6 displays a relative molecular mass of 22,000 when compared with the actual mass of 28 kDa (Fig. 1), but the relative mass drops to 19,000 at a 12% polyacrylamide concentration (Fig. 6). Subunit 6 is located in the periphery of the ATP synthase in contact with the membranous rotor with which it forms the proton channel. It belongs to the peripheral stalk of the ATP synthase and likely interacts with other components of the stalk such as subunits 4 (b), i, and 8, as shown by cross-linking experiments (29, 43, 44). In this work, we produced a disulfide bridge between two subunits 6 via their Cys23 upon oxidation of either mitochondria or the solubilized supramolecular forms of ATP synthase. Subunit 6 Cys23 is unique and is not conserved. We show here that such oxidation has no effect on ATPase activity and conclude that if such oxidation of the intermembrane space-located subunit 6 Cys23 occurs in vivo, it will not alter the function of the yeast enzyme. Because the stoichiometry of subunit 6 is 1 per ATP synthase monomer (15), cross-linking demonstrates the proximity of two subunits 6, thus indicating the existence of a contact between the two subunits 6 of the ATP synthase dimer. On the basis of its homology with the E. coli a-subunit, Cys23 should be located at the N terminus of the TMH1 of subunit 6. We show that the disulfide bridge between two subunit 6 Cys23 residues occurs at 30 °C, whereas the disulfide bridge between subunits e through their Cys28 occurs at 4 °C (24). It can be concluded that subunit 6 Cys23 is partially inaccessible (the two cysteines involved in the disulfide bridge might be buried in the numerous bundles of TMHs present at the interface between the two membrane sectors). The poor efficiency of Cu(1,10-phenanthroline)2SO4 to cross-link subunit 6 Cys23 (Fig. 1) at the protein interface when compared with Cu2+ could result from the size of this oxidative agent that renders it less accessible to cysteines than copper ion. Such a difference in the efficiency of the two oxidative agents has already been described (52). The E. coli subunit a has five TMHs. The proton channel activity is provided mainly by the TMH4 and TMH5 at the interface with the c-ring (53). We postulate that the N-terminal part of subunit 6 including the membrane-spanning segment TMH1 is involved in the interface between ATP synthase monomers, but on the other side of the interface with the c-ring (Fig. 7). The model of the interface between ATP synthase monomers presented in Fig. 7 is based on the known intersubunit cross-links of F0 stator subunits of either yeast or bovine mitochondria (29, 43, 44, 54).
FIGURE 7.
Proposed model of the interface between two ATP synthase monomers. The c10-ring is drawn with the subunit c Glu59 side chain in ball-and-stick. The five TMHs of subunit 6 are black cylinders with subunit 6 Arg176 and subunit 6 Cys23 in ball-and-stick. The model has been enhanced with the two transmembrane domains of subunit 4 (b) and with the unique transmembrane domains of subunits f, i, 8, e, and g. The location of F0 stator subunits is based on known intersubunit cross-linking. The F0 domain is presented from the intermembrane space. The subunit 6 Arg176 is a conserved residue that is important for proton translocation. The scale bar represents 50 Å.
In the wild-type context, the CuCl2-induced dimerization of subunit 6 in solubilized supramolecular forms of ATP synthase indicates that the interface between ATP synthase monomers involving subunit 6 was not destroyed or strongly modified (Fig. 3, C and D) despite the solubilization with digitonin or Triton X-100. Subunit 6 may be a major component involved at the monomer-monomer interface between ATP synthases. Some dimerization of subunit 6 was expected in CuCl2-treated ΔTIM11 mitochondria because cross-links between two subunits i and two subunits h were already reported in the null mutant ΔTIM11, which is devoid of subunits e and g (23), thus indicating again that the supernumerary subunits e and g are not essential for the formation of dimeric forms of ATP synthases (22). Our data indicate that in the absence of subunit e, ATP synthase monomers are still close enough in the inner mitochondrial membrane to form a disulfide bridge between their subunits 6 (Fig. 6) and that this proximity is maintained in detergent extract (Fig. 6A), although we cannot fully exclude a cross-linking between ΔTIM11 ATP synthase monomers resulting from the Brownian motion at the temperature used in our experiments. In fact, in the absence of subunits e and g, a faint amount of ΔTIM11 ATP synthase was detected under a dimeric form by CN-PAGE in the control experiment, and the amount of dimer was slightly increased upon oxidation (Fig. 6B). As a result, we cannot exclude the presence of a higher amount of dimeric enzymes in the inner mitochondrial membrane of the ΔTIM11 mutant strain and cannot exclude that they are destabilized during CN-PAGE. These data are in favor of a fragile association of ATP synthase monomers, at least in the dimeric form, in the inner mitochondrial membrane in the absence of supernumerary subunits e and g.
Acknowledgments
We are grateful to Dr. Marie-France Giraud for critical reading of the manuscript and Dr. Ray Cooke for the contribution to the editing of the manuscript. Mass spectrometry analyses were performed at the Pôle Protéomique, Centre de Génomique Fonctionnelle Bordeaux, Université Victor Segalen Bordeaux 2, Bordeaux, France.
This work was supported by grants from the CNRS, the Université Victor Segalen, Bordeaux, the Conseil Régional d'Aquitaine, and the Agence Nationale de la Recherche (ANR-06-PCV-0016).
- TMH
- transmembrane helix
- CN-PAGE
- clear-native polyacrylamide gel electrophoresis
- NEM
- N-ethyl maleimide.
REFERENCES
- 1. Devenish R. J., Prescott M., Rodgers A. J. (2008) Int. Rev. Cell Mol. Biol. 267, 1–58 [DOI] [PubMed] [Google Scholar]
- 2. Stock D., Leslie A. G., Walker J. E. (1999) Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 3. Dautant A., Velours J., Giraud M. F. (2010) J. Biol. Chem. 285, 29502–29510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Watt I. N., Montgomery M. G., Runswick M. J., Leslie A. G., Walker J. E. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16823–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lau W. C., Baker L. A., Rubinstein J. L. (2008) J. Mol. Biol. 382, 1256–1264 [DOI] [PubMed] [Google Scholar]
- 6. Arnold I., Pfeiffer K., Neupert W., Stuart R. A., Schägger H. (1998) EMBO J. 17, 7170–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eubel H., Jänsch L., Braun H. P. (2003) Plant Physiol. 133, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krause F., Reifschneider N. H., Goto S., Dencher N. A. (2005) Biochem. Biophys. Res. Commun. 329, 583–590 [DOI] [PubMed] [Google Scholar]
- 9. Minauro-Sanmiguel F., Wilkens S., García J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12356–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dudkina N. V., Heinemeyer J., Keegstra W., Boekema E. J., Braun H. P. (2005) FEBS Lett. 579, 5769–5772 [DOI] [PubMed] [Google Scholar]
- 11. Dudkina N. V., Sunderhaus S., Braun H. P., Boekema E. J. (2006) FEBS Lett. 580, 3427–3432 [DOI] [PubMed] [Google Scholar]
- 12. Thomas D., Bron P., Weimann T., Dautant A., Giraud M. F., Paumard P., Salin B., Cavalier A., Velours J., Brèthes D. (2008) Biol. Cell 100, 591–601 [DOI] [PubMed] [Google Scholar]
- 13. Couoh-Cardel S. J., Uribe-Carvajal S., Wilkens S., García-Trejo J. J. (2010) J. Biol. Chem. 285, 36447–36455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cano-Estrada A., Vázquez-Acevedo M., Villavicencio-Queijeiro A., Figueroa-Martínez F., Miranda-Astudillo H., Cordeiro Y., Mignaco J. A., Foguel D., Cardol P., Lapaille M., Remacle C., Wilkens S., González-Halphen D. (2010) Biochim. Biophys. Acta 1797, 1439–1448 [DOI] [PubMed] [Google Scholar]
- 15. Spannagel C., Vaillier J., Arselin G., Graves P. V., Grandier-Vazeille X., Velours J. (1998) Biochim. Biophys. Acta 1414, 260–264 [DOI] [PubMed] [Google Scholar]
- 16. Paumard P., Arselin G., Vaillier J., Chaignepain S., Bathany K., Schmitter J.M., Brèthes D., Velours J. (2002) Biochemistry 41, 10390–10396 [DOI] [PubMed] [Google Scholar]
- 17. Gavin P. D., Prescott M., Luff S. E., Devenish R. J. (2004) J. Cell Sci. 117, 2333–2343 [DOI] [PubMed] [Google Scholar]
- 18. Buzhynskyy N., Sens P., Prima V., Sturgis J. N., Scheuring S. (2007) Biophys. J. 93, 2870–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strauss M., Hofhaus G., Schröder R. R., Kühlbrandt W. (2008) EMBO J. 27, 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dudkina N. V., Oostergetel G. T., Lewejohann D., Braun H. P., Boekema E. J. (2010) Biochim. Biophys. Acta 1797, 272–277 [DOI] [PubMed] [Google Scholar]
- 21. Nicastro D., Frangakis A. S., Typke D., Baumeister W. (2000) J. Struct. Biol. 129, 48–56 [DOI] [PubMed] [Google Scholar]
- 22. Gavin P. D., Prescott M., Devenish R. J. (2005) J. Bioenerg. Biomembr. 37, 55–66 [DOI] [PubMed] [Google Scholar]
- 23. Fronzes R., Weimann T., Vaillier J., Velours J., Brèthes D. (2006) Biochemistry 45, 6715–6723 [DOI] [PubMed] [Google Scholar]
- 24. Arselin G., Giraud M. F., Dautant A., Vaillier J., Brèthes D., Coulary-Salin B., Schaeffer J., Velours J. (2003) Eur. J. Biochem. 270, 1875–1884 [DOI] [PubMed] [Google Scholar]
- 25. Bustos D. M., Velours J. (2005) J. Biol. Chem. 280, 29004–29010 [DOI] [PubMed] [Google Scholar]
- 26. Wittig I., Velours J., Stuart R., Schägger H. (2008) Mol. Cell Proteomics 7, 995–1004 [DOI] [PubMed] [Google Scholar]
- 27. Valiyaveetil F. I., Fillingame R. H. (1998) J. Biol. Chem. 273, 16241–16247 [DOI] [PubMed] [Google Scholar]
- 28. Cox G. B., Fimmel A. L., Gibson F., Hatch L. (1986) Biochim. Biophys. Acta 849, 62–69 [DOI] [PubMed] [Google Scholar]
- 29. Spannagel C., Vaillier J., Chaignepain S., Velours J. (1998) Biochemistry 37, 615–621 [DOI] [PubMed] [Google Scholar]
- 30. Rak M., Tetaud E., Godard F., Sagot I., Salin B., Duvezin-Caubet S., Slonimski P. P., Rytka J., di Rago J. P. (2007) J. Biol. Chem. 282, 10853–10864 [DOI] [PubMed] [Google Scholar]
- 31. Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chateaubodeau G. A., Guérin M., Guérin B. (1976) Biochimie 58, 601–610 [DOI] [PubMed] [Google Scholar]
- 33. Guérin B., Labbe P., Somlo M. (1979) Methods Enzymol. 55, 149–159 [DOI] [PubMed] [Google Scholar]
- 34. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 35. Velours J., Vaillier J., Paumard P., Soubannier V., Lai-Zhang J., Mueller D. M. (2001) J. Biol. Chem. 276, 8602–8607 [DOI] [PubMed] [Google Scholar]
- 36. Paul M. F., Velours J., Arselin de Chateaubodeau G., Aigle M., Guerin B. (1989) Eur. J. Biochem. 185, 163–171 [DOI] [PubMed] [Google Scholar]
- 37. Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 38. Arselin G., Vaillier J., Graves P. V., Velours J. (1996) J. Biol. Chem. 271, 20284–20290 [DOI] [PubMed] [Google Scholar]
- 39. Schägger H., Cramer W. A., von Jagow G. (1994) Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 40. Yoshida M., Sone N., Hirata H., Kagawa Y. (1975) J. Biol. Chem. 250, 7910–7916 [PubMed] [Google Scholar]
- 41. Grandier-Vazeille X., Guérin M. (1996) Anal. Biochem. 242, 248–254 [DOI] [PubMed] [Google Scholar]
- 42. Ansorge W. (1983) in Electrophoresis '82: Advanced Methods Biochemical and Clinical Applications (Stathakos D. ed) pp. 235–242, Walter de Gruyter, Berlin, Germany [Google Scholar]
- 43. Paumard P., Vaillier J., Napias C., Arselin G., Brèthes D., Graves P. V., Velours J. (2000) Biochemistry 39, 4199–4205 [DOI] [PubMed] [Google Scholar]
- 44. Stephens A. N., Khan M. A., Roucou X., Nagley P., Devenish R. J. (2003) J. Biol. Chem. 278, 17867–17875 [DOI] [PubMed] [Google Scholar]
- 45. Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., Brèthes D., di Rago J. P., Velours J. (2002) EMBO J. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wittig I., Schägger H. (2008) Biochim. Biophys. Acta 1777, 592–598 [DOI] [PubMed] [Google Scholar]
- 47. Soubannier V., Vaillier J., Paumard P., Coulary B., Schaeffer J., Velours J. (2002) J. Biol. Chem. 277, 10739–10745 [DOI] [PubMed] [Google Scholar]
- 48. Weimann T., Vaillier J., Salin B., Velours J. (2008) Biochemistry 47, 3556–3563 [DOI] [PubMed] [Google Scholar]
- 49. Fearnley I. M., Walker J. E. (1986) EMBO J. 5, 2003–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Michon T., Galante M., Velours J. (1988) Eur. J. Biochem. 172, 621–625 [DOI] [PubMed] [Google Scholar]
- 51. Rais I., Karas M., Schägger H. (2004) Proteomics 4, 2567–2571 [DOI] [PubMed] [Google Scholar]
- 52. Hastrup H., Sen N., Javitch J. A. (2003) J. Biol. Chem. 278, 45045–45048 [DOI] [PubMed] [Google Scholar]
- 53. Moore K. J., Fillingame R. H. (2008) J. Biol. Chem. 283, 31726–31735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Belogrudov G. I., Tomich J. M., Hatefi Y. (1996) J. Biol. Chem. 271, 20340–20345 [DOI] [PubMed] [Google Scholar]