Background: Rubisco activase has been linked to the inhibition of net photosynthesis upon warming.
Results: The structure of the C-terminal domain adopts an unusually elongated shape.
Conclusions: Reactivation of Rubisco may involve movement of a paddle-like extension.
Significance: This work will aid in gaining a better understanding of Rubisco regulation.
Keywords: ATPases, Bioenergetics, Chloroplast, Crystal Structure, Rubisco, AAA, Carbon Fixation
Abstract
The rapid release of tight-binding inhibitors from dead-end ribulose-bisphosphate carboxylase/oxygenase (Rubisco) complexes requires the activity of Rubisco activase, an AAA+ ATPase that utilizes chemo-mechanical energy to catalyze the reactivation of Rubisco. Activase is thought to play a central role in coordinating the rate of CO2 fixation with the light reactions of photosynthesis. Here, we present a 1.9 Å crystal structure of the C-domain core of creosote activase. The fold consists of a canonical four-helix bundle, from which a paddle-like extension protrudes that entails a nine-turn helix lined by an irregularly structured peptide strand. The residues Lys-313 and Val-316 involved in the species-specific recognition of Rubisco are located near the tip of the paddle. An ionic bond between Lys-313 and Glu-309 appears to stabilize the glycine-rich end of the helix. Structural superpositions onto the distant homolog FtsH imply that the paddles extend away from the hexameric toroid in a fan-like fashion, such that the hydrophobic sides of each blade bearing Trp-302 are facing inward and the polar sides bearing Lys-313 and Val-316 are facing outward. Therefore, we speculate that upon binding, the activase paddles embrace the Rubisco cylinder by placing their hydrophobic patches near the partner protein. This model suggests that conformational adjustments at the remote end of the paddle may relate to selectivity in recognition, rather than specific ionic contacts involving Lys-313. Additionally, the superpositions predict that the catalytically critical Arg-293 does not interact with the bound nucleotide. Hypothetical ring-ring stacking and peptide threading models for Rubisco reactivation are briefly discussed.
Introduction
In recent years, interest in understanding the structure and function of Rubisco2 activase has intensified, propelled in part by its purported role in modulating the thermotolerance of photosynthesis (1). Activase is a chemo-mechanical motor protein that regulates photosynthesis by controlling the activity of the CO2-fixing enzyme, Rubisco (2). Without activase, catalytic misfire reactions and unproductive substrate binding generate dead-end complexes of Rubisco that are catalytically impaired. Activase reactivates inhibited Rubisco sites by using the energy derived from ATP hydrolysis to facilitate conformational changes that restore Rubisco sites to catalytic competence (3, 4). Activase is primarily regulated by the ambient ATP/ADP ratio in the chloroplast stroma (5); however, redox regulation occurs in some plants due to the co-expression of two isoforms. The longer, 418-residue α-form (46 kDa) is highly redox-sensitive as it bears 2 conserved C-terminal cysteine residues, whereas the shorter 379-residue β-form (42 kDa) lacks these residues and is unresponsive to environmental oxidation (4, 6).
The catalytic competence of Rubisco in the fixation of atmospheric carbon is thought to be a critical parameter in heat-related limitations on net photosynthesis (7). In this process, the thermolabile nature of activase has been proposed to play a significant role (1, 8, 9). As the temperature rises, the active sites of Rubisco increasingly partition into their closed, inactivated forms (10, 11), whereas reactivation by activase appears to be stalled (1, 8). Studies on plants engineered to express reduced levels of activase have demonstrated that the amount of activase correlates tightly with Rubisco carboxylation activity (12). Directed evolution experiments on Arabidopsis activase have suggested that a rise in the stability of the enzyme may be linked to improved biomass production at warmer temperatures (13). Therefore, activase may be a valuable engineering target in the development of plant varieties better adapted to rising global temperatures (14).
To devise a knowledge-based protein engineering strategy, atomic resolution structural data are highly desirable. Activase belongs to the AAA+ superfamily of P-loop ATPases (15), and some structural information may be gleaned from other members of this group. The 200–250-residue AAA+ module consists of an αβα N-domain bearing the Walker A and B motifs followed by a more divergent C-domain with a conserved structural core composed of two helical hairpins with a left-handed twist (16–18). The nucleotide binding site is formed by a groove between the two domains, which are linked by a short peptide bridge thought to serve as a pivot point for domain rearrangements upon ATP hydrolysis (see Fig. 1). Rubisco activase has been assigned to the extended classic clade of AAA+ modules typically involved in protein disaggregation, unfolding, and remodeling, where closed hexameric rings appear to be the default assembly state (16, 17).
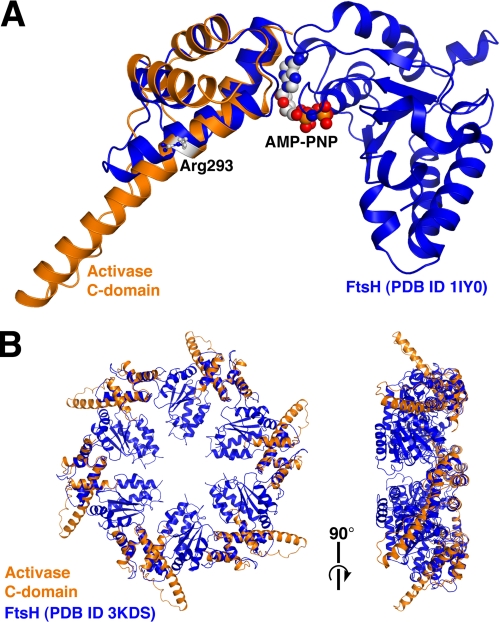
FIGURE 1.
Superposition of the activase C-domain with FtsH. A, structural overlay of the C-domain (orange) with a monomer of the FtsH AAA+ module (blue). Shown in space-filling and ball-and-stick representations are the nucleotide of FtsH and activase Arg-293, respectively. AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate. B, left, top view of the FtsH hexamer (blue) with six superimposed C-domains (orange) looking down the six-fold axis. Right, side view.
The physiologically relevant oligomeric form of activase remains poorly determined as molecular weight estimates vary broadly (2, 9, 19, 20). Further, the mode of interaction with Rubisco continues to be enigmatic, although several activase regions have been identified that are involved in reactivation (21, 22). Importantly, some progress has been made in elucidating the molecular basis of recognition. Activases from plants in the Solanaceae family such as tobacco are incapable of activating Rubiscos from non-Solanaceae plants such as spinach or creosote or from the green alga Chlamydomonas reinhardtii (23). Some years ago, the species specificity was analyzed by targeting select large subunit (LS) surface residues in Chlamydomonas Rubisco (24, 25). A mutational analysis indicated that the residues Pro-89 and Asp-94 (Glu-94 in spinach) are essential in protein-protein recognition as either P89R or D94K resulted in a reversal of species specificity. These residues are located near the Rubisco midsection on an exposed loop of the LS N-domain, remote from the active site by about 30 Å. Conformational changes within this loop region were proposed to underlie selectivity (25).
A subsequent investigation of the activase C-domain identified residues 313 (Asp-313 in tobacco, Lys-313 in spinach) and 316 (Leu-316 in tobacco, Val-316 in spinach) as critical sites in species selectivity (26). (For clarity, residue numbering is based on creosote α-activase; see supplemental Fig. S1). A Lys in position 313 was shown to be essential for the interaction with spinach Rubisco, whereas an Asp at this site was shown to play a role in the interaction with tobacco Rubisco. Residue 316 was demonstrated to augment the primary selectivity introduced by position 313. Extensive complementation studies with the mutant Chlamydomonas Rubiscos P89R and D94K led to a binding model in which activase-313 contacts Rubisco-94 and activase-316 contacts Rubisco-89 (26). This model suggests that selectivity in non-Solanaceae plants may be based on formation of an intermolecular glutamate-lysine salt bridge. However, the data cannot distinguish between a direct interaction and more indirect effects such as conformational changes.
To engineer activases that maintain appropriate cross-species reactivity, a structural understanding of recognition is imperative. Here, we describe the atomic resolution structure of a core portion of the activase C-domain. We find that the drumstick-like appearance allows for presentation of the recognition motif around the periphery of the toroidal assembly.
EXPERIMENTAL PROCEDURES
LtAα(250–351) Cloning and Protein Expression
Blunt-ended DNA coding for the activase fragment LtAα(250–351) was PCR-amplified from a pET23a(+) expression plasmid containing the gene for full-length α-isoform creosote (Larrea tridentata) activase. All procedures for PCR amplification, directional “TOPO” cloning of the PCR product into a linear pET151/D-TOPO vector (Invitrogen), and transformation into One Shot TOP10 competent cells (Invitrogen) were carried out according to the manufacturer's recommendations. Plasmid was prepared from individual transformant colonies using the QIAprep kit (Qiagen). The pET151/D-TOPO vector containing LtAα(250–351) was transformed into Escherichia coli BL21*(DE3) cells (Invitrogen). Single colonies were cultured overnight at 37 °C in 25 ml of Luria-Bertani (LB) media with 100 μg/ml carbenicillin, used to inoculate 1 liter of LB/carbenicillin, and cultured at 37 °C until the A600 reached 0.6. The cultures were cooled to 25 °C, 100 mg of isopropyl-β-d-thio-galactoside was added, and growth continued for 8 h at reduced rpm and 25 °C. After centrifugation, the cells were frozen at −80 °C. Selenomethionyl protein was also expressed in BL21*(DE3) cells; however, media were modified according to published protocols (27).
Protein Purification
Cell paste was suspended in 50 ml of 25 mm HEPES, pH 8.0, 20 mm imidazole, 150 mm NaCl, 10% glycerol, 1 mm EDTA, 1 mm PMSF, 1 mm DTT, and 0.1% Triton X-100 and then disrupted by sonication. The lysate was centrifuged, and the supernatant was passed through a 0.2-μm syringe filter before loading onto a nickel-nitrilotriacetic acid column (Qiagen). His-tagged protein was purified using an imidazole step gradient in 25 mm HEPES, pH 8.0, 150 mm NaCl, 1 mm DTT. Fractions containing the His-tagged protein were pooled, 1.2 mg of tobacco etch virus protease was added, and the sample was dialyzed overnight at 10 °C against 1 liter of 25 mm Tris, pH 8.0, 150 mm NaCl, 1 mm DTT, 1 mm EDTA. The dialysate was reapplied to a nickel-nitrilotriacetic acid column, and LtAα(250–351) was collected in the early fractions. Protein was concentrated, buffer-exchanged into 20 mm HEPES, pH 7.0, 100 mm NaCl, and 0.04% NaN3, and passed through a 0.1-μm spin filter (Millipore). Prior to crystallization, the protein concentration was determined by UV absorbance to be ∼62 mg/ml. The same procedure was used for purification of selenomethionyl protein, and efficient incorporation of selenomethionine was confirmed by MALDI-TOF.
Crystallization and Diffraction Data Collection
Using hanging drop vapor diffusion, native crystals of LtAα(250–351) were obtained in 4-μl drops containing a 3:1 protein:reservoir solution ratio. The reservoir contained 0.5 ml of 0.1 m Tris, pH 8.5, 0.2 m trimethylamine N-oxide dihydrate, and 20% (w/v) PEG monomethyl ether 2000. The best selenomethionyl crystals were obtained when the reservoir contained 0.1 m sodium/potassium tartrate and 20% PEG 3350. Crystals with dimensions of 450 × 300 × 150 μm were grown at 10 °C over 1–2 weeks. Prior to storage in liquid nitrogen, single crystals were soaked in mother liquor plus 20% glycerol for 30 min at 10 °C. Diffraction data were collected at 100 K at the Advanced° Photon Source (Argonne, IL) beamline 19-ID-D, using the Area Detector Systems Corp. Q315r 315 × 315-mm mosaic CCD detector. Native data were collected at a wavelength of 0.97921 Å. For multiple anomalous dispersion (MAD) experiments, data were collected based on the results of an HKL3000 (28) fluorescence scan of the selenium absorption edge (see Table 1).
TABLE 1.
Crystallographic statistics
| Data collection | |||||
|---|---|---|---|---|---|
| Data set | Native | Peak | Inflection | Remote | |
| Wavelength (Å) | 0.97921 | 0.97918 | 0.979291 | 0.97151 | |
| Cell dimensions (Å, degrees) | a = b = 71.9, c = 151.7, γ = 120 | a = b = 71.5, c = 153.0, γ = 120 | a = b = 71.3, c = 154.3, γ = 120 | a = b = 71.3, c = 153.4, γ = 120 | |
| Total reflections | 123,313 | 52,772 | 36,510 | 55,164 | |
| Unique reflections | 12,496 | 7727 | 3554 | 4844 | |
| Resolutiona (Å) | 48.1–1.88 (1.98–1.88) | 30.4–2.19 (2.31–2.19) | 48.2–2.88 (3.04–2.88) | 48.1–2.59 (2.73–2.59) | |
| R-mergea | 0.092 (0.536) | 0.058 (0.355) | 0.092 (0.326) | 0.061 (0.348) | |
| I/σ(I)a | 22.1 (4.4) | 16.8 (5.6) | 19.6 (5.5) | 22.5 (6.7) | |
| Completenessa (%) | 99.6 (97.2) | 97.2 (95.7) | 98.6 (93.1) | 98.6 (94.2) | |
| Redundancya | 9.9 (7.2) | 6.8 (7.0) | 10.3 (5.9) | 11.4 (11.3) | |
| Phasing/refinement | |||||
| Patterson figure of merit | 100.8 | ||||
| Resolution | 48.1–1.88 | ||||
| R-factor | 0.217 | ||||
| Rfree | 0.238 | ||||
| Number of protein atoms | 746 | ||||
| Number of solvent atoms | 42 | ||||
| Average B-factors (Å2) (TLS residuals) | 22.3 | ||||
| Protein atoms (Å2) | 21.5 | ||||
| Solvent atoms (Å2) | 29.1 | ||||
| r.m.s.d. bond lengths (Å2) | 0.016 | ||||
| r.m.s.d. bond angles (degrees) | 1.742 | ||||
| B-factor correlations (Å2) | 7.2 | ||||
a The values in parentheses indicate statistics for the highest resolution shell.
Data Reduction and Structure Determination
Native and selenomethionine diffraction data were integrated using the program XDS (29) and scaled and merged with SCALA (28). SHELXC/D/E (30) was employed to obtain and modify initial phases using the MAD method, and two heavy atom sites were located. A preliminary model consisting of 96 residues was built into the experimental electron density map using ARP/wARP (31). The model was completed using iterative cycles of model building with COOT (32) and refinement with REFMAC5 with inclusion of translation-libration screw parameters (33). Water molecules were placed in regions with significant positive difference density within reasonable distance to a hydrogen bond partner.
RESULTS AND DISCUSSION
In this work, we describe the atomic resolution structure of a 97-residue fragment of higher plant Rubisco activase derived from the creosote bush, L. tridentata, a common shrub in desert habitats. The crystallographic model spans the core of the C-domain bearing several elements important for recognition, specificity, and catalysis.
Construct Selection, Crystallization, and Structure Solution
All Rubisco activases examined to date display a complex spread of oligomeric assembly states (2), a feature that has rendered crystallization of the holoenzyme difficult. To identify constructs with reduced size polydispersity, we have cloned and expressed over 50 different activase fragments derived from a variety of plant species. Soluble constructs were purified, characterized by size-exclusion chromatography and ThermoFluor stability assays, and subjected to crystallization trials. In this way, a monodisperse fragment of creosote α-activase was identified that comprised residues 250–351, LtAα(250–351). The N-terminal boundary was selected based on its proximity to the predicted AAA+ hinge region (Fig. 1), and the C-terminal boundary was based on trial and error. Crystals of LtAα(250–351) grew in space group R32 with unit cell dimensions a = b = 71.9 Å, c = 151.7 Å, γ = 120°. Molecular replacement using MOLREP (34) with known AAA+ C-domain structures as search models (sequence identities <20%) failed to produce useful solutions. Therefore, the structure of LtAα(250–351) was solved by MAD phasing using data collected on a selenomethionyl-derivatized crystal that diffracted to 2.2 Å. The native LtAα(250–351) data diffracted to 1.88 Å resolution, and refinement provided Rwork/Rfree = 0.217/0.238. Crystallographic statistics are provided in Table 1. Each asymmetric unit comprises one molecule with a packing coefficient Vm = 3.1 Å3/Da. The final model consists of residues 248–346, 42 water molecules, and one molecule of glycerol. Residues 248 and 249 are cloning site remnants, and the C-terminal 5 residues were not modeled due to a lack of electron density. A Ramachandran plot (32) places 96.8% of residues into “preferred regions,” 3.2% into “allowed regions,” and none into “outlier regions.”
The C-domain Fold Features a Paddle-like Protrusion Extending Outward from the Core
As expected, the structural core of the creosote C-domain consists of a four-helix bundle. However, the overall fold adopts a surprisingly elongated shape spanning about 50 Å end to end (Fig. 2). The salient feature of the drumstick-like appearance appears to be an α-helix comprising about nine turns, helix 3 (residues 284–314), which extends outward from the knob-like arrangement of the two helical hairpins. The more compact portion of the structure encloses a distinct hydrophobic core, with the N-terminal part of helix 3 packed against helix 1 (residues 250–261) and helix 2 (269–276) packed against helix 4 (333–343), each pair arranged in nearly parallel fashion. The flat protrusion consists of the five C-terminal turns of helix 3 packed side-by-side against an extended peptide strand of irregular secondary structure (residues 318–331).
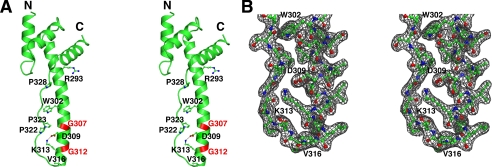
FIGURE 2.
Structure of the activase C-domain. A, stereo image depicting the C-domain fold with select side chains shown in ball-and-stick model. The salt bridge between Asp-309 and Lys-313 is represented by a dashed line, and the positions of Gly-307 and Gly-312 are highlighted in red. B, stereo image of the 2Fo − Fc electron density (1 σ) for the remote end of the paddle-like extension.
The irregular strand bears three proline residues in trans conformation (322, 323, and 328), likely providing rigidity and conformational restriction to the narrow part of the structure (Fig. 2). Two hydrogen bonds connect this strand to helix 3, one near the tip of the paddle between the Lys-313 carbonyl and the Ser-318 amide and the other near the center of the paddle between the Trp-302 indole nitrogen and the Pro-323 carbonyl. To a large degree, packing of helix 3 against the more extended part of the chain involves an intercalation of hydrophobic side chains, with van der Waals contacts formed by Tyr-295, Val-299, Trp-302, Val-303, Val-306, Pro-322, and Phe-325. The tip of the protrusion consists of a type-1 β-turn, in which the expected main-chain H-bond between the 1st and 4th residue Lys-314 and Asn-317 is only weakly formed (3.4 Å). The 2nd and 3rd residue of the turn consist of the highly conserved Leu-315 and the recognition element Val-316 (see below), a surprising feature as the β-turn propensity is particularly low for these residues. Interestingly, all of the hydrophobic side chains along the protrusion are at least partially surface-exposed, generating a large hydrophobic patch centered around Trp-302 on one face of the paddle (Fig. 2 and supplemental Fig. S3). In the crystal, molecular contacts between adjoining molecules appear to stabilize these exposed groups, suggesting a possible interaction surface for Rubisco. The hydrophobic face is lined by 2 basic residues (Lys-314 and Lys-319) at the remote end of the paddle. The opposite face is dominated by acidic residues Glu-305, Asp-309, Glu-320, and Glu-326 (supplemental Fig. S3) but also bears the recognition elements Lys-313 and Val-316 described below.
The Specificity Elements Are Located near the Remote End of the Paddle
The specificity of Rubisco recognition appears to be mediated by activase residues Lys-313 and Val-316 (26). Intriguingly, we found that Val-316 is located at the very tip of the protrusion within the β-turn connecting helix 3 to the irregular strand (Fig. 2). Lys-313 precedes Val-316 by one helical turn, and both side chains line the acidic face of the paddle-like extension without protruding from the surface. The last two turns of helix 3 appear to be stabilized by a salt bridge between the ϵ-amino group of Lys-313 and the carboxylate of Asp-309. This appears significant in light of the peculiar location of the helix-destabilizing but highly conserved residues Gly-307 and Gly-312 flanking Asp-309 (Fig. 2). In Solanaceae such as tobacco, the equivalent residues are Asp-313 and Glu-309, which are unable to form an ionic bond with each other. However, the carboxylate of site 309 is not always conserved (supplemental Fig. S1); therefore, the Lys-313-Asp-309 interaction in and of itself cannot account for selectivity. Although in higher plants, the charge in position 313 correlates tightly with selectivity (26), the conformational rearrangements and electrostatic effects of a K313D or D313K substitution are currently unknown. Regardless, based on the data presented here, it appears unlikely that Lys-313 would make direct contact with Rubisco Asp-94 (Glu-94 in spinach) as proposed (26) as the formation of a strong intermolecular ionic bond would require disruption of the internal salt bridge. Importantly, binding to Rubisco is predicted to orient the polar face bearing Lys-313 away from Rubisco (see below).
Superpositions onto AAA+ Hexameric Rings Suggest a Fan-like Appearance of the Assembly
A DALI (35) structural homology search with the LtAα(250–351) x-ray model as query revealed numerous structural neighbors belonging to the AAA+ superfamily. The highest similarity scores were obtained for spastin, VPS4, and the metallo-protease FtsH, all members of the extended classic clade to which activase has been assigned (16). Superpositions onto all three proteins demonstrate that the LtAα(250–351) helix 3 extends by an additional four turns (Fig. 1). In contrast, FtsH contains a canonical, compact four-helix bundle (36). A structural overlay of LtAα(250–351) onto a six-fold symmetric FtsH x-ray model (Protein Data Bank (PDB) code 3KDS) provides an r.m.s.d. of 2.3 Å for 69 aligned α-carbons at 12% sequence identity (Fig. 1). This comparison indicates that a 26-residue peptide insert is responsible for the paddle-like extension of creosote activase (supplemental Fig. S2). The insert starts with Trp-302 and bears the recognition elements Lys-313 and Val-316, as well as Pro-322 and Pro-323. Other members of the classic clade have been shown to incorporate insertions between C-domain helices 3 and 4, e.g. spastin, VPS4, ClpD1, and p97 (17). However, the inserted elements exhibit a broad variety of different structural features with no obvious relationship to activase.
The Sensor-2 Arg-293 May Not Interact with the Nucleotide
The overlays reveal an interesting feature of activase Arg-293, shown to be critical for ATP hydrolysis and Rubisco reactivation (20). This residue is located on helix 3 close to the structural core (Fig. 2). Surprisingly, the superpositions place its side chain ∼22 Å from the phosphate tail of the bound nucleotide, rendering a direct contact unlikely (Fig. 1). Arg-293 has been proposed to be equivalent to a sensor-2 arginine that is known to interact directly with the γ-phosphate of ATP (20), is conserved among a subset of non-classic AAA+ modules, and is thought to serve as a mediator between ATP hydrolysis and domain rearrangements (38, 39). Perhaps the apparent contradiction between the functional importance of Arg-293 and its predicted location remote from the active site could be explained by a rearrangement in subunit packing.
Hypothetical Models for Rubisco Reactivation Are Ring-Ring Stacking and Peptide Threading
Currently, the mode of interaction of activase with its partner protein is largely unknown, although it appears likely that the subunits self-associate to form a hexameric ring with the recognition domains mounted onto the periphery (Fig. 1). Rubisco binding may involve a molecular contact between the activase C-domains and the Rubisco LS N-domains (26). We speculate that the fan-like extensions embrace the Rubisco cylinder, such that the hydrophobic faces of the paddles are brought into proximity to Rubisco. Upon ATP hydrolysis and associated domain rotations, the paddles reorient, thereby applying mechanical force onto the Rubisco LS N-domain. A well characterized subtle movement of the LS N-domain is known to involve loop 89–94 (25) and is thought to be coupled to the opening of Rubisco active sites by dislocation of loop 6 and the C-terminal tail (37).
In analogy with other classic AAA+ proteins, a hypothetical binding mode may involve the stacking of an activase toroidal assembly onto the four-fold ring of L8S8 Rubisco, such that the two axes defined by the respective central pores are aligned with each other. Accordingly, the fan-like protrusions would surround the top edge of the Rubisco cylinder. Ample precedent exists for the stacking of closed hexameric AAA+ rings onto multisubunit rings of partner proteins with either matched or mismatched stoichiometries, such as in HslU-HslV, FtsH, ClpAP, and the proteasomal ATPase.
Another, equally speculative model may involve peptide threading through the central pore formed by the αβα domain of the AAA+ assembly. An appropriate peptide target may consist of the Rubisco LS C-terminal tail involved in the transition from the closed to the open form (37). According to this idea, the activase toroid would align its central pore over one of the Rubisco active sites. Alternatively, peptide targets may consist of loop regions lining the entrance to the Rubisco solvent channel, suggesting a geometric arrangement as in the ring-ring stacking model. Either way, sequestration of disordered segments of proteins into the central AAA+ pore occurs in a number of assemblies, such as FtsH, the proteasomal ATPase, spastin, and ClpA/B. We hope that the activase recognition domain will serve as a structural framework to test these and other possible models for the physical interaction between activase and Rubisco.
Supplementary Material
Acknowledgment
Crystallographic data were collected at the Advanced Photon Source Beamline 19-ID-D, operated for the Department of Energy by the University of Chicago Argonne, LLC.
This work was supported by the United States Department of Energy Office of Basic Energy Sciences Photosynthetic Systems Grants DE-FG02-09-ER16123 (to R. M. W.) and DE-FG02-08ER-20268 (to M. E. S.).
The atomic coordinates and structure factors (code 3THG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- LS
- large subunit
- MAD
- multiple anomalous dispersion
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Crafts-Brandner S. J., Salvucci M. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13430–13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Portis A. R., Jr. (2003) Photosynth. Res. 75, 11–27 [DOI] [PubMed] [Google Scholar]
- 3. Wang Z. Y., Portis A. R., Jr. (1992) Plant Physiol. 99, 1348–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Portis A. R., Jr., Li C., Wang D., Salvucci M. E. (2008) J. Exp. Bot. 59, 1597–1604 [DOI] [PubMed] [Google Scholar]
- 5. Wang D., Portis A. R., Jr. (2006) Photosynth. Res. 88, 185–193 [DOI] [PubMed] [Google Scholar]
- 6. Zhang N., Kallis R. P., Ewy R. G., Portis A. R., Jr. (2002) Proc. Natl. Acad. Sci 99, 3330–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spreitzer R. J., Salvucci M. E. (2002) Annu. Rev. Plant Biol. 53, 449–475 [DOI] [PubMed] [Google Scholar]
- 8. Salvucci M. E., Crafts-Brandner S. J. (2004) Plant Physiol. 134, 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barta C., Dunkle A. M., Wachter R. M., Salvucci M. E. (2010) Arch. Biochem. Biophys. 499, 17–25 [DOI] [PubMed] [Google Scholar]
- 10. Feller U., Crafts-Brandner S. J., Salvucci M. E. (1998) Plant Physiol. 116, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharkey T. D., Badger M. R., von Caemmerer S., Andrews T. J. (2001) Photosynth. Res. 67, 147–156 [DOI] [PubMed] [Google Scholar]
- 12. Salvucci M. E., DeRidder B. P., Portis A. R., Jr. (2006) J. Exp. Bot. 57, 3793–3799 [DOI] [PubMed] [Google Scholar]
- 13. Kurek I., Chang T. K., Bertain S. M., Madrigal A., Liu L., Lassner M. W., Zhu G. (2007) Plant Cell 19, 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mueller-Cajar O., Whitney S. M. (2008) Photosynth. Res. 98, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999) Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 16. Snider J., Houry W. A. (2008) Biochem. Soc. Trans. 36, 72–77 [DOI] [PubMed] [Google Scholar]
- 17. Ammelburg M., Frickey T., Lupas A. N. (2006) J. Struct. Biol. 156, 2–11 [DOI] [PubMed] [Google Scholar]
- 18. Erzberger J. P., Mott M. L., Berger J. M. (2006) Nat. Struct. Mol. Biol. 13, 676–683 [DOI] [PubMed] [Google Scholar]
- 19. Wang Z. Y., Ramage R. T., Portis A. R., Jr. (1993) Biochim. Biophys. Acta 1202, 47–55 [DOI] [PubMed] [Google Scholar]
- 20. Li C., Wang D., Portis A. R., Jr. (2006) Arch. Biochem. Biophys. 450, 176–182 [DOI] [PubMed] [Google Scholar]
- 21. van de Loo F. J., Salvucci M. E. (1996) Biochemistry 35, 8143–8148 [DOI] [PubMed] [Google Scholar]
- 22. Esau B. D., Snyder G. W., Portis A. R., Jr. (1996) Arch. Biochem. Biophys. 326, 100–105 [DOI] [PubMed] [Google Scholar]
- 23. Wang Z. Y., Snyder G. W., Esau B. D., Portis A. R., Jr., Ogren W. L. (1992) Plant Physiol. 100, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larson E. M., O'Brien C. M., Zhu G., Spreitzer R. J., Portis A. R., Jr. (1997) J. Biol. Chem. 272, 17033–17037 [DOI] [PubMed] [Google Scholar]
- 25. Ott C. M., Smith B. D., Portis A. R., Jr., Spreitzer R. J. (2000) J. Biol. Chem. 275, 26241–26244 [DOI] [PubMed] [Google Scholar]
- 26. Li C., Salvucci M. E., Portis A. R., Jr. (2005) J. Biol. Chem. 280, 24864–24869 [DOI] [PubMed] [Google Scholar]
- 27. Doublié S., Kapp U., Aberg A., Brown K., Strub K., Cusack S. (1996) FEBS Lett. 384, 219–221 [DOI] [PubMed] [Google Scholar]
- 28. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 29. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sheldrick G. M. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 35. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niwa H., Tsuchiya D., Makyio H., Yoshida M., Morikawa K. (2002) Structure 10, 1415–1423 [DOI] [PubMed] [Google Scholar]
- 37. Taylor T. C., Andersson I. (1996) Nat. Struct. Biol. 3, 95–101 [DOI] [PubMed] [Google Scholar]
- 38. Ogura T., Whiteheart S. W., Wilkinson A. J. (2004) J. Struct. Biol. 146, 106–112 [DOI] [PubMed] [Google Scholar]
- 39. Erzberger J. P., Berger J. M. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.