Background: In Moco biosynthesis, sulfur is transferred from l-cysteine to MPT synthase, catalyzing the conversion of cPMP to MPT.
Results: The rhodanese-like protein YnjE is a novel protein involved in Moco biosynthesis.
Conclusion: YnjE enhances the rate of conversion of cPMP to MPT and interacts with MoeB and IscS.
Significance: To understand the mechanism of sulfur transfer and the role of rhodaneses in the cell.
Keywords: Enzyme Mechanisms, Enzyme Purification, Iron-Sulfur Protein, Molybdenum, Sulfur, l-Cysteine Desulfurase, Molybdenum Cofactor, Persulfide, Rhodanese, Sulfurtransferase, Moco Biosynthesis, Sulfur Transfer
Abstract
In the second step of the molybdenum cofactor (Moco) biosynthesis in Escherichia coli, the l-cysteine desulfurase IscS was identified as the primary sulfur donor for the formation of the thiocarboxylate on the small subunit (MoaD) of MPT synthase, which catalyzes the conversion of cyclic pyranopterin monophosphate to molybdopterin (MPT). Although in Moco biosynthesis in humans, the thiocarboxylation of the corresponding MoaD homolog involves two sulfurtransferases, an l-cysteine desulfurase, and a rhodanese-like protein, the rhodanese-like protein in E. coli remained enigmatic so far. Using a reverse approach, we identified a so far unknown sulfurtransferase for the MoeB-MoaD complex by protein-protein interactions. We show that YnjE, a three-domain rhodanese-like protein from E. coli, interacts with MoeB possibly for sulfur transfer to MoaD. The E. coli IscS protein was shown to specifically interact with YnjE for the formation of the persulfide group on YnjE. In a defined in vitro system consisting of MPT synthase, MoeB, Mg-ATP, IscS, and l-cysteine, YnjE was shown to enhance the rate of the conversion of added cyclic pyranopterin monophosphate to MPT. However, YnjE was not an enhancer of the cysteine desulfurase activity of IscS. This is the first report identifying the rhodanese-like protein YnjE as being involved in Moco biosynthesis in E. coli. We believe that the role of YnjE is to make the sulfur transfer from IscS for Moco biosynthesis more specific because IscS is involved in a variety of different sulfur transfer reactions in the cell.
Introduction
The biosynthesis of the molybdenum cofactor (Moco)2 is an evolutionarily conserved pathway present in bacteria, archaea, and eukaryotes (1). In Escherichia coli, Moco biosynthesis is divided into four steps: 1) conversion of GTP into cyclic pyranopterin monophosphate (cPMP); 2) insertion of two sulfur atoms into cPMP by molybdopterin (MPT) synthase; 3) insertion of molybdenum to form Moco; and 4) additional modification by covalent addition of GMP or CMP to the C4′ phosphate of MPT, forming either the molybdopterin guanine dinucleotide cofactor or the molybdopterin cytosine dinucleotide cofactor (2, 3). Two molybdopterin guanine dinucleotide cofactor moieties are ligated to the molybdenum atom and form the bis-molybdopterin guanine dinucleotide cofactor (4), which is characteristic for the majority of molybdoenzymes in E. coli, like nitrate reductase. For the formation of MPT from cPMP, two sulfur atoms are incorporated to the C1′ and C2′ positions of cPMP, a reaction catalyzed by MPT synthase (5). The E. coli MPT synthase is a heterotetrameric enzyme consisting of two MoaE and two MoaD subunits (5, 6). MoaD shares structural similarities with ubiquitin, and in its active form, it contains a C-terminal thiocarboxylate group that acts as a direct sulfur donor for the synthesis of the dithiolene group of Moco (5, 7, 8). For the synthase to act catalytically, it is necessary to regenerate its transferable sulfur, a reaction for which the MoeB protein and ATP are required (5). MoeB exhibits significant sequence similarities to two segments of the activating enzyme (E1) for ubiquitin (Uba1). However, biochemical data and the crystal structure of the MoeB-MoaD complex revealed that the interaction of MoeB with MoaD resembles only the first step of the ubiquitin-targeted degradation of proteins (8, 9). It was shown that MoeB solely activates the C terminus of MoaD by formation of an acyl-adenylate, and no thioester intermediate between MoaD and MoeB was identified (9). Subsequently, the activated MoaD acyl-adenylate is converted to a thiocarboxylate by action of a persulfide-containing protein (10). Recently, the l-cysteine desulfurase IscS in its persulfide-bound form was identified as the primary physiological sulfur-donating enzyme for the generation of thiocarboxylate on MPT synthase (11). Proteins homologous to E. coli MoeB in eukaryotes, including humans, named MOCS3 (12), and some bacteria contain an additional C-terminal domain exhibiting homologies to rhodaneses (13). Recently, in humans, the l-cysteine desulfurase Nfs1 was shown to act as a direct sulfur donor for the formation of a persulfide group on MOCS3 (14). Thus, the sulfur transfer for Moco biosynthesis in humans involves two sulfurtransferases, an l-cysteine desulfurase and a rhodanese-like protein (14). In general, rhodaneses are widespread enzymes that catalyze in vitro the transfer of a sulfur atom from thiosulfate to cyanide via a protein-bound persulfide intermediate. Rhodanese-like proteins are either composed of a single catalytic rhodanese domain or fusions of two rhodanese domains, with their C-terminal domains containing the catalytic cysteine (15). Furthermore, catalytic or inactive rhodanese domains are also found in other proteins, such as ThiI (16), or in Cdc25 phosphatases (17). In the E. coli genome, eight genes have been identified coding for proteins containing a rhodanese-like protein domain bearing a conserved cysteine residue as a potential active site for persulfide formation (15, 18). Three of them code for the proteins GlpE, PspE, and YgaP that are composed of only a single rhodanese domain. ThiI, YbbB, and YceA contain a single rhodanese module in the context of a larger protein construct. SseA (3-mercaptopyruvate sulfurtransferase) is composed of two rhodanese domains, whereas YnjE contains three rhodanese domains. In each case, only the C-terminal domain contains the catalytic cysteine residue. The crystal structure of YnjE has been solved recently (19). YnjE can efficiently be persulfurated by IscS while having only a residual activity with thiosulfate as substrate (19). The role of YnjE so far remained enigmatic for E. coli. In total, with the exception of ThiI and YbbB, which are required for thiamine/thiouridine and selenouridine biosynthesis (18, 20), respectively, little is known about the in vivo role of the other rhodaneses. It was of interest to identify whether, by analogy to Moco biosynthesis in humans, a rhodanese-like protein acts as a mediator between IscS and MoaD for the formation of the thiocarboxylate group of MPT synthase in conjunction with MoeB.
In this report, the tandem affinity purification (TAP) method was used to identify proteins that interact with MoeB in E. coli. This approach revealed that the three-domain rhodanese-like protein YnjE interacts with TAP-tagged MoeB. Further characterization of YnjE showed that IscS is the sulfur donor for YnjE. Surface plasmon resonance studies showed that IscS, MoaD, and MoeB specifically bind to YnjE. In a defined in vitro system consisting of MPT synthase, MoeB, Mg-ATP, IscS, and l-cysteine, YnjE was shown to enhance the rate of the conversion of added cPMP to MPT. This is the first report identifying the rhodanese-like protein YnjE to be involved in Moco biosynthesis in E. coli. Our studies suggest that YnjE has a mediator role in the thiocarboxylation reaction of MoaD in Moco biosynthesis and makes the sulfur transfer from IscS to MoaD more specific because IscS is involved in a variety of different sulfur transfer reactions in the cell.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, Media, and Growth Conditions
Strains and plasmids used in this study are listed in Table 1. E. coli strain BL21(DE3) (Novagen) was used for homologous expression of the proteins YnjE, SufE, CsdE, IscS, MoeB, MoaD, MoaE, and MPT synthase. The DE3 lysogens of E. coli strains MC1061 (21), CL100 (ΔiscS) (21), JLD42301 (ΔynjE), and PJ18 (ΔiscS/ΔynjE), were used for various assays. Bacterial cultures were generally grown in LB medium under aerobic conditions at 30 °C. When required, 150 μg/ml ampicillin, 25 μg/ml kanamycin, or 50 μg/ml chloramphenicol were added to the medium.
TABLE 1.
E. coli strains and plasmids used in this study
| Plasmid or strain | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| Plasmids | ||
| pJD10 | sufS gene cloned into NdeI and BglII sites of pACYCDuet1, CMR | This study |
| pJD11 | csdA gene cloned into NdeI and BglII sites of pACYCDuet1, CMR | This study |
| pJD12 | sufS gene cloned into NdeI and BamHI sites of pET11b, AmpR | This study |
| pJD13 | csdA gene cloned into NdeI and BamHI sites of pET11b, AmpR | This study |
| pAU1 | ynjE gene cloned into XhoI and XhoI sites of pEB327, AmpR | This study |
| pAU2 | ynjE gene without its periplasmic leader sequence (YnjEΔ1-21) cloned into BamHI and NotI sites of pACYCDuet1, CMR | Ref. 19 |
| pAU3 | ynjEΔ1-21C385A cloned into BamHI and NotI sites of pACYCDuet1, CMR | Ref. 19 |
| pPJ15 | ynjE cloned into EcoRI and XhoI sites of pT7-7 | This study |
| pAF40 | moaD gene cloned into SalI and HindIII sites of pEB327, AmpR | This study |
| pAF41 | moeB gene cloned into SalI and HindIII sites of pEB327, AmpR | This study |
| pET-Ehis | pET22b expressing SufE-His6, AmpR | Ref. 30 |
| pLEC-E | pET22b expressing CsdE-His6, AmpR | Ref. 29 |
| pSL209 | iscS gene cloned into NcoI and BamHI sites of pET15b, AmpR | Ref. 10 |
| pSL213 | sufS gene cloned into NdeI and BamHI sites of pET15b, AmpR | Ref. 10 |
| pSL215 | csdA gene cloned into NdeI and BamHI sites of pET15b, AmpR | Ref. 10 |
| pSL219 | iscS gene cloned into BamHI and BglII sites of pACYC184, CmR | This study |
| pGG110 | moaE gene cloned into NcoI and BglII sites of pQE60, AmpR | Ref. 7 |
| pMW15eB | moeB gene cloned into NcoI and BamHI sites of pET15b, AmpR | Ref. 10 |
| pMW15aD | moaD gene cloned into NcoI and BamHI sites of pET15b, AmpR | Ref. 49 |
| pMW15aDaE | moaDE genes cloned into NcoI and BamHI sites of pET15b, AmpR | Refs. 9 and 49 |
| Strains | ||
| MC1061(DE3) | F−araD139 Δ(ara leu)7696 Δ(lacY74) galU galK hsdR hsdM+ rpsL150(DE3) | Ref. 21 |
| JLD42301 | MC1061 ΔynjE | This study |
| CL100(DE3) | MC1061 ΔiscS | Ref. 21 |
| PJ18(DE3) | MC1061 ΔynjE ΔiscS | This study |
| BL21(DE3) | F−ompT hsdSB(rB−mB−) gal dcm (DE3) | Novagen |
Construction of ΔynjE Strains
A plasmid in which the ynjE gene was replaced by a kanamycin resistance cassette was constructed by cloning DNA fragments that flank ynjE into pFRT-K (18) on either side of the Kmr FRT cassette. The 5′-flanking (0.3 kb) and 3′-flanking regions (0.7 kb) of ynjE were obtained by PCR using the appropriate primers and cloned into the BamHI-EcoRI and HindIII-XhoI sites of pFRT-K, respectively, yielding pPJ20 in which codons 4–445 of the ynjE gene were deleted. The 2.5-kb BamHI-XhoI fragment of pPJ20 containing ΔynjE::Kmr FRT was cloned into pKO3 (yielding pPJ24), and the method of Link et al. (22) was used for replacing the wild-type ynjE allele of strain TL524 (18) with the ΔynjE::Kmr FRT allele to create strain PJ2. The ynjE deletion was verified by PCR using chromosomal DNA of strain PJ2 as template. In addition, the Kmr cassette was mapped to the expected chromosomal location by demonstrating that it was 85% cotransducible with the zdj-276::Tn10 allele of strain CAG18464 and 19% cotransducible with the zea-225::Tn10 allele of strain CAG18465 during phage P1-mediated transduction. These Tn10 insertions are located ∼3 kb counterclockwise and ∼32 kb clockwise from ynjE, respectively (23). Bacteriophage P1-mediated transduction (24) was used to introduce the ΔynjE::Kmr FRT into strains MC1061 and CL100 (ΔiscS) by using strain PJ2 as the P1 donor and selection for kanamycin resistance. The resulting strains were named JLD42301 and PJ18, respectively (Table 1). Strains containing multiple mutations affecting the other sulfurtransferases were constructed by cloning appropriate DNA fragments on either side of the Kmr cassette of pFRT-K. Mutations were introduced into the chromosome using a strain expressing λ Red recombinase (25). Intermediate kanamycin-sensitive strains were obtained by excising the resistance cassette with FLP recombinase, as described for glpE and pspE mutations (26, 27).
Expression and Purification of Proteins
His6-YnjEΔ1–21 and His6-YnjEΔ1–21C385A were expressed and purified as described previously (19). YnjE-His6 was expressed from plasmid pPJ15 and purified by nickel-NTA chromatography.
For measurement of various enzyme activities, the E. coli proteins MoaDE (MPT synthase), MoaD, MoaE, MoeB, IscS, CsdA, and SufS were expressed and purified as described previously (7–10). SufE and CsdE were expressed from plasmids pET-Ehis and pLEC-E, respectively, and purified using published procedures (28–30).
TAP with TAP-MoeB and TAP-YnjEΔ1-21
The TAP tag consists of two IgG binding domains of protein A and a calmodulin binding (CB) peptide separated by a tobacco etch virus protease cleavage site, which allows the purification of proteins under mild conditions so that protein complexes are retained. For purification of proteins that interact with MoeB, the moeB gene was cloned into the SalI-HindIII site of vector pEB327 (31), resulting in a fusion of MoeB with an N-terminal TAP tag, and the plasmid was named pAF41. N-terminal TAP-tagged MoeB was expressed in E. coli strain CL100(DE3) (ΔiscS) in 2 liters of LB medium. For purification of proteins that interact with YnjE, 5′-truncated ynjE was cloned into the XhoI site of vector pEB327 (31), resulting in a fusion of YnjEΔ1–21 with an N-terminal TAP tag, which was named pAU1. N-terminal TAP-tagged YnjEΔ1–21 was expressed in the ΔynjE mutant strain JLD42301(DE3) in 2 liters of LB medium. Expression of TAP-MoeB or TAP-YnjEΔ1–21 was induced by the addition of 100 mg/liter arabinose. Cells were grown at 30 °C for 4 h, and purification was achieved as described previously (31) using IgG-Sepharose 6 Fast Flow and calmodulin-Sepharose 4B (GE Healthcare). Eluted protein complexes were analyzed by SDS-PAGE and/or immunodetection using YnjE and MoeB antisera. Specific protein bands were excised from the gels, digested with trypsin, analyzed by MALDI-peptide mapping, and confirmed by MS/MS analyses. In addition, for test of functionality of the TAP method, the E. coli moaD gene was cloned into the SalI-HindIII sites of vector pEB327, resulting in plasmid pAF40. N-terminal TAP-tagged MoaD was expressed in an E. coli moaD mutant strain (9) in 2 liters of LB medium. Expression of TAP-MoaD and its purification were achieved as described above for TAP-YnjE. Eluted protein complexes were analyzed by SDS-PAGE and immunodetection with MoeB antisera. Specific protein bands were excised from SDS-PAGE, digested with trypsin, analyzed by MALDI-peptide mapping, and confirmed by MS/MS analyses.
Co-purification of Proteins Interacting with a YnjE Antibody Coupled to Protein G-Sepharose
A polyclonal murine YnjE antibody was bound to Protein G-Sepharose (GE Healthcare), and the column was subsequently washed with 100 mm Tris, pH 7.2. A mixture of 40 μm YnjEΔ1–21, 50 μm IscS, 50 μm MoeB, 50 μm MoaD, 50 μm MoaE, 2.5 mm ATP, and 2.5 mm MgCl2 in 100 mm Tris, pH 7.2, was added and incubated overnight at 4 °C. After washing with 100 mm Tris (pH 7.2), bound protein complexes were eluted with 500 mm sodium citrate (pH 2). Eluted proteins were separated by 15% SDS-PAGE and detected by staining with Coomassie Brilliant Blue.
Enzyme Assays
Nitrate reductase activity was determined as described by Jones and Garland (32). The strains MC1061, CL100 (ΔiscS) (21), JLD42301 (ΔynjE), and PJ18 (ΔiscS/ΔynjE) were grown anaerobically for 10 h in 15 ml of LB medium at 37 °C in the presence of 20 mm KNO3. A 4.5-ml portion of the culture was harvested, resuspended in 50 mm Tris-HCl (pH 7.5), and sonified. Aliquots of 200 μl of cell lysate were analyzed for NR activity in a total volume of 4 ml of an assay mixture containing 0.3 mm benzylviologen, 10 mm KNO3, and 20 mm Tris-HCl (pH 6.8). The reaction was initiated by the addition of sodium dithionite. The change in absorbance at 600 nm was measured for 1 min, and the units of activity were in mmol of nitrate reduced/min/A600 (33).
For a test of functional complementation, plasmids pAU2 (His6-YnjEΔ1–21), pACYCDuet1, pPJ15 (YnjE-His6), and pET15b, respectively, were transformed into PJ18(DE3) (ΔiscS ΔynjE), and nitrate reductase activity was determined as described above.
l-Cysteine desulfurase activity of IscS, CsdA, and SufS was measured by determination of the rate of sulfide production as described previously (34). l-Cysteine desulfurases (0.5–2 μm) were mixed in a 1:2 ratio with SufE, CsdE, MoeB-MoaD, or YnjE in a total volume of 800 μl containing 50 mm Tris-HCl, 200 mm NaCl, 10 μm pyridoxal phosphate, and 2 mm dithiothreitol, pH 8.0, and incubated for 10 min at 30 °C. The reactions were stopped by the addition of 100 μl of 20 mm N,N-dimethyl-p-phenylenediamine in 7.2 m HCl and 100 μl of 30 mm FeCl3 in 1.2 m HCl. After an incubation time of 20 min, precipitated protein was removed by centrifugation, and methylene blue was measured at 670 nm. A standard curve was generated using known amounts of sodium sulfide.
MPT Synthase Reaction
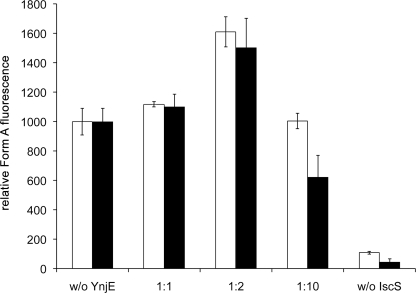
MPT synthase reactions were performed at room temperature in a total volume of 400 μl of 100 mm Tris-HCl (pH 7.2). The MPT produced was oxidized to Form A and quantified following published procedures (35, 36). The reaction mixtures contained a 5 μm concentration of the inactive form of MPT synthase, 5 μm cPMP, 5 μm MoeB, 2.5 mm Mg-ATP, 0.5 mm l-cysteine, and 2.5 μm IscS dimer. l-Cysteine desulfurase, His6-YnjEΔ1–21, or His6-YnjEΔ1–21C385A was added in increasing concentrations as indicated in the legend to Fig. 4 (active site ratio between 1:1 and 1:10). The reaction was initiated with the addition of cPMP, which was purified by using published procedures (37).
FIGURE 4.
Effect of YnjE on the sulfur transfer from IscS for in vitro formation of MPT. Mixtures containing a 5 μm concentration of the inactive form of MPT synthase, cPMP (in excess), 5 μm MoeB, 2.5 mm Mg-ATP, 0.5 mm l-cysteine, and 2.5 μm IscS dimers were incubated in the absence and presence of 2.5, 5, and 25 μm His6-YnjEΔ1–21 (white bars) and His6-YnjEΔ1–21C385A (black bars) as indicated. Mixtures were incubated for 15 min at room temperature, and the amount of MPT formed during the reaction was quantified as Form A using the peak area. Fluorescence was monitored with excitation at 383 nm and emission at 450 nm. Error bars, S.E.
Detection of the Total MPT Content
The total MPT content in E. coli cell extracts of the strains MC1061, CL100 (ΔiscS), JLD42301 (ΔynjE), and PJ18 (ΔiscS/ΔynjE) was determined as follows. 50-ml LB cultures of each strain were grown to stationary phase, harvested by centrifugation, and resuspended in 3 ml of 100 mm Tris-HCl, pH 7.2. The lysate was obtained by sonification. MPT was converted to its fluorescent derivative Form A by adjusting the pH of the supernatant to 2.5 with HCl and heating at 95 °C for 30 min in the presence of iodine (35). Excess iodine was removed by the addition of 55 μl of 1% (w/v) ascorbic acid, and the sample was adjusted with 1 m Tris to pH 8.3. Form A was obtained from Form A-phospho by the addition of 40 mm MgCl2 and 1 unit of calf intestine alkaline phosphatase. Form A was isolated with 10 mm acetic acid on a QAE ion exchange column (Sigma), which was equilibrated in H2O. Form A was identified and quantified by HPLC analysis with a C18 reversed phase HPLC column (4.6 × 250-mm ODS Hypersil; particle size 5 μm) with 5 mm ammonium acetate, 15% (v/v) methanol at an isocratic flow rate of 1 ml/min. In-line fluorescence was monitored by an Agilent 1100 series detector with an excitation at 383 nm and emission at 450 nm.
Surface Plasmon Resonance (SPR) Measurements
All binding experiments were performed with the SPR-based instrument BiacoreTM 2000 on CM5 sensor chips at a temperature of 25 °C and a flow rate of 10 μl/min, using the control software 2.1 and evaluation software 3.0 (Biacore AB, Uppsala, Sweden). Immobilization of YnjEΔ1–21, MoaD, MoeB, and BSA on a CM5 sensor chip was performed according to the surface thiol method. Proteins (1 mg/ml in PBS buffer) were incubated with 0.5 mg of phosphodimethylethanolamine and 5 μl of 0.4 m EDC for 1 h on ice to introduce reactive disulfides. Afterwards, modified proteins were dialyzed in 10 mm acetate, pH 4.0 (for MoeB and MoaD) and pH 5.7 (for YnjEΔ1–21), respectively. Briefly, the biosensor surface was primed with PBS buffer and then activated by a 3-min injection of a solution of 0.2 m EDC and 0.05 m NHS. Cystamine dihydrochloride (40 mm in 0.1 m boric acid (pH 8.5)) was then injected (1.5 min) to introduce disulfides, followed by a 1.5-min injection of 0.1 m DTT-reducing solution (in 0.1 m boric acid, pH 8.5) to reduce the dithio-bridges of the immobilized cystamine. The proteins were then injected over the modified biosensor surface of the measuring cell (3 min) in order to immobilize them. A 2-min injection of phosphodimethylethanolamine/NaCl solution (20 mm phosphodimethylethanolamine and 1 m NaCl in 0.1 m acetate buffer) followed to block remaining unreacted active thiol groups in the flow cells. The autosampler racks containing the sample vials were cooled to 4 °C. Immobilization of proteins yielded the following resonance units (RU) per flow cell: BSA, 194 RU; YnjEΔ1–21, 190 RU; MoeB, 198 RU; MoaD, 183 RU.
As running buffer, 20 mm phosphate, 150 mm NaCl, 0.005% (v/v) Tween 20, pH 7.4, was used. YnjEΔ1–21, MoeB, MoaD, MoaE, IscS, SufS, and CsdA with concentrations of 0.8, 1.6, 3.1, 6.3, 12.5, and 25 μm were injected for 4.5 min at a flow rate of 30 μl/min followed by 15 min of dissociation using the kinject command and regeneration of the sensor surface with 50 mm HCl for 1 min. As a control, BSA was used as ligand. Binding curves were corrected by subtraction of buffer injection curves for all four flow cells.
Detection of the Persulfide Group on YnjE with 1,5-I-AEDANS
For coexpression of YnjE-His6, the l-cysteine desulfurases CsdA and SufS were cloned into the NdeI/BglII restriction sites of pACYC-Duet1 (Novagen), resulting in plasmids pJD10 (SufS) and pJD11 (CsdA). For the co-expression with His6-YnjEΔ1–21, CsdA and SufS were cloned into the NdeI/BamHI restriction sites of pET11b (Novagen), resulting in plasmids pJD12 (SufS) and pJD13 (CsdA). Different protein pairs were coexpressed in E. coli strain CL100 (ΔiscS) (DE3). YnjE was purified by nickel-NTA chromatography and analyzed for its sulfuration level by a procedure that was described earlier (38, 39). 10 μm YnjE was incubated with 0.2 mm 1,5-I-AEDANS for 1 h on ice. The excess of unbound 1,5-I-AEDANS was removed by buffer exchange using a Centricon concentrator with a 30-kDa molecular mass cut-off (Millipore) to avoid nonspecific fluorescence. The addition of 2.5 mm DTT resulted in the release of 1,5-I-AEDANS molecules from proteins containing an YnjE-bound cysteinyl persulfide. The released 1,5-I-AEDANS molecules were identified and quantified by HPLC analysis on a ZORBAX-G250 gel filtration HPLC column (Agilent) equilibrated in 100 mm Tris-HCl, 200 mm NaCl, pH 7.5, at a flow rate of 1 ml/min. In-line fluorescence was monitored by an Agilent 1100 series detector with excitation at 337 nm and emission at 498 nm.
Cellular Localization of YnjE
The cellular localization of endogenous YnjE or His6-YnjEΔ1–21 from plasmid pAU2 was determined in strain BL21(DE3) grown on LB medium. In addition, endogenous YnjE and the periplasmic maltose-binding protein (MBP) were determined in strain MC1061(DE3) grown on LB medium supplemented with 0.2% (w/v) maltose. The cytoplasmic and periplasmic subcellular fractions were isolated according to Rouvière and Gross (40). The cell pellet was resuspended in 20 mm Tris, pH 7.5, 100 mm NaCl containing 20% (w/v) saccharose. The outer membrane was broken by incubation with 20 μg/ml lysozyme and 20 μm MgSO4. After centrifugation in 50% (w/v) sucrose, the spheroplasts were separated from the periplasmic fraction. The spheroplasts were thoroughly washed with 20 mm Tris, resuspended in 100 mm Tris, pH 7.2, and lysed by sonification. The volume of the periplasmic fraction was reduced until a comparable concentration of total protein was obtained. To localize YnjE, cytoplasmic and periplasmic fractions were analyzed by immunodetection using polyclonal YnjE antisera. As a control, the periplasmic MBP was detected with a monoclonal MBP antibody (Sigma) in the same periplasmic and cytoplasmic fractions.
In-gel Tryptic Digestion of Protein Bands
Gel bands were excised, washed, reduced, carboxamidomethylated, and tryptically digested using standard protocols.
MALDI-TOF MS
Peptide mapping was performed on a Bruker ULTRAFLEX time-of-flight (TOF/TOF) instrument in the positive mode using the reflectron for enhanced resolution and a matrix of α-cyano-4-hydroxycinnamic acid. For MS/MS analyses, selected parent ions were subjected to laser-induced dissociation, and the resulting fragment ions were separated by the second TOF stage of the instrument. Samples of 1 μl and an approximate concentration of 1–10 pmol/μl were mixed with equal amounts of matrix. This mixture was spotted onto a stainless steel target and dried at room temperature before analysis.
RESULTS
Co-purification of MoeB Protein Complexes by TAP
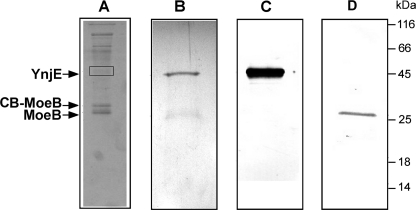
To identify protein interaction partners for MoeB, we chose the TAP method, which has been successfully used for the purification of native protein complexes from E. coli extracts (31). Biochemical purification in combination with mass spectroscopy allows identification of interacting partners. Using the TAP method with MoaD fused to an N-terminal TAP tag, we were able to co-purify MoeB (data not shown), which provided the first co-purification of this complex from E. coli cells and demonstrated that the method is suitable for our purposes. To purify proteins that interact with MoeB, we constructed a TAP fusion to the N terminus of MoeB, and MoeB-associated proteins were purified from a 2-liter culture of the E. coli CL100(DE3) (ΔiscS) mutant strain. Co-purified proteins were concentrated, fractionated by SDS-PAGE, and visualized by Coomassie Blue staining (Fig. 1A). The bands indicated were excised from the gel, and the proteins were identified by MALDI peptide fingerprinting. The two protein bands around 26 kDa were found to contain native MoeB and CB-fused MoeB, so that endogenous MoeB was co-purified by the TAP-tagged MoeB and obviously formed heterodimers. The protein band around 47 kDa was found to contain YnjE, an E. coli rhodanese-like protein of unknown function. The bands with a higher molecular mass were identified as GroEL, EF-TU, cold shock-like protein, glyceraldehyde-3-phosphate dehydrogenase, or with a smaller molecular mass as 30 S ribosomal subunit proteins that are generally co-purified by this method. We thus considered these interactions to be nonspecific. To verify the interaction of YnjE with MoeB, an N-terminal fusion of YnjE to the TAP tag was also constructed. Because YnjE contains an N-terminal leader peptide for export to the periplasm, the first 21 amino acids were deleted before fusion to the N-terminal TAP tag. YnjE-associated proteins purified from a 2-liter culture of an E. coli ynjE-deficient strain were concentrated, fractionated by SDS-PAGE, and detected by Coomassie Blue staining (Fig. 1B). The bands indicated were excised from the gel, and the proteins were again identified by MALDI peptide fingerprinting. Additionally, we detected the proteins by immunodetection using YnjE (Fig. 1C) and MoeB (Fig. 1D) antisera. As shown in Fig. 1, by Western blotting and immunodetection, the 47 kDa protein band was detected as TAP-tagged YnjE using YnjE antisera (C), and the 26 kDa protein band was detected as MoeB using MoeB antisera (D). This result, together with the reverse approach, shows that MoeB interacts with YnjE in vivo.
FIGURE 1.
Tandem affinity purification. Lane A, tandem affinity purification with TAP-tagged MoeB. N-terminal TAP-tagged MoeB was expressed from pAF41 in strain CL100(DE3) (ΔiscS). Extracts were purified as described under “Experimental Procedures.” Eluted protein complexes were analyzed by 12% SDS-PAGE. The indicated bands were excised from the gel, digested with trypsin, and analyzed by MALDI peptide mapping. The 47 kDa band was identified as YnjE, the 26 kDa band was identified as MoeB, and the 31 kDa band was identified as calmodulin-binding peptide CB-MoeB (indicated by arrows). Lanes B–D, tandem affinity purification with N-terminal TAP-tagged YnjEΔ1–21. N-terminal TAP-tagged YnjEΔ1–21 was expressed from pAU1 in strain JLD42301(DE3) (ΔynjE). Extracts were purified as described under “Experimental Procedures.” Eluted protein complexes were analyzed by SDS-PAGE followed by Coomassie Blue staining (B) and immunodetection after transfer to a PVDF membrane (C and D). Lane B, 12% SDS-polyacrylamide gel of the eluted proteins after IgG-Sepharose and calmodulin-Sepharose. The stained bands were excised from the gel, digested with trypsin, analyzed by MALDI peptide mapping, and confirmed by MS/MS analyses. The 47 kDa band was identified as TAP-tagged YnjEΔ1–21, and the 26 kDa band was identified as MoeB (indicated by arrows). Lane C, TAP-tagged YnjEΔ1–21 detected with polyclonal YnjE antiserum. Lane D, MoeB detected with a polyclonal MoeB antiserum.
Co-purification of Proteins That Interact with an YnjE Antibody Bound to Protein G-Sepharose
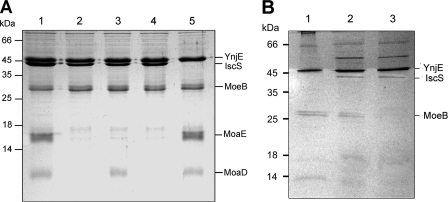
It had been suggested before that IscS acts as primary sulfur donor for the sulfuration of YnjE (19). The TAP purification method showed that YnjE also interacts with MoeB in Moco biosynthesis. To analyze these interactions further, a coimmunoprecipitation approach was employed. Evidence for the formation of a complex between IscS, YnjE, MoeB, and MoaD was obtained from co-purification experiments using purified IscS, MoeB, MoaD, MoaE, His6-YnjEΔ1–21, and a YnjE antibody (Fig. 2). The YnjE antibody was bound to Protein G-Sepharose. When mixtures of MoeB, IscS, His6-YnjEΔ1–21, MoaD, and MoaE were incubated in the presence of Mg-ATP and applied to the YnjE antibody-coupled Protein G-Sepharose, His6-YnjEΔ1–21, MoeB, IscS, MoaD, and MoaE were co-purified as shown after separation on SDS-polyacrylamide gels (Fig. 2A, lane 1). When Mg-ATP was omitted from the mixture, MoaD and MoaE were not coeluted with the other proteins, showing that MoaD binds to MoeB only in the presence of ATP (Fig. 2A, lane 2). The binding of MoaE was dependent on the presence of MoaD (Fig. 2A, lane 4), and IscS was not essential for the binding of MoeB to YnjE. Co-purifications of combinations of YnjE-IscS and YnjE-MoeB showed that IscS and MoeB bind independently to YnjE (Fig. 2B). Control experiments showed that MoaD, MoaE, MoeB, or IscS alone did not interact with the Protein G-Sepharose (data not shown).
FIGURE 2.
Co-purification of proteins that interact with YnjEΔ1–21 by purification using Protein G-Sepharose coupled with a YnjE antibody. A polyclonal antibody raised against YnjEΔ1–21 was bound to Protein G-Sepharose (GE Healthcare). A, His6-YnjEΔ1–21 (40 μm) was incubated with 50 μm MoaD, 50 μm IscS, 50 μm MoeB, 50 μm MoaE, and 100 μm Mg-ATP (lane 1); Mg-ATP was omitted (lane 2); MoaE was omitted (lane 3); MoaD was omitted (lane 4); and IscS was omitted (lane 5). B, His6-YnjEΔ1–21 (40 μm) was incubated with 30 μm MoeB (lane 1); 30 μm MoeB and 30 μm IscS (lane 2); and 30 μm IscS (lane 3). The mixtures in 100 mm Tris-HCl, pH 7.2, were applied to the beads, and after extensive washing steps, proteins were eluted with 500 mm citrate buffer, pH 2. Eluted proteins were concentrated and separated by 15% SDS-PAGE.
Analysis of Protein-Protein Interactions by Surface Plasmon Resonance Measurements
To further confirm the formation of a complex of IscS, YnjE, MoeB, and MPT synthase in Moco biosynthesis in vitro and to determine their dissociation constants, SPR measurements were employed for real-time detection of specific interactions using the purified proteins. His6-YnjEΔ1–21, MoeB, and MoaD were immobilized on the CM5 chip via surface thiol coupling. Successful coupling was confirmed by analysis of the interaction between MoaD and MoaE (KD of 0.29 μm) in addition to MoeB and MoaD (KD of 8.71 μm), which generally confirmed the values reported before by isothermal titration calorimetry measurements (36).
The results obtained by SPR measurements for the protein pairs listed in Table 2 showed the strongest interaction between His6-YnjEΔ1–21 and MoeB, with a KD value of 0.58 μm. IscS bound to immobilized His6-YnjEΔ1–21 with a KD value of 1.36 μm, whereas there was no apparent interaction between His6-YnjEΔ1–21 and the other two E. coli l-cysteine desulfurases, SufS and CsdA. The same result was obtained for MoaD, which interacted with His6-YnjEΔ1–21 (KD of 1.49 μm) and IscS (KD of 0.54 μm) but not with SufS or CsdA, confirming the data of Zhang et al. (11) reported previously. Immobilized His6-YnjEΔ1–21 interacted only with MoaE and not with MoaD, showing that His6-YnjEΔ1–21 was immobilized in a conformation that inhibited this interaction but not the interaction with MoeB. In general, the results obtained from the coimmuniprecipitation analysis were confirmed by SPR, showing that IscS, YnjE, MoeB, and MPT synthase form a complex.
TABLE 2.
Analysis of protein-protein interactions between YnjE and different protein partners by SPR measurements
| Immobilized proteina | RUb | Protein partnerc | KDd | c2 |
|---|---|---|---|---|
| μm | ||||
| YnjEΔ1–21 | 190 | BSA | NDe | —f |
| YnjEΔ1–21 | 190 | IscS | 1.36 | 1.89 |
| YnjEΔ1–21 | 190 | SufS | ND | — |
| YnjEΔ1–21 | 190 | CsdA | ND | — |
| YnjEΔ1–21 | 190 | MoeB | 0.58 | 0.91 |
| YnjEΔ1–21 | 190 | MoaE | 10.6 | 0.31 |
| YnjEΔ1–21 | 190 | MoaD | ND | — |
| MoaD | 183 | BSA | ND | — |
| MoaD | 183 | YnjEΔ1-21 | 1.49 | 0.38 |
| MoaD | 183 | MoaE | 0.29 | 2.00 |
| MoaD | 183 | IscS | 0.54 | 0.74 |
| MoaD | 183 | SufS | ND | — |
| MoaD | 183 | CsdA | ND | — |
| MoeB | 198 | MoaD | 8.71 | 0.27 |
| MoeB | 198 | IscS | 0.16 | 0.46 |
| MoeB | 198 | BSA | ND | — |
a Proteins were immobilized via surface thiol coupling (see “Experimental Procedures”).
b Resonance units.
c Proteins were injected using the KINJECT protocol, injecting samples in a concentration range of 0.8–25 μm. Cells were regenerated by injection of 20 mm HCl.
d KD values were obtained by global fitting procedures for a 1:1 binding.
e ND, no binding detectable.
f —, not calculated.
Analysis of the Activity of Nitrate Reductase and Detection of MPT Levels in ynjE-deficient E. coli Strains
So far, we obtained evidence for a role of YnjE in Moco biosynthesis by analysis of in vivo and in vitro protein-protein interactions. To further analyze the influence of YnjE in this pathway, the activity of nitrate reductase and the overall MPT level were determined in ynjE-deficient E. coli strains.
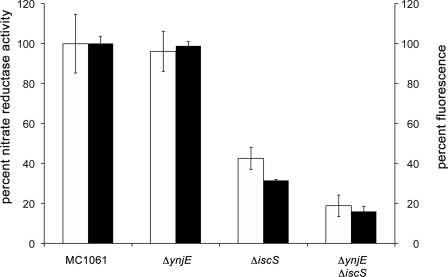
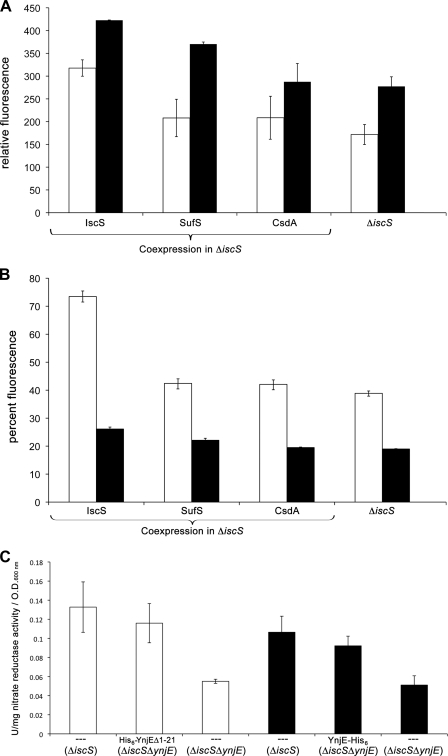
The mutant strains JDL42301(DE3) (ΔynjE), CL100(DE3) (ΔiscS), and PJ18(DE3) (ΔiscSΔynjE) were grown anaerobically in the presence of 20 mm nitrate and tested for the activity of nitrate reductase. As shown in Fig. 3, nitrate reductase activities were the same in lysates of the ΔynjE and wild-type strain. However, the nitrate reductase activity decreased by 58% in the ΔiscS strain. In the ΔiscSΔynjE double mutant strain, the activity of nitrate reductase was even lower, decreasing by 81% relative to the wild type (Fig. 3).
FIGURE 3.
Nitrate reductase activity and total MPT content of different E. coli sulfurtransferase mutant strains. Nitrate reductase activity was determined by using cell lysates of the mutant strains CL100 (ΔiscS), JLD42301 (ΔynjE), and PJ18 (ΔiscS/ΔynjE), which were grown anaerobically in the presence of 20 mm nitrate (white bars). Reduced benzyl viologen was used as electron donor. The activity of the wild type strain was set to 100%. Shown is total MPT content of the strains CL100 (ΔiscS), JLD42301 (ΔynjE), and PJ18 (ΔiscS/ΔynjE). MPT was quantified after conversion to its fluorescence derivative Form A and is shown as relative fluorescence quantified from the peak areas (black bars). The fluorescence obtained from the wild type strain was set to 100%. Fluorescence was monitored with excitation at 383 nm and emission at 450 nm. Nitrate reductase activities and fluorescence were normalized to the optical density of cell suspensions determined at 600 nm. Error bars, S.E.
In addition, the overall MPT levels were analyzed in these mutant strains by conversion of MPT to its fluorescent derivate Form A using the cell extracts. As shown in Fig. 3, the overall MPT levels correlated well with the obtained nitrate reductase activities. The ΔynjE strain showed the same MPT levels as the corresponding wild-type strain, whereas in the ΔiscS strain, the MPT level was 69% decreased, and in the ΔiscSΔynjE strain, the MPT level was 84% decreased in comparison with the wild type extract. These results are consistent with previous results, which showed that the ΔiscS strain contained only a level of 10% Moco in comparison with the corresponding wild-type strain (10), and confirm involvement of IscS in the sulfuration of MoaD (11). However, the results also show that the effect of the deletion of ynjE is only visible in conjunction with the absence of IscS, implying that IscS can fully substitute for the role of YnjE in a ΔynjE mutant and suggesting that YnjE can only partially take over the role of IscS in the ΔiscS mutant strain. This also shows that both proteins work in conjunction in Moco biosynthesis.
YnjE Has an Enhancing Effect on the Synthesis of MPT in Vitro
To verify the role of YnjE for Moco biosynthesis, we used a fully defined in vitro system, in which the thiocarboxylated form of MPT synthase can be formed by incubation of MoeB, Mg-ATP, IscS, and l-cysteine as sulfur source (10). The activity of MPT synthase was determined by testing its influence on the conversion of cPMP to MPT. MPT formation was monitored after its conversion to the stable fluorescent degradation product Form A (35). The addition of His6-YnjEΔ1–21 to this incubation mixture resulted in 61% increased levels of MPT production (IscS dimer/YnjE ratio, 1:2; active site ratio, 1:1) in comparison with the incubation mixture containing only IscS (Fig. 4). However, the same result was obtained using the His6-YnjEΔ1–21C385A variant, in which the catalytically active cysteine residue was exchanged by an alanine. This shows that YnjE has an effect on Moco biosynthesis in conjunction with IscS, a role that is based on making the interaction of both proteins tighter rather than having a direct influence on the sulfur transfer to MoaD when IscS is present.
YnjE Is Not an Enhancer of l-Cysteine Desulfurase Activity
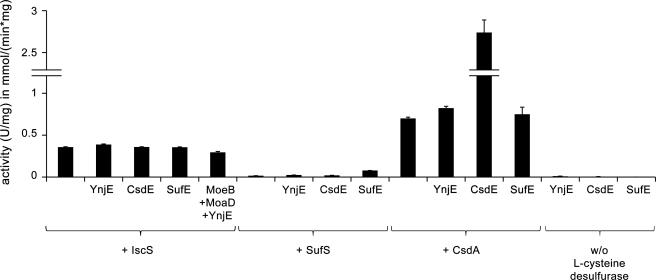
SufE and CsdE act as sulfur transfer proteins that accept sulfur from the E. coli l-cysteine desulfurases SufS and CsdA, thereby stimulating their activities. SufSE and CsdAE consequently form l-cysteine desulfurases with two different subunits (28, 29). To determine if YnjE has a similar stimulating effect on the l-cysteine desulfurase activity for IscS, the l-cysteine desulfurase activity of IscS was analyzed in the presence or absence of His6-YnjEΔ1–21. As controls, the influence of His6-YnjE on the activity of SufS and CsdA was analyzed and compared with the enhancers SufE and CsdE. As shown in Fig. 5, YnjE was not able to enhance the l-cysteine desulfurase activity of IscS, SufS, or CsdA, even when MoeB and MoaD were included. In comparison, SufE and CsdE were able to enhance the activity of SufS and CsdA, respectively, but not of IscS, as shown before (28, 29). Thus, the effect of YnjE on Moco biosynthesis shown above is not based on an enhanced IscS activity but rather based on a better rate of conversion of cPMP to MPT.
FIGURE 5.
Influence of YnjE on l-cysteine desulfurase activity of CsdA, SufS, or IscS in comparison with the enhancers CsdE and SufE. l-Cysteine desulfurase activity was measured by determination of total sulfide produced. l-Cysteine desulfurase CsdA, SufS, or IscS (1–2 μm) was mixed in a 1:2 ratio with His6-YnjEΔ1–21, SufE, or CsdE, respectively, and incubated for 10 min at 30 °C, or 2 μm IscS was mixed in a 1:2 ratio with His6-YnjEΔ1–21, MoeB, or MoaD, respectively, and incubated for 20 min at 30 °C. One unit is defined as the amount of enzyme producing 1 μmol of sulfide/min. Error bars, S.E.
IscS Is the Preferred Sulfur Donor for YnjE in Vivo
Because YnjE was shown to be efficiently persulfurated by the l-cysteine desulfurase IscS in vitro (19), we wanted to elucidate whether IscS or one of the other cysteine desulfurases, SufS or CsdA, serves as the sulfur donor for YnjE in vivo. YnjE was co-expressed with IscS, SufS, or CsdA in a ΔiscS strain, and the sulfuration level of YnjE was then determined. Because YnjE contains an N-terminal leader peptide for the Sec-dependent transport to the periplasm, we coexpressed N-terminal His6-tagged YnjEΔ1–21 (His6-YnjEΔ1–21) in addition to a construct of YnjE with a C-terminal His6 tag, where the Sec-leader peptide was present (YnjE-His6).
After purification of YnjE from CL100(DE3), the sulfuration level of YnjE was analyzed in a defined in vitro assay for the conversion of cPMP to MPT, consisting of YnjE, MoeB, MPT synthase, and cPMP. The produced MPT was quantified after conversion to Form A. The results in Fig. 6A show that overall, YnjE was purified in a sulfurated form from all strains, showing that additional sulfur donors for YnjE exist, which might be thiosulfate, as shown before (19). The results in Fig. 6A also show that the lowest activity was obtained with YnjE purified from the ΔiscS mutant strain. The activity increased about 52% after coexpression with IscS. SufS also was able to substitute for IscS in the ΔiscS mutant strain because the activity was 34% increased using YnjE-His6 purified from this strain. Overall, the same results were obtained with His6-YnjEΔ1–21 and YnjE-His6.
FIGURE 6.
Detection of the sulfuration level of YnjE. To determine the sulfuration level of YnjE, His6-YnjEΔ1–21 (white bars) or YnjE-His6 (black bars) was co-expressed with SufS, CsdA, or IscS in ΔiscS mutant strain CL100(DE3) and purified after nickel-NTA chromatography. A, the sulfuration level of YnjE was analyzed in a defined in vitro assay for the conversion of cPMP to MPT, consisting of 5 μm YnjE, 5 μm MoeB, 5 μm inactive MPT synthase, 2.5 mm Mg-ATP, and cPMP (in excess). After 45 min, the reactions were stopped by the addition of acidic iodine, and MPT was quantified as Form A. B, the persulfide group was directly quantified by the fluorescence of 1,5-I-AEDANS. His6-YnjEΔ1–21 or YnjE-His6 (10 μm each, white or black bars, respectively) was incubated with 0.2 mm 1,5-I-AEDANS. After 1 h, the 1,5-I-AEDANS molecules bound to the persulfide group on YnjE were released by the addition of 2.5 mm DTT and quantified by HPLC as described under “Experimental Procedures.” His6-YnjEΔ1–21 sulfurated by IscS with l-cysteine in vitro was set to 100%. C, functional complementation of the strains PJ18(DE3) (ΔiscSΔynjE) and CL100 (ΔiscS) with plasmid pAU2 (encoding cytoplasmic His6-YnjEΔ1–21), or pPJ15 (encoding YnjE-His6), as indicated. Cells were grown anaerobically in the presence of 0.4% (w/v) glucose, 100 μm isopropyl β-d-thiogalactopyranoside, and 20 mm nitrate. Nitrate reductase activity of lysates from strains containing YnjE-His6 (black bars) and His6-YnjEΔ1–21 (white bars) was determined. Reduced benzyl viologen was used as electron donor. The units of activity are calculated in mmol of nitrate reduced/min for each sample normalized to A600 nm. Error bars, S.E.
Additionally, we analyzed the persulfide group present on YnjE directly by using the fluorescent alkylating reagent 1,5-I-AEDANS (38). 1,5-I-AEDANS is able to bind to exposed thiol groups of protein cysteinyl residues, resulting in the formation of either thioesters at free SH groups or disulfide bonds in the case of persulfides. 1,5-I-AEDANS can only be released from YnjE by reducing agents, such as DTT, when linked to the protein by disulfide bonds. Analysis of the fluorescence of the persulfide-bound 1,5-I-AEDANS molecules purified from the different strains showed the highest fluorescence for His6-YnjEΔ1–21 or YnjE-His6 after coexpression with IscS. The lowest sulfuration level was obtained from proteins expressed in the ΔiscS mutant. Consistent with the data for MPT production, YnjE was purified in a sulfurated form from all strains (Fig. 6B). However, the position of the His6 tag seems to have an influence on the sulfuration level.
The data also show that both YnjE variants are sulfurated by IscS, including the construct with the N-terminal leader peptide for export to the periplasm. Because IscS is a cytoplasmic protein, sulfuration is only possible when YnjE and IscS interact in the cytoplasm. To confirm a role of YnjE in the cytoplasm, we introduced the plasmids encoding cytoplasmic His6-YnjEΔ1–21 and YnjE-His6 (containing Sec-leader peptide) into the ΔiscSΔynjE double mutant and tested for functional complementation of the nitrate reductase. As shown in Fig. 6C, overexpression of both YnjE variants restored nitrate reductase activities in the double mutant (1.8-fold increased nitrate activity for overexpression of YnjE-His6 and 2.1-fold increased nitrate activity for overexpression of His6-YnjEΔ1–21) to the level of the ΔiscS mutant strain. Because the cytoplasmic His6-YnjEΔ1–21 was able to functionally complement the ΔiscSΔynjE double mutant to the same extent as the YnjE-His6 construct, this clearly shows that YnjE has a role in the cytoplasm.
Cellular Localization of YnjE
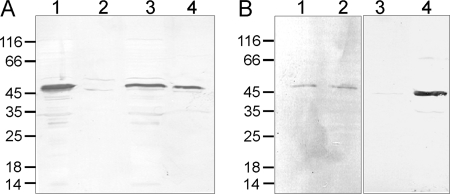
Analyses of the amino acid sequence of YnjE using prediction programs, such as SignalP (41), showed that YnjE contains a Sec-type N-terminal signal peptide for its export to the periplasm. Because Moco biosynthesis in E. coli occurs in the cytosol, it was of interest to determine the cellular localization of YnjE. The periplasmic and the cytosolic fractions of strains BL21(DE3) and MC1061(DE3) were isolated, and YnjE was detected in the separate fractions using an YnjE antiserum. Fig. 7, A (lanes 3 and 4) and B (lanes 1 and 2), shows that YnjE was readily detected both in the periplasm and in the cytoplasm of both strains. As control for a cytoplasmic protein, we used the His6-YnjEΔ1–21 protein, where the N-terminal leader was replaced by a His6 tag. Fig. 7A (lanes 1 and 2) shows that YnjEΔ1–21 was only detected in the cytoplasmic fraction. As control for a periplasmic protein, the E. coli MBP protein was detected using a monoclonal MBP antibody. Fig. 7B (lanes 3 and 4) shows that MBP was only detected in the periplasmic fraction. Localization of the two control proteins in their expected fractions demonstrates that the fractionation method effectively isolated the periplasmic and cytoplasmic fractions with minimal cross-contamination.
FIGURE 7.
Cellular localization of YnjE. Strains BL21(DE3), pAU2 in BL21(DE3), and MC1061(DE3) were used to determine the cellular localization of YnjE. E. coli cells were grown in LB medium in the presence or absence of 0.2% (w/v) maltose. After centrifugation, the cells were separated into cytoplasmic and periplasmic fractions. A, BL21(DE3) (pAU2) cells were grown in LB medium without isopropyl β-d-thiogalactopyranoside induction and fractionated, and cytoplasmic and periplasmic fractions were loaded in lanes 1 and 2, respectively. BL21(DE3) cells were grown in LB and fractionated, and cytoplasmic and periplasmic fractions were loaded in lanes 3 and 4, respectively. His6-tagged YnjEΔ1–21 and endogenous YnjE were detected by using polyclonal YnjE antisera. B, MC1061(DE3) cells were grown in LB medium containing 0.2% (w/v) maltose, and YnjE and MBP (which is a periplasmic protein) were detected using polyclonal YnjE (lanes 1 and 2) and MBP (lanes 3 and 4) antisera, respectively. Lanes 1 and 3, cytoplasmic fraction; lanes 2 and 4, periplasmic fraction.
DISCUSSION
The studies presented here identify the proteins involved in the sulfur transfer for the formation of the dithiolene moiety of Moco in E. coli. Earlier work demonstrated that any of three NifS-like sulfurtransferases, namely IscS, CsdA, or SufS, is capable of mobilizing and transferring sulfur from l-cysteine to MoaD in vitro (10). However, IscS was identified recently as the specific sulfur donor for Moco biosynthesis in vivo (11). Its involvement in Moco biosynthesis added another biosynthetic pathway to the versatile role of IscS, acting as sulfur donor for the biosynthesis of FeS clusters, biotin, thiamine, lipoic acid, and sulfur-containing bases in tRNA (42). Recent studies by Shi et al. (43) showed that the binding sites for the different proteins interacting with IscS are different and that the acceptor proteins approach the active site Cys-328 from different directions, which suggested that the conformational plasticity of a long loop containing this cysteine is essential for the ability of IscS to transfer sulfur to multiple acceptor proteins. However, this also provides a basis for competition of binding sites around the active site loop and possibly directing sulfur transfer to the right interaction partner when the sulfur-accepting protein is present in sufficient amounts.
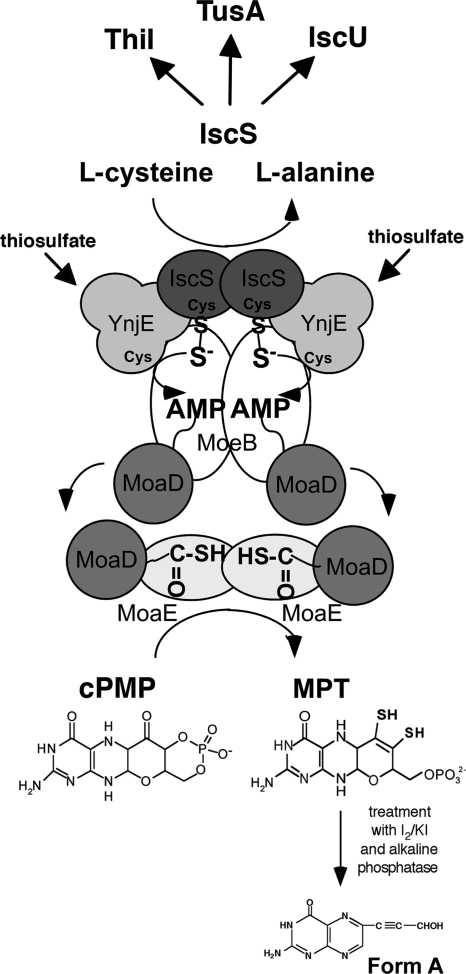
Our studies identified a new interaction partner of IscS, the rhodanese-like protein YnjE, which directs IscS toward Moco biosynthesis. Several independent methods for detecting protein-protein interactions revealed that YnjE directly interacts with IscS, MoeB, and MoaD, showing that these proteins form a complex during Moco biosynthesis in the cell. MoaD subsequently interacts with MoaE, which is also part of the complex, concomitantly converting cPMP to MPT (Fig. 8). However, studies of Moco biosynthesis using a ΔynjE stain revealed that YnjE is not essential for Moco biosynthesis under the conditions tested. An effect on Moco biosynthesis was only visible when iscS and ynjE were both deleted. This implied that under normal growth conditions in LB medium, IscS can fully substitute for the role of YnjE, whereas YnjE can partially take over the role of IscS in a ΔiscS mutant strain. We believe that the effect of YnjE is likely to be more apparent under conditions where one of the other sulfur-containing cofactors competes for the role of IscS. To determine these conditions when YnjE is required is planned for future studies. Our studies also showed that YnjE does not directly enhance the sulfurtransferase activity of IscS, as previously reported for the enhancers SufE and CsdE for their partners SufS and CsdA, respectively (28, 29). However, YnjE was shown to enhance the level of MoaD sulfuration in an in vitro system containing IscS and l-cysteine as sulfur source, possibly by making the interaction between the MoeB and IscS more specific because IscS is also involved in numerous sulfur transfer reactions in the cell. A protein component similar to YnjE has been reported previously for the biosynthesis of thiamine in E. coli. In thiamine biosynthesis, ThiS, a protein that is structurally similar to MoaD, carries the sulfur for synthesis of the thiazole moiety in the form of a C-terminal thiocarboxylate group (44). The minimum requirements for the activation of ThiS in vitro were shown to be ThiF (a protein similar in sequence to MoeB), IscS, Mg-ATP, and l-cysteine. ThiI, a protein possessing a C-terminal rhodanese domain, was not essential in this assay, but it was reported that ThiI stimulated the formation of ThiS-COSH, an effect that was believed to be necessary for sufficient ThiS-COSH production in vivo (21). Thus, mechanistically, the biosynthesis of Moco and thiamine are very similar in E. coli.
FIGURE 8.
Model for the biosynthesis of MPT in E. coli. The sulfur transfer pathway for the formation of the thiocarboxylate group on MoaD involves two sulfurtransferase proteins, IscS and YnjE, and l-cysteine as sulfur source. Prior to the sulfuration, MoaD is adenylated by MoeB. After sulfur transfer, thiocarboxylated MoaD dissociates from MoeB and reassociates with MoaE, forming active MPT synthase, which in turn converts cPMP to MPT. We believe that the proteins form a transient complex in the cell.
The crystal structure of YnjE has been reported recently (19). Amino acid sequence comparisons classified YnjE as a 3-mercaptopyruvate sulfurtransferase (17). However, structural comparisons and activity analyses showed that YnjE acts as a thiosulfate:cyanide sulfurtransferase (19). IscS was shown to be the likely primary sulfur donor to YnjE. However, because YnjE was purified in a sulfurated form from ΔiscS mutant strain, this implies that other sulfur donors might exist for YnjE. This sulfur donor might either be another rhodanese-like protein or thiosulfate as direct sulfur donor. We believe that backup systems for the sulfur transfer pathway exist in the cell. The ΔynjE/iscS double mutant also was able to produce residual amounts of MPT and an active nitrate reductase. Thus, other systems are able to substitute weakly for YnjE and IscS in their specific roles to provide the sulfur for the generation of the dithiolene group in Moco. When IscS is absent, one of the two remaining NifS-like l-cysteine desulfurases (CsdA or SufS) or one of the seven rhodanese-like proteins might be able to transfer sulfur to YnjE or directly to MoaD to ensure a low level of MPT formation in the cell. Analysis of a mutant strain in which seven of the eight rhodanese genes were deleted (ybbB was not deleted) revealed that this strain showed MPT synthase activity comparable with that of the wild-type strain (data not shown). Unfortunately, an E. coli mutant strain lacking all l-cysteine desulfurases is not viable, making it difficult to determine which protein is able to replace the YnjE/IscS pair in its function. Because IscS is involved in a variety of different sulfur transfer reactions in the cell, we propose two different roles for YnjE in respect to Moco biosynthesis: 1) YnjE acts as mediator to direct IscS toward Moco biosynthesis, and 2) YnjE can act as sulfur donor because it was found to be sulfurated in a ΔiscS mutant.
Phylogenetic analyses showed that in bacteria, two different types of proteins homologous to MoeB exist; some bacteria, like E. coli, contain solely a MoeB-like protein, some contain a MoeB homologue with a fused C-terminal rhodanese-like domain, and some contain both (13). In contrast, all eukaryotes contain a MoeB homologue with the C-terminal rhodanese-like domain. The well characterized human MOCS3 has been shown to catalyze both the adenylation and the subsequent generation of a thiocarboxylate group at the C terminus of MOCS2A during Moco biosynthesis (12). The sulfur is mobilized via a persulfide at the catalytic active site cysteine residue of the MOCS3 rhodanese-like domain, transferring sulfur from thiosulfate to MOCS2A in vitro. However, the low activity of these proteins with thiosulfate as sulfur donor in addition to mutagenesis studies of the active site loop of MOCS3 showed that thiosulfate is not the physiological sulfur source for Moco biosynthesis in eukaryotes (13). It was shown recently that in humans, the l-cysteine desulfurase NFS1 acts as a direct sulfur donor for MOCS3 in the cytosol (14). Thus, in humans and E. coli, the same protein components participate in the sulfur transfer pathway to MPT synthase during Moco biosynthesis (Fig. 8); l-cysteine is the direct sulfur donor for an l-cysteine desulfurase, which transfers the sulfur to the adenylated small subunit of MPT synthase with the aid of a rhodanese-like protein. In total, there seems to be an evolutionary advantage for eukaryotes to fuse the rhodanese-like protein to the C terminus of MoeB, probably making this reaction more specific.
YnjE is an enigmatic protein because it appears to have a dual role in E. coli; cytoplasmic YnjE is involved in Moco biosynthesis, whereas the function of periplasmic YnjE still remains obscure. So far, several examples for the dual localization of proteins exist in eukaryotes (45). In eukaryotes, several proteins, like Nfs1, were shown to be targeted to more than one subcellular compartment. Consequently, these proteins participate in different biochemical pathways and have several physiological functions (45). One mechanism for dual localization was suggested to be an incomplete translocation (e.g. the protein is folded before it is translocated) (45). Such a mechanism also remains possible in prokaryotes. Proteins with periplasmic leader sequences are transported to the periplasm either by the Sec system (46) or by the Tat system (47) to perform a specific role in the periplasm. In the case of YnjE, a 23-amino acid leader peptide has been identified that is cleaved during expression of the protein during its transport to the periplasm. Surprisingly, we showed that a major part of YnjE remains in the cytoplasm and presumably performs a specific role in the sulfur transfer for Moco biosynthesis. The functional complementation of the ΔiscSΔynjE mutant with YnjE where the Sec leader was truncated clearly confirmed the role of YnjE in the cytoplasm and also showed that YnjE is fully functional in the cell. The mechanism(s) that govern the dual localization of YnjE, along with determination of the physiological role of YnjE in the periplasm, must be investigated in future studies. It is possible that a portion of YnjE escapes the SecB-specific targeting to the membrane, if a part of YnjE is folded before SecB binds to it. Because we performed our experiments with YnjE purified from the cytoplasm, YnjE is correctly folded in this compartment. Also, the crystal structure was solved of the protein purified from the cytoplasm (19). The role of YnjE is also comparable with that of a protein identified recently in the formation of 2-thiouridine in tRNA of eukaryotes. The eukaryotic Tum1 protein was identified to be a two-domain rhodanese-like protein of the mercaptopyruvate sulfurtransferase family that possesses a mitochondrial leader sequence (48). By ribonucleome analysis in Saccharomyces cerevisiae, Tum1p was shown to participate in thiolation of 5-carboxycarbonylmethyl-2-thiouridine in cytoplasmic tRNA. It was shown that Tum1p stimulates the l-cysteine desulfurase activity of Nfs1p, the eukaryotic IscS homologue that is located in mitochondria (48). Tum1p accepts the persulfide sulfur from Nfs1p and is able to transfer this sulfur further onto the rhodanese-like domain of the MOCS3 homologue Uba4p in yeast, which is a cytoplasmic protein. Uba4p was also able to accept the sulfur directly from Nfs1p but at a slower rate (48). A dual localization for Tum1p in both mitochondria and the cytosol has been suggested. Thus, Tum1p acts as a mediator in the sulfur transfer reaction from Nfs1p to Uba4p to enhance the sulfur transfer rate in this reaction. Both the dual role of Tum1p in sulfur transfer and the dual localization of Tum1p correspond closely to the novel role of the YnjE protein identified in this study. Future studies might shed light on the dual localization of persulfide-containing proteins in different compartments in the cells of eukaryotes and prokaryotes and how these proteins are shuttled through the different membranes.
Acknowledgments
We thank Frederic Barras (CNRS Marseille, France) for providing plasmids for the expression of SufE and CsdE, Emanuelle Bouveret (CNRS Marseille, France) for providing plasmid pEB327, Charles Lauhon (University of Wisconsin) for providing strains CL100 and CL102, and Burkhard Micheel (University of Potsdam) for producing YnjE antisera. We also thank Dennis Dean (Virginia Tech) for longstanding support.
This work was supported by Deutsche Forschungsgemeinschaft Grant LE1171/5-3 (to S. L.), and by a grant from the Thomas F. Jeffress and Kate Miller Jeffress Memorial Trust (to T. J. L.).
- Moco
- molybdenum cofactor
- MPT
- molybdopterin
- cPMP
- cyclic pyranopterin monophosphate
- TAP
- tandem affinity purification
- CB
- calmodulin binding
- SPR
- surface plasmon resonance
- RU
- resonance units
- MBP
- maltose-binding protein
- 1,5-I-AEDANS
- N-(iodoacetaminoethyl)-1-naphtylamine-5′-sulfonic acid
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride
- PDEA
- 2-(2-pyridinyldithio) ethaneamine.
REFERENCES
- 1. Leimkühler S., Wuebbens M. M., Rajagopalan K. V. (2011) Coord. Chem. Rev. 255, 1129–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rajagopalan K. V. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhardt F. C. ed) pp. 674–679, American Society for Microbiology Press, Washington, D. C [Google Scholar]
- 3. Neumann M., Mittelstädt G., Seduk F., Iobbi-Nivol C., Leimkühler S. (2009) J. Biol. Chem. 284, 21891–21898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hilton J. C., Rajagopalan K. V. (1996) Arch. Biochem. Biophys. 325, 139–143 [DOI] [PubMed] [Google Scholar]
- 5. Pitterle D. M., Johnson J. L., Rajagopalan K. V. (1993) J. Biol. Chem. 268, 13506–13509 [PubMed] [Google Scholar]
- 6. Pitterle D. M., Rajagopalan K. V. (1989) J. Bacteriol. 171, 3373–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gutzke G., Fischer B., Mendel R. R., Schwarz G. (2001) J. Biol. Chem. 276, 36268–36274 [DOI] [PubMed] [Google Scholar]
- 8. Rudolph M. J., Wuebbens M. M., Rajagopalan K. V., Schindelin H. (2001) Nat. Struct. Biol. 8, 42–46 [DOI] [PubMed] [Google Scholar]
- 9. Leimkühler S., Wuebbens M. M., Rajagopalan K. V. (2001) J. Biol. Chem. 276, 34695–34701 [DOI] [PubMed] [Google Scholar]
- 10. Leimkühler S., Rajagopalan K. V. (2001) J. Biol. Chem. 276, 22024–22031 [DOI] [PubMed] [Google Scholar]
- 11. Zhang W., Urban A., Mihara H., Leimkühler S., Kurihara T., Esaki N. (2010) J. Biol. Chem. 285, 2302–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matthies A., Rajagopalan K. V., Mendel R. R., Leimkühler S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5946–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krepinsky K., Leimkühler S. (2007) FEBS J. 274, 2778–2787 [DOI] [PubMed] [Google Scholar]
- 14. Marelja Z., Stöcklein W., Nimtz M., Leimkühler S. (2008) J. Biol. Chem. 283, 25178–25185 [DOI] [PubMed] [Google Scholar]
- 15. Spallarossa A., Donahue J. L., Larson T. J., Bolognesi M., Bordo D. (2001) Structure 9, 1117–1125 [DOI] [PubMed] [Google Scholar]
- 16. Mueller E. G., Palenchar P. M. (1999) Protein Sci. 8, 2424–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bordo D., Bork P. (2002) EMBO Rep. 3, 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolfe M. D., Ahmed F., Lacourciere G. M., Lauhon C. T., Stadtman T. C., Larson T. J. (2004) J. Biol. Chem. 279, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 19. Hänzelmann P., Dahl J. U., Kuper J., Urban A., Müller-Theissen U., Leimkühler S., Schindelin H. (2009) Protein Sci. 18, 2480–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Begley T. P., Xi J., Kinsland C., Taylor S., McLafferty F. (1999) Curr. Opin. Chem. Biol. 3, 623–629 [DOI] [PubMed] [Google Scholar]
- 21. Lauhon C. T., Kambampati R. (2000) J. Biol. Chem. 275, 20096–20103 [DOI] [PubMed] [Google Scholar]
- 22. Link A. J., Phillips D., Church G. M. (1997) J. Bacteriol. 179, 6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nichols B. P., Shafiq O., Meiners V. (1998) J. Bacteriol. 180, 6408–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silhavy T. J., Berman M. L., Enquist L. W. (1984) Experiments with Gene Fusions, pp. 107–112, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng H., Donahue J. L., Battle S. E., Ray W. K., Larson T. J. (2008) Open Microbiol. J. 2, 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cherepanov P. P., Wackernagel W. (1995) Gene 158, 9–14 [DOI] [PubMed] [Google Scholar]
- 28. Loiseau L., Ollagnier-de-Choudens S., Nachin L., Fontecave M., Barras F. (2003) J. Biol. Chem. 278, 38352–38359 [DOI] [PubMed] [Google Scholar]
- 29. Loiseau L., Ollagnier-de Choudens S., Lascoux D., Forest E., Fontecave M., Barras F. (2005) J. Biol. Chem. 280, 26760–26769 [DOI] [PubMed] [Google Scholar]
- 30. Ollagnier-de-Choudens S., Lascoux D., Loiseau L., Barras F., Forest E., Fontecave M. (2003) FEBS Lett. 555, 263–267 [DOI] [PubMed] [Google Scholar]
- 31. Gully D., Moinier D., Loiseau L., Bouveret E. (2003) FEBS Lett. 548, 90–96 [DOI] [PubMed] [Google Scholar]
- 32. Jones R. W., Garland P. B. (1977) Biochem. J. 164, 199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glaser J. H., DeMoss J. A. (1972) Mol. Gen. Genet. 116, 1–10 [DOI] [PubMed] [Google Scholar]
- 34. Urbina H. D., Silberg J. J., Hoff K. G., Vickery L. E. (2001) J. Biol. Chem. 276, 44521–44526 [DOI] [PubMed] [Google Scholar]
- 35. Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. (1984) J. Biol. Chem. 259, 5414–5422 [PubMed] [Google Scholar]
- 36. Schmitz J., Wuebbens M. M., Rajagopalan K. V., Leimkühler S. (2007) Biochemistry 46, 909–916 [DOI] [PubMed] [Google Scholar]
- 37. Wuebbens M. M., Rajagopalan K. V. (1995) J. Biol. Chem. 270, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 38. Zheng L., White R. H., Cash V. L., Dean D. R. (1994) Biochemistry 33, 4714–4720 [DOI] [PubMed] [Google Scholar]
- 39. Heidenreich T., Wollers S., Mendel R. R., Bittner F. (2005) J. Biol. Chem. 280, 4213–4218 [DOI] [PubMed] [Google Scholar]
- 40. Rouvière P. E., Gross C. A. (1996) Genes Dev. 10, 3170–3182 [DOI] [PubMed] [Google Scholar]
- 41. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 42. Marquet A. (2001) Curr. Opin. Chem. Biol. 5, 541–549 [DOI] [PubMed] [Google Scholar]
- 43. Shi R., Proteau A., Villarroya M., Moukadiri I., Zhang L., Trempe J. F., Matte A., Armengod M. E., Cygler M. (2010) PLoS Biol. 8, e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang C., Xi J., Begley T. P., Nicholson L. K. (2001) Nat. Struct. Biol. 8, 47–51 [DOI] [PubMed] [Google Scholar]
- 45. Regev-Rudzki N., Pines O. (2007) BioEssays 29, 772–782 [DOI] [PubMed] [Google Scholar]
- 46. de Keyzer J., van der Does C., Driessen A. J. (2003) Cell Mol. Life. Sci. 60, 2034–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berks B. C., Palmer T., Sargent F. (2005) Curr. Opin. Microbiol. 8, 174–181 [DOI] [PubMed] [Google Scholar]
- 48. Noma A., Sakaguchi Y., Suzuki T. (2009) Nucleic Acids Res. 37, 1335–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wuebbens M. M., Rajagopalan K. V. (2003) J. Biol. Chem. 278, 14523–14532 [DOI] [PubMed] [Google Scholar]