Abstract
Newly synthesized proteins and lipids are transported in vesicular carriers along the secretory pathway. Arfs (ADP-ribosylation factors), a family of highly conserved GTPases within the Ras superfamily, control recruitment of molecular coats to membranes, the initial step of coated vesicle biogenesis. Arf1 and coatomer constitute the minimal cytosolic machinery leading to COPI vesicle formation from Golgi membranes. Although some functional redundancies have been suggested, other Arf isoforms have been poorly analyzed in this context. In this study, we found that Arf1, Arf4, and Arf5, but not Arf3 and Arf6, associate with COPI vesicles generated in vitro from Golgi membranes and purified cytosol. Using recombinant myristoylated proteins, we show that Arf1, Arf4, and Arf5 each support COPI vesicle formation individually. Unexpectedly, we found that Arf3 could also mediate vesicle biogenesis. However, Arf3 was excluded from the vesicle fraction in the presence of the other isoforms, highlighting a functional competition between the different Arf members.
Keywords: G Proteins, Golgi, Intracellular Trafficking, Membrane Trafficking, Protein Purification, ADP-ribosylation Factors, COPI Vesicles, Reconstitution Assay
Introduction
In eukaryotic cells, newly synthesized secretory and membrane proteins as well as lipids are transported along the secretory pathway from the endoplasmic reticulum (ER)2 through the Golgi apparatus to their final destination. Most of the trafficking events are served by spherical carriers that bud from a donor compartment, travel through the cytoplasm and fuse with an acceptor compartment. Vesicle formation is mainly controlled by GTPase-regulated recruitment of molecular coats, protein complexes able to select cargo to be included in the transport carrier and to deform membranes. The COPII coat contributes to export from the ER (1), whereas coatomer forms COPI-coated vesicles, involved in retrograde transport back to the ER as well as different trafficking steps within the Golgi (2). At the trans-Golgi network (TGN), clathrin coats mediate sorting of cargos toward the plasma membrane and different internal compartments (3).
Small GTPases regulate the recruitment of molecular coats to membranes. Sar1 controls the association of COPII with ER membranes (4), whereas Arf1 recruits Mints (5), coatomer and the clathrin adaptor proteins (APs)-1 and -3 and Golgi-localized γ-ear-containing Arf-binding proteins (GGAs) (6) to Golgi membranes as well as AP-4 to membranes of the late secretory pathway (6). Arfs constitute a family of highly conserved proteins, with multiple family members found in all eukaryotes. The Arf family is functionally divided into Arfs and Arf-like (Arl) proteins, with the latter demonstrating more divergent sequences and functions. Mammals express six isoforms of Arfs, namely Arf1–6 (with Arf2 being absent in human), divided into three classes (7). Class I is made up of Arf1–3, Arf4, and Arf5 comprise class II, and Arf6, the most divergent member of the family, is the sole member of class III. Arfs cycle between an inactive, soluble GDP-loaded form, and an active, membrane associated GTP-loaded form. Upon activation by an Arf guanine exchange factor (ArfGEF), Arfs undergo a conformational change, exposing their myristoylated N-terminal amphipathic helix, which inserts into the membrane (8). With the exception of Arf6 that functions at the plasma membrane (9–11), Arfs localize primarily to the Golgi complex (12), though also act at endosomes (13, 14).
In COPI vesicle biogenesis, Arf1-GDP is first recruited to Golgi membranes by a transmembrane receptor: p23 (and probably p24) (15, 16) and/or membrin (17). Nucleotide exchange by the ArfGEF GBF1 (Golgi-specific Brefeldin A resistance factor) induces the activation of Arf1 and the recruitment of coatomer (18, 19). After vesicle formation, hydrolysis of GTP by Arf1 upon stimulation by an Arf-GTPase-activating protein (ArfGAP) precedes the release of the coat from the membranes (20–22).
Liang and Kornfeld (23) showed that in addition to Arf1, Arf5, and Arf6 could also recruit coatomer to Golgi membranes in an in vitro assay. In the living cell, knockdown of only one Arf isoform did not lead to any phenotype, whereas simultaneous knockdown of two isoforms resulted in specific trafficking defects, highlighting redundancies between Arf isoforms (14). Altogether, these data suggest that Arfs other than Arf1 could support COPI vesicle formation. In this study we found that Arf1, Arf4 and Arf5 but not Arf3 and Arf6 associate with COPI vesicles generated in vitro with cytosol. Using recombinant proteins, we show that Arf1, Arf4, Arf5, but not Arf6 can induce COPI vesicle formation individually. Unexpectedly, we found that Arf3 can also mediate vesicle biogenesis. However, Arf3 is excluded from the vesicle fraction in the presence of the other isoforms, highlighting a potential functional competition between the different Arf proteins.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: the rabbit polyclonal anti-δCOP (877) (24), anti-p27 (25), anti-Arf1 (C1) (20), anti-Arf3 (R-1023), anti-Arf4 (R-891), anti-Arf5 (R-1525) (11), anti-Arf6 (26), and mouse monoclonal anti-clathrin heavy chain (BD Transduction Laboratories).
Purification of Recombinant Arfs
N-myristoylated human wild type Arf1 (27) and Arf5 (28) were expressed and purified as described. For N-myristoylated human wild-type Arf3 and Arf4, bacterial pellets were resuspended in lysis buffer (50 mm Tris/HCl, pH 8.0, 1 mm MgCl2, 1 mm GDP, 1 mm DTT, 1.5 μg/ml leupeptin, 2.5 μg/ml antipain, 25 μg/ml trypsin inhibitor, 12.5 μg/ml benzamidin, 6.25 μg/ml pefabloc, 1.25 μg/ml aprotinin, 5 μg/ml chymostatin, 2.5 μg/ml pepstatin A) and passed through a microfluidizer. Cellular debris was removed by centrifugation (100,000 × g, 1 h, 4 °C). Arfs were precipitated by gradual addition of ammonium sulfate up to 40% saturation over 45 min, and then stirred for 1 h. After centrifugation (10,000 × g, 20 min, 4 °C), the pellets were resuspended in resuspension buffer (same as lysis buffer with only 20 mm Tris/HCl). After ultracentrifugation (100,000 × g, 15 min, 4 °C) to remove residual aggregates, samples were loaded on a Superdex 75 (26/60) column and eluted with gel filtration buffer (10 mm Tris/HCl, pH 8.0, 50 mm KCl, 1 mm MgCl2, 10% glycerol, 1 mm DTT). N-myristoylated human wild type Arf6 was purified as a partially GTP-loaded form according to a modification of the protocol from Michel Franco (29). Bacterial pellets were resuspended in lysis buffer (50 mm Tris/HCl, pH 8.0, 1 mm EDTA, 200 μm GTP, 1 mm DTT, 1.5 μg/ml leupeptin, 2.5 μg/ml antipain, 25 μg/ml trypsin inhibitor, 12.5 μg/ml benzamidin, 6.25 μg/ml pefabloc, 1.25 μg/ml aprotinin, 5 μg/ml chymostatin, 2.5 μg/ml pepstatin A). The bacterial suspension was incubated for 1 h in the presence of lysozyme (0.5 mg/ml). Then 0.05% sodium deoxycholate, 3 mm MgCl2, and 0.1 mg/ml DNase I were added and incubated for 30 min on ice. Cellular debris was removed by centrifugation (10,000 × g, 20 min, 4 °C). Arf6 was precipitated by gradual addition of ammonium sulfate up to 37.5% saturation over 45 min, and then stirred for 1 h. After centrifugation (10,000 × g, 20 min, 4 °C), the pellet was resuspended in resuspension buffer (50 mm Tris/HCl, pH 8.0, 1 mm MgCl2, 2 μm GTP, 1 mm DTT). After ultracentrifugation (100,000 × g, 15 min, 4 °C) to remove residual aggregates, samples were loaded on a Superdex 200 (26/60) column and eluted with gel filtration buffer (50 mm Tris/HCl, pH 8.0, 1 mm MgCl2, 1 mm DTT). Loading of Arf6 with GDP was performed by incubating the protein for 4 h on ice with 2 mm EDTA and 5 mm GDP followed by addition of 2 mm MgCl2. GDP-loaded Arf6 was separated from free nucleotide by gel filtration on a PD-10 column.
Rat Liver Golgi, Cytosol, and Rabbit Liver Coatomer
Golgi membranes were purified from rat liver homogenates as described (30), and cytosol was dialyzed against 25 mm HEPES/KOH pH 7.2, 50 mm KCl, 1 mm DTT. Rabbit liver coatomer was purified as described (31).
Golgi Binding Assay
Prior to each experiment, proteins were centrifuged for 20 min at 100,000 × g to remove aggregates. Rat liver Golgi membranes (10 μg) were incubated for 10 min at 37 °C with rat liver cytosol (330 μg), supplemented with protein inhibitor mixture (Roche), or 0.8 μg (for Fig. 7, 350 ng Arf1, 120 ng Arf3, 63 ng Arf4, and 63 ng Arf5 were used; see text for more details) of recombinant myr-Arf in the presence of 100 μm GTPγS. Reactions were performed in a total volume of 42 μl in assay buffer (25 mm HEPES/KOH pH 7.2, 2.5 mm MgAc, 20 mm KCl, 200 mm sucrose, 0.25 mm DTT), and 6% of the input was taken for Western blotting. Samples were laid on top of sucrose cushions of 17% (150 μl) and 50% (15 μl) sucrose (% by weight) in a Beckman SW60-mini tube. After centrifugation for 20 min at 17,000 × g, Golgi membranes were collected at the 17/50% sucrose interphase, and 25% was used for Western blotting.
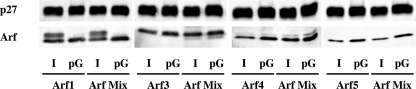
FIGURE 7.
Individual versus collective binding of Arf isoforms to Golgi membranes. Individual Arfs or a mixture of Arf1, Arf3, Arf4, and Arf5 were subjected according to their endogenous ratios (350 ng Arf1, 120 ng Arf3, 63 ng Arf4, 63 ng Arf5) to Golgi-enriched membranes and incubated for 10 min at 37 °C in the presence of GTPγS. Primed-Golgi was purified by centrifugation through two sucrose cushions. 6% of the initial reaction mix (I for Input) and 25% of primed-Golgi (pG) were subjected to SDS-PAGE and Western blot analysis.
Vesicle Formation Assays
Prior to each experiment, protein preparations were centrifuged (10 min, 16,000 × g) to remove aggregates. Rat liver Golgi membranes (60 μg) were incubated for 10 min at 37 °C with rat liver cytosol (2 mg), supplemented with protein inhibitor mixture (Roche), or 25 μg of rabbit liver coatomer and 5 μg recombinant myr-Arf (for Fig. 8, 2.1 μg Arf1, 0.72 μg Arf3, 0.38 μg Arf4, and 0.38 μg Arf5 were used; see text for more details) in the presence of 100 μm GTPγS. Reactions were performed in a total volume of 250 μl in assay buffer (25 mm HEPES/KOH pH 7.2, 2.5 mm MgAc, 20 mm KCl, 200 mm sucrose, 0.25 mm DTT), and 1% of the input was taken for Western blotting. Samples were then subjected to 250 mm KCl to dissociate tethered COPI vesicles from the donor Golgi membranes, and were then pelleted by centrifugation twice for 5 min each, at 16,000 × g and 4 °C. The supernatant, containing COPI vesicles, was laid on top of sucrose cushions of 37.5% (50 μl) and 50% (5 μl) sucrose (% by weight) in a Beckman SW60-mini tube. After centrifugation for 50 min at 100,000 × g, the vesicles were collected at the 37.5/50% sucrose interphase and the collected material was analyzed by Western blotting and negative staining electron microscopy.
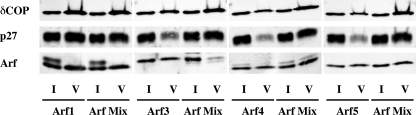
FIGURE 8.
Vesicle preparation in the presence of individual or a mixture of Arf isoforms. Individual Arf or a mixture of Arf1, Arf3, Arf4 and Arf5 were subjected according to their endogenous ratios (2.1 μg Arf1, 0.72 μg Arf3, 0.38 μg Arf4, 0.38 μg Arf5) to Golgi-enriched membranes and incubated for 10 min at 37 °C in the presence of GTPγS. COPI vesicle were purified by centrifugation through two sucrose cushions. 1% of the initial reaction mix (I for Input) and 50% of the vesicle fraction (V) were subjected to SDS-PAGE and Western blot analysis.
Mass Spectrometry
Six COPI vesicles preparations were pooled, diluted with 2 volumes of assay buffer and centrifuged for 1 h at 100,000 × g at 4 °C. The pellet was resuspended in 8 μl of 20 mm ammonium bicarbonate buffer (pH 7.0) and submitted to peptide mass fingerprinting analysis. The samples were reduced, alkylated, and digested by either trypsin, chymotrypsin, or both proteases. After in gel digestion, peptides were purified and enriched using C18-ZipTips (Millipore Corp., Billerica, MA), resuspended in an aqueous solution containing 5% acetonitrile and 0.1% formic acid, and loaded on reversed phase columns (trapping column: particle size 5 μm, C18, L = 20 mm; analytical column: particle size 3 or 5 μm, C18, L = 15 cm; NanoSeparations, Nieuwkoop, The Netherlands) on a nano- high pressure liquid chromatography (HPLC) (Proxeon easy-nLC). Peptides were eluted in gradients of water (0.1% formic acid, buffer A) and acetonitrile (0.1% formic acid, buffer B). Typically, gradients were ramped from 5% to 70% B in 95 min at flow rates of 300 nl/min (extended gradients: 5% to 65% B in 160 min). Peptides eluting from the column were ionized online using a Bruker Apollo ESI-source with a nanoSprayer emitter and analyzed in a quadrupole time-of-flight mass spectrometer (Bruker maXis, Bruker Daltonics, Bremen, Germany). Mass spectra were acquired over the mass range 50–2200 m/z, and sequence information was acquired by computer-controlled, data-dependent automated switching to MS/MS using collision energies based on mass and charge state of the candidate ions.
The data sets were processed using a standard proteomics script with the software Bruker DataAnalysis 4.0 (Service Pack 1 Build 253) and exported as Mascot generic files. Spectra were internally recalibrated on autoproteolytic trypsin fragments when applicable.
Proteins were identified by matching the derived mass lists against the NCBI nr protein data base on a local Mascot server (Version 2.2.2, Matrix Science, UK). In general, a mass tolerance ± 0.05 Da for parent ion and fragment spectra, three missed cleavages, oxidation of Met and fixed modification of carbamidomethyl cysteine were selected as matching parameters in the search program. The proteomics data associated with this report were uploaded to the PRIDE online repository (62).
Quantification of Arfs in Cytosol
Concentrations of purified recombinant Arf preparations were determined using the Bradford protein assay (32). To compensate for differences in purity of different Arf preparations, concentrations were compared by SDS-PAGE and Coomassie staining using BSA as standard and adjusted accordingly. The amount of each Arf isoform in cytosol preparations was quantified by Western blot analysis using recombinant purified Arfs as standards.
Analysis of Nucleotides
Purified Arfs (33 μg of Arf4 and 43 μg of Arf6) were incubated 1 min at 95 °C. Denatured proteins were pelleted by centrifuging 5 min, 16,000 × g, room temperature. Supernatant was analyzed by reversed-phase HPLC as already described (33).
RESULTS
Arf1, Arf4, and Arf5, but Not Arf2, Arf3, or Arf6 Are Found in COPI Vesicles
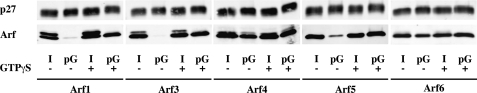
We first assessed whether other Arf members besides Arf1 can associate with COPI vesicles. Rat liver Golgi membranes were incubated with rat liver cytosol, as a source of coatomer and Arfs, in the presence of GTPγS, a slowly hydrolyzing analog of GTP. Upon binding GTPγS, Arfs are retained in their active form and remain attached to membranes. Primed Golgi (Golgi after incubation with cytosol and GTPγS) (Fig. 1) and COPI-coated vesicles (Fig. 2) were then purified by isopycnic centrifugation through sucrose gradients and analyzed by Western blot using isoform-specific antibodies (11, 26, 34). It is of note that vesicle fractions were devoid of clathrin transport carrier (Fig. 2).
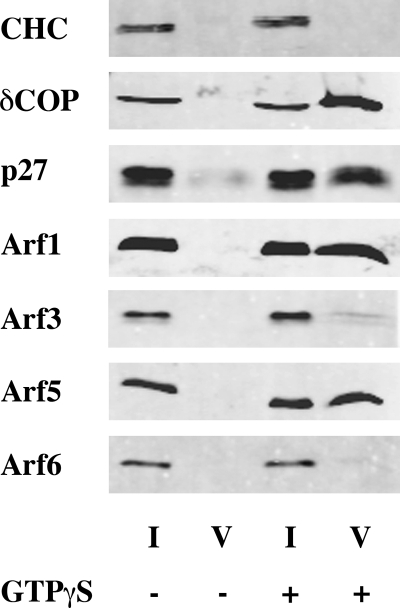
FIGURE 1.
Arf1, Arf3, Arf5, and Arf6 bind to Golgi membranes. Rat liver Golgi-enriched membranes were incubated with rat liver cytosol in the presence or absence of GTPγS and purified by centrifugation through two sucrose cushions. 6% of the initial reaction mix (I for Input) and 25% of primed-Golgi (pG) were subjected to SDS-PAGE and Western blot analysis.
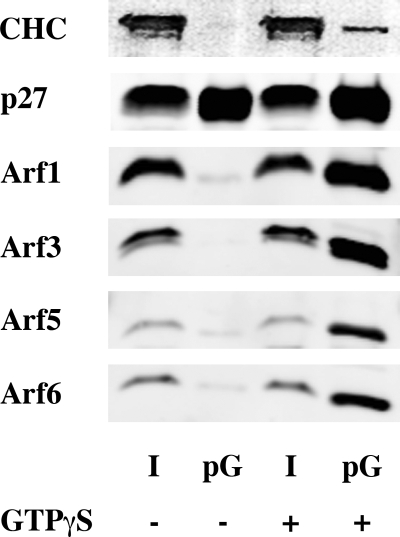
FIGURE 2.
Arf1 and Arf5 but not Arf3 nor Arf6 are present in COPI vesicles. COPI vesicles were generated in vitro by adding rat liver cytosol to rat liver Golgi-enriched membranes in the presence or absence of GTPγS, and purified by centrifugation through two sucrose cushions. 1% of the initial reaction mix (I for Input) and 50% of the vesicle fraction (V) were subjected to SDS-PAGE and Western blot analysis.
Each of the Arf isoforms monitored by Western blot (Arf1, 3, 5, and 6) was able to associate with the Golgi-enriched membrane fraction in a GTPγS-dependent manner (Fig. 1). By contrast, only Arf1 and Arf5 were observed in the vesicle fraction (Fig. 2). As the isoform-specific antibodies were raised against human proteins, we were not able to detect Arf2, which does not exist in humans, or Arf4, whose sequence in rat differs from humans. To circumvent this issue, COPI vesicles were analyzed by mass spectrometry. Several unique peptides corresponding to Arf1 (data not shown), Arf4 and Arf5 (Fig. 3) were detected in the vesicle fraction in a GTPγS-dependent manner. No peptide unique for Arf2, Arf3, or Arf6 could be detected under these conditions. Failure to find any unique peptides from a protein by mass spectrometry analysis is typically considered a weak argument that the protein is absent from the sample. However, the similarities in protein sequences and predicted behaviors in gels, trypsin digests, HPLC (high pressure liquid chromatography), and ionization and the failure to identify unique peptides after a specific search for them, strengthen our conclusion that they are simply not present. Taken together, these data suggest that although each of the Arf isoforms could associate with Golgi-enriched membranes, only Arf1, Arf4, and Arf5, but not Arf2, Arf3, or Arf6, were efficiently incorporated into COPI vesicles.
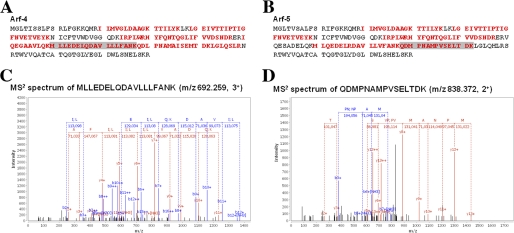
FIGURE 3.
Arf4 and Arf5 are present in COPI vesicles. COPI vesicles were generated in vitro by adding rat liver cytosol to rat liver Golgi-enriched membranes in the presence or absence of GTPγS, and purified by centrifugation through two sucrose cushions. Samples were then submitted to peptide mass fingerprinting analysis. Arf4 and Arf5 were identified by multiple unique peptides (see “Experimental Procedures”): sequence coverage (bold red) obtained in both tryptic and chymotryptic digests (A and B) and fragmentation spectra of representative unique peptides for Arf4 (C) and Arf5 (D).
Recombinant Arf1, Arf3, Arf4, and Arf5 Support in Vitro Formation of COPI Vesicles
Arf1, in combination with coatomer, constitutes the minimal cytosolic machinery able to induce in vitro COPI vesicle formation (20, 35). The presence of class II Arfs in the vesicles could thus be either a consequence of budding from Golgi membrane areas initially containing bound Arf4 and Arf5, or reflect an active participation of these Arf isoforms in COPI vesicle biogenesis. To discriminate between these two possibilities, myristoylated human Arfs were expressed and purified from bacteria and analyzed independently. All purified recombinant proteins were functional and could be recruited to Golgi-enriched membranes (Fig. 4). For Arf1, Arf3, and Arf5, a clear GTPγS-dependent binding was observed. Interestingly, Arf4 and Arf6, although initially present in their GDP-loaded state (supplemental Fig. S1), could associate, at least partially, to membranes in absence of GTPγS. This is consistent with previous studies showing membrane binding of GDP-loaded Arf4 (12, 36). Arf6 have been reported to be recruited to membranes in GTP-dependent (23, 37) and independent (11, 38) manners. In our hands, purified recombinant Arf6 could partially bind to Golgi in the absence of GTPγS (Fig. 4) whereas cytosolic Arf6 could be detected on membranes only in presence of GTPγS (Fig. 1). These data suggest that some cytosolic factors would participate in coupling the GDP/GTP cycle of Arf6 to its membrane association.
FIGURE 4.
Recombinant Arf1, Arf3, Arf4, Arf5, and Arf6 bind to Golgi-enriched membranes. Rat liver Golgi-enriched membranes were incubated with each of the recombinant Arf isoforms and purified rabbit liver coatomer in the presence or absence of GTPγS, and then purified by centrifugation through two sucrose cushions. 6% of the initial reaction mix (I for Input) and 25% of primed-Golgi (pG) were subjected to SDS-PAGE and Western blot analysis.
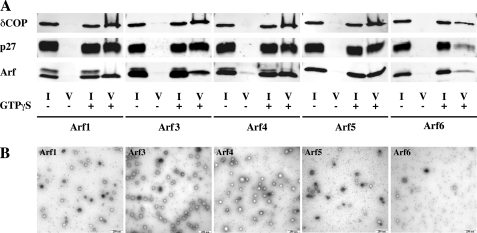
Five micrograms of each individual purified Arf were incubated with 60 μg of Golgi-enriched membranes, purified coatomer and GTPγS. Consistent with the results shown in Fig. 2, Arf1, Arf4, and Arf5 could efficiently support COPI vesicle formation, whereas only few vesicles could be detected in the presence of Arf6 (Fig. 5). Electron microscopic analysis of the vesicle fractions confirmed the presence of ∼80 nm spherical coated structures characteristic of COPI vesicles (Fig. 5B). Interestingly, Arf3 was also able to induce COPI vesicle formation (Fig. 5), which was unexpected given its absence from COPI vesicles generated in the presence of cytosol (Fig. 2). Note that no vesicle could be detected in the absence of Arfs (supplemental Fig. S2). Thus, whereas only Arf1, Arf4, and Arf5 associated with COPI vesicles generated with the endogenous pool of Arf isoforms present in the cytosol, individual recombinant Arf1, Arf4, Arf5, but also Arf3 supported COPI vesicle formation independently.
FIGURE 5.
Arf1, Arf3, Arf4, Arf5, but not Arf6 can generate COPI vesicle in vitro. COPI vesicles were generated in vitro from purified rabbit liver coatomer and recombinant myristoylated Arf1, Arf4, or Arf5 and rat liver Golgi-enriched membranes in the presence of GTPγS, and purified by centrifugation through two sucrose cushions. A, 1% of the initial reaction mix (I for Input) and 50% of the vesicle fraction (V) were subjected to SDS-PAGE and Western blot analysis. B, sample of the vesicle fractions were analyzed by negative stain electron microscopy. Scale bar: 250 nm.
Competition between Arf3 and the Other Arf Isoforms
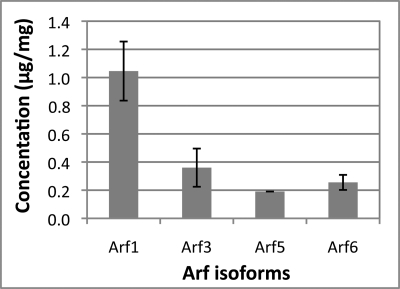
Arf3 alone supports COPI vesicle formation, however, was not detected in vesicles generated from cytosol. A possible explanation for this apparent contradiction would be a competition between Arf3 and the other Arf isoforms, either alone or in combination. To analyze an interplay between the various isoforms, we first determined the concentrations of each Arf in our rat liver cytosol preparation. This was performed by Western blot quantifications using purified recombinant Arfs as standards (Fig. 6). In agreement with the literature, Arf1 is the most abundant isoform, present at just under 0.1% of total cytosolic protein, whereas Arf5 is present at the lowest concentration with 0.02% of total cytosol protein. Arf3 is present at an intermediate level of 0.035% of total cytosol protein, at just over 35% the level of Arf1 and 190% the level of Arf5. Because of the lack of an antibody specific to rat Arf4, we were not able to determine its concentration using this approach. It is however known that, in different human tissues, Arf4 is, like Arf5, much less abundant than Arf1 (11). We thus decided, in a first approximation, to consider that Arf4 and Arf5 are present in similar concentrations.
FIGURE 6.
Quantification of Arf isoforms in rat liver cytosol. Defined aliquots of rat liver cytosol were subjected to SDS-gel electrophoresis and Western blotting. Blots were developed with antibodies directed against Arf isoforms as indicated, and quantified with reference to recombinant Arf isoforms. For details, see “Experimental Procedures.”
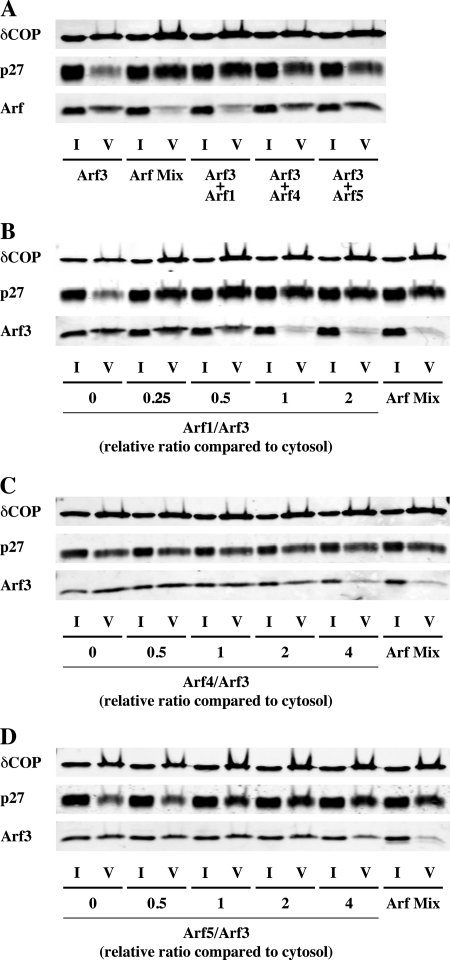
We then analyzed the behavior of Arf1, Arf3, Arf4, and Arf5 alone, or in the context of a mixture of all of them (Arf Mix). For this purpose, we used them according to their cytosolic concentrations (Fig. 6). Each individual Arf bound to Golgi membranes with similar efficiencies when they were alone or in the presence of the other isoforms (Fig. 7). The amounts of Arf1, Arf4, and Arf5 in the vesicle fraction were similar in both conditions (Fig. 8). However, although Arf3 alone did induce COPI vesicle formation and was found in purified vesicles, it was excluded from the vesicle fraction in the presence of the other Arf isoforms (Fig. 8), highlighting a competition between Arf3 and other Arfs for inclusion into COPI vesicles.
To further characterize this competition, we analyzed the interplay between Arf3 and each of the other isoforms at endogenous concentrations. Arf1 alone was able to exclude Arf3 from the vesicle fraction as efficiently as Arf1, Arf4, and Arf5 together, whereas neither Arf4 nor Arf5 competed with Arf3 under these conditions (Fig. 9A). Additionally, we performed competition assays using different ratios between Arf3 and the other isoforms (Fig. 9, B–D). Ratios are expressed as normalized values with regard to cytosol, i.e. a ratio of 1 corresponds to the endogenous ratio. As we are lacking knowledge of the exact amount of Arf4 in rat-liver cytosol, normalized ratios for Arf4 have been calculated assuming that Arf4 and Arf5 are expressed at similar levels. Arf1 efficiently competed with Arf3 at a cytosolic ratio (Fig. 9B). Arf4, and to a lesser extent Arf5, were also able to compete with Arf3 at high ratios (Fig. 9, C and D). Without knowing the cytosolic concentration of Arf4, we cannot rule out partial interplay between this isoform and Arf3 under endogenous conditions. We nevertheless considerer unlikely that Arf4 could be expressed at high enough levels for competing with Arf3. Altogether, these data offer a simple explanation for why Arf3 was not detectable on COPI vesicles generated from cytosol. In conclusion, it seems that at the concentrations prevailing in the living cell, a competition exists between Arf3 and the other Arf isoforms with regard to stable incorporation into COPI vesicles. This interplay seems mainly driven by Arf1 and suggests a more complicated functional interplay between Arf isoforms in the cell than previously thought.
FIGURE 9.
Characterization of the competition between Arf3 and the other Arf isoforms. A, Arf3 (0.72 μg) was mixed to each other Arf isoforms separately of all together (ArfMix) according to their endogenous ratios (2.1 μg Arf1, 0.38 μg Arf4, 0.38 μg Arf5) before being subjected to Golgi-enriched membranes and incubated for 10 min at 37 °C in the presence of GTPγS. COPI vesicles were purified by centrifugation through two sucrose cushions. 1% of the initial reaction mix (I for Input) and 50% of the vesicle fraction (V) were subjected to SDS-PAGE and Western blot analysis. In B–D, Arf3 (0.72 μg) has been mixed with Arf1 (B), Arf4 (C), and Arf5 (D) according to different ratios. Ratios are expressed as normalized values with regards to cytosol, i.e. a ratio of 1 corresponds to the endogenous ratio. For Arf4, normalized ratios have been calculated considering Arf4 and Arf5 to be expressed at similar levels.
DISCUSSION
Arf1, together with coatomer, represents the minimal cytosolic machinery required for in vitro COPI vesicle biogenesis. In this study, we show that additional Arf isoforms can regulate this process. While all isoforms bound to Golgi-enriched membranes, only Arf1, Arf4, and Arf5, but not Arf3 or Arf6, could be detected in COPI vesicles generated in the presence of cytosol and GTPγS. Using recombinant proteins, we showed that Arf1, Arf4, and Arf5 could support COPI vesicle formation as individual proteins. Unexpectedly, we found that Arf3 could also induce vesicle formation and incorporate into mature vesicles when the sole source of Arf, but was specifically excluded from vesicles when other Arfs, especially Arf1, were present. These results highlight a competition between Arfs resulting in the exclusion of Arf3 from COPI vesicles when the isoforms are present at ratios found in cytosol.
Involvement of Several Arfs in COPI Vesicle Formation
Several lines of evidence suggested that Arf isoforms other than Arf1 could be involved in COPI vesicle formation. In the living cell, knockdown of a single Arf isoform did not induce any phenotype, whereas the simultaneous depletion of two isoforms induced specific intracellular trafficking defects, highlighting functional redundancies between the different Arfs (14). Class I (Arf1–3) and class II (Arf4–5) Arfs localize to the Golgi and the ERGIC (ER-Golgi Intermediate Compartment), respectively (12, 36). Depletion of different pairs of these Arf isoforms inhibit transport at the ER-Golgi interface (14), where COPI vesicles operate. In vitro, Arf1 and Arf5, and to a lesser extent Arf6, can recruit coatomer to Golgi-enriched membranes (23). All these data are in agreement with our finding that Arf4 and Arf5, in addition to Arf1, can serve COPI vesicle formation.
Interestingly, several Arf isoforms have also been related to the clathrin system, a vesicular coat with homologies to coatomer (23, 39, 40). The coat of clathrin-coated vesicles is composed of an outer layer of clathrin heavy and light chains, linked to the vesicular membrane by an inner layer of adaptor proteins. Various isoforms of hetero-tetrameric APs and monomeric GGAs allow the formation of different types of clathrin vesicles with different cargo compositions (41). The coat protein complex of COPI vesicles is composed of seven subunits, and can experimentally be resolved into two sub-complexes: a trimer and a tetramer (31). Structural determinations revealed partial homologies between the trimeric coatomer sub-complex and clathrin, and the tetrameric coatomer subcomplex and APs (42–46). In the clathrin system, Arf1 and Arf5 have been implicated in the recruitment of AP-1 (23), AP-3 (39, 47) and GGAs (40) to Golgi membranes.
The various APs interact differently with Arf isoforms. Cross-linking experiments revealed an interaction of AP-1 with Arf1, Arf3 and Arf6, whereas AP-3 interacted only with Arf1 and Arf6 (48). The σ subunit of AP-3 exists in two isoforms (σ3A and σ3B). Interestingly, whereas Arf5 stimulated, in vitro, recruitment to membranes of both isoforms, Arf1 showed a preference for the σ3A isoform of AP-3 (47). Such differential interactions would provide a way to discriminate the recruitment of these different adaptors to the Golgi membranes, prerequisite to the formation of distinct clathrin-coated vesicles. The tetrameric sub-complex of coatomer, a structural homolog of APs, also exists in different isoforms (49), and COPI vesicles have been suggested to be involved in different trafficking steps at the ER-Golgi interface as well as in intra-Golgi transport (2). It is thus tempting to speculate that different Arf isoforms would specifically recruit different coatomer isoforms, generating different subpopulations of COPI vesicles involved in different trafficking steps. Such models will clearly require additional testing.
Differences between Arf Isoforms
Arf1 has been characterized quite extensively in the context of COPI vesicle formation. Amino acids I46, I49, and Y167 mediate contact between Arf1 and coatomer (50), whereas Y35 is involved in dimerization of Arf1, which in turn is a prerequisite for the biogenesis of COPI vesicles (51). These amino acids are well conserved among the different Arf isoforms. The only exception is I49 that is not present in Arf6. However, although Arf6 was found to be able to recruit coatomer to Golgi membranes in vitro (albeit with only low efficiency) (23), its physiological functions are restricted to the plasma membrane (9–11). In agreement with this, we did not find Arf6 associated with COPI vesicles generated from cytosol, and recombinant Arf6 only supported very limited levels of COPI vesicle formation.
Surprisingly, Arf3, the isoform the most related to Arf1 (sharing >96% identity), did not associate with COPI vesicles generated from cytosol, whereas the more distantly related members Arf4 and Arf5 did. A recent study identified differences in the mode of recruitment of Arf1 and Arf3 to membranes (52). Arf1 is recruited to the cis-Golgi by the ArfGEF GBF1 (Golgi-specific Brefeldin A-resistance factor 1), whereas Arf3 associates, upon activation by the ArfGEF BIG (Brefeldin A-inhibited guanine nucleotide exchange factor), to the trans-Golgi network (TGN) (52). These findings suggest a correlation between localization and involvement into a budding process. Arf1 and class II Arfs localize to the cis-Golgi and ERGIC, respectively. In this area, transmembrane machinery proteins of the p24 family that are required for COPI vesicle biogenesis are available (53). By contrast, Arf3, localized to the TGN, would be locally separated from these machinery components and not able to participate in COPI vesicle formation.
The protein cores of the various Arf isoforms are well conserved, and all Arf proteins crystallized so far present very similar structures (54, 55, 56, PDB data base). Most of the differences between these small GTPases reside within the sequences of their N and C termini. In their GDP-loaded state, both termini form α helices close to each other, forming an interface with the membrane prior to Arf anchoring upon activation. It is thus likely that this interface mediates the interaction between Arf-GDP and its membrane receptor (and possibly aided by the phospholipids present in the microenvironment of each membrane). As a result, sequence differences in this region would allow the different Arf isoforms to bind different receptors, and thus to localize to different membranes. Transmembrane proteins p23, p24 (16, 57), and membrin (17) bind Arf1-GDP. Receptors for other Arf-GDP isoforms still remain to be identified.
By contrast, regulators and effectors bind to sites on Arf facing the cytosol, at the opposite side of the membrane interface. This is notably the case for Sec7 domains, the catalytic domain of ArfGEFs (58). As the amino acids of this area are very well conserved, it is unlikely that ArfGEFs can differentiate between substrates based on the sequences of the Arfs, though it is possible that other regions of (particularly the large) Arf GEFs may interact with other regions of Arfs and play a role in specificity determination. This again would argue in favor of a spatial discrimination. Arfs would thus be first recruited to a specific membrane and then activated by the locally present ArfGEF. This principle is illustrated by Arf3: a single mutation in the C terminus of Arf3 simultaneously triggers a re-localization of the protein and a change in the activating ArfGEF, although the interface between Arf3 and Sec7 is most likely identical within the wild-type form and the mutant (52).
In this study, we highlighted competition as an additional mechanism ensuring functional specificities of Arf isoforms. Although Arf3 could support COPI vesicle formation, it was not detected in vesicles when other Arf isoforms were present, suggesting that Arf3 either got excluded from the budding sites or was used but dissociated more rapidly than the other Arfs. Both Arf1 and Arf3 contain a MXXE motif, a signal involved in the recruitment to the early Golgi by the SNARE membrin (17). However, in the living cells, only Arf1 localizes to the cis-Golgi whereas Arf3 was found to be localized to the trans-Golgi (52). This differential localization could be the cellular reflection of the competition mechanism between Arf1 and Arf3 identified here.
Together, these data suggest that the different roles of the different Arf isoforms are due more to differences in membrane localization rather than intrinsic functionalities. However, another way to look at these results is that there is considerable redundancy built into the actions of Arfs1–5 at the Golgi. This diversity in Arfs emerged in evolution and therefore confers some advantage to the organism. Two possibilities to explain this include it may provide cells with protection against loss or mutation of any Arf isoforms or it allows for Arfs to play roles in development as the timing and tissue expression of the Arfs seems independently regulated (59–61).
In summary, we have obtained further insights into the mechanisms of COPI vesicle formation, and of functional specificities of the different Arf isoforms. Until now, COPI vesicle formation has exclusively been characterized with Arf1. Identification of Arf4 and Arf5 as additional components of the COPI machinery broaden our view of this coating system. This will be of special interest for our understanding of how COPI vesicles can be involved in different trafficking steps at the ER-Golgi interface. On the other hand, we have highlighted a competition mechanism between different Arf isoforms that contributes to a functional discrimination between the different members of Arf family. Further studies will be needed for our complete understanding of how different Arfs are targeted to different membranes and what are their detailed specific roles.
Supplementary Material
Acknowledgments
We thank Dr. Paul Melancon (University of Alberta, Alberta, Canada) and Dr. Michel Franco (Institut de Pharmacologie Moléculaire et Cellulaire, Valbonne-Sophia-Antipolis, France) for providing Arf5 cDNA and Arf6 cDNA, respectively, Imke Wüllenweber for technical assistance and Hartmut Michel for supporting this work, and Dan Cassel (Technion-Israel Institute of Technology, Haifa 32000, Israel) for critical discussions and reading the manuscript.
This work was supported by the German Research Council, SFB 638, A10, the ESFRI-INSTRUCT program of the European Union, and the Max Planck Gesellschaft.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ER
- endoplasmic reticulum
- TGN
- trans-Golgi network
- ERGIC
- ER-Golgi intermediate compartment
- GDP
- guanosine diphosphate
- GTP
- guanosine triphosphate
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- Arf
- ADP-ribosylation factor
- Arl
- Arf-like
- ArfGAP
- ArfGTPase-activating protein
- ArfGEF
- Arf guanine exchange factor
- GBF1
- Golgi-specific Brefeldin A resistance factor
- BIG
- Brefeldin A-inhibited guanine nucleotide exchange factor
- GGA
- Golgi localized γ-ear-containing Arf-binding protein
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1. Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. (2004) Annu. Rev. Cell Dev. Biol. 20, 87–123 [DOI] [PubMed] [Google Scholar]
- 2. Beck R., Rawet M., Wieland F. T., Cassel D. (2009) FEBS Lett. 583, 2701–2709 [DOI] [PubMed] [Google Scholar]
- 3. McMahon H. T., Mills I. G. (2004) Curr. Opin. Cell Biol. 16, 379–391 [DOI] [PubMed] [Google Scholar]
- 4. Matsuoka K., Orci L., Amherdt M., Bednarek S. Y., Hamamoto S., Schekman R., Yeung T. (1998) Cell 93, 263–275 [DOI] [PubMed] [Google Scholar]
- 5. Hill K., Li Y., Bennett M., McKay M., Zhu X., Shern J., Torre E., Lah J. J., Levey A. I., Kahn R. A. (2003) J. Biol. Chem. 278, 36032–36040 [DOI] [PubMed] [Google Scholar]
- 6. D'Souza-Schorey C., Chavrier P. (2006) Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 7. Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. (2006) J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 9. D'Souza-Schorey C., Li G., Colombo M. I., Stahl P. D. (1995) Science 267, 1175–1178 [DOI] [PubMed] [Google Scholar]
- 10. Peters P. J., Hsu V. W., Ooi C. E., Finazzi D., Teal S. B., Oorschot V., Donaldson J. G., Klausner R. D. (1995) J. Cell Biol. 128, 1003–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cavenagh M. M., Whitney J. A., Carroll K., Zhang C., Boman A. L., Rosenwald A. G., Mellman I., Kahn R. A. (1996) J. Biol. Chem. 271, 21767–21774 [DOI] [PubMed] [Google Scholar]
- 12. Chun J., Shapovalova Z., Dejgaard S. Y., Presley J. F., Melançon P. (2008) Mol. Biol. Cell 19, 3488–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lenhard J. M., Kahn R. A., Stahl P. D. (1992) J. Biol. Chem. 267, 13047–13052 [PubMed] [Google Scholar]
- 14. Volpicelli-Daley L. A., Li Y., Zhang C. J., Kahn R. A. (2005) Mol. Biol. Cell 16, 4495–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gommel D. U., Memon A. R., Heiss A., Lottspeich F., Pfannstiel J., Lechner J., Reinhard C., Helms J. B., Nickel W., Wieland F. T. (2001) EMBO J. 20, 6751–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Majoul I., Sohn K., Wieland F. T., Pepperkok R., Pizza M., Hillemann J., Söling H. D. (1998) J. Cell Biol. 143, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Honda A., Al-Awar O. S., Hay J. C., Donaldson J. G. (2005) J. Cell Biol. 168, 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao X., Lasell T. K., Melançon P. (2002) Mol. Biol. Cell 13, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Niu T. K., Pfeifer A. C., Lippincott-Schwartz J., Jackson C. L. (2005) Mol. Biol. Cell 16, 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reinhard C., Schweikert M., Wieland F. T., Nickel W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Randazzo P. A. (1997) Biochem. J. 324, 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu X., Zhang C., Xing G., Chen Q., He F. (2001) FEBS Lett. 490, 79–83 [DOI] [PubMed] [Google Scholar]
- 23. Liang J. O., Kornfeld S. (1997) J. Biol. Chem. 272, 4141–4148 [DOI] [PubMed] [Google Scholar]
- 24. Faulstich D., Auerbach S., Orci L., Ravazzola M., Wegchingel S., Lottspeich F., Stenbeck G., Harter C., Wieland F. T., Tschochner H. (1996) J. Cell Biol. 135, 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jenne N., Frey K., Brugger B., Wieland F. T. (2002) J. Biol. Chem. 277, 46504–46511 [DOI] [PubMed] [Google Scholar]
- 26. Lay D. L., Grosshans B., Heid H., Gorgas K., Just W. W. (2005) J. Biol. Chem. 280, 34489–34499 [DOI] [PubMed] [Google Scholar]
- 27. Franco M., Chardin P., Chabre M., Paris S. (1996) J. Biol. Chem. 271, 1573–1578 [DOI] [PubMed] [Google Scholar]
- 28. Claude A., Zhao B. P., Kuziemsky C. E., Dahan S., Berger S. J., Yan J. P., Armold A. D., Sullivan E. M., Melançon P. (1999) J. Cell Biol. 146, 71–84 [PMC free article] [PubMed] [Google Scholar]
- 29. Chavrier P., Franco M. (2001) Methods Enzymol. 329, 272–279 [DOI] [PubMed] [Google Scholar]
- 30. Hui N., Nakamura N., Slusarewicz P., Warren G. (1998) Cell Biology: A Laboratory Handbook, 2nd Ed., pp. 46–55, Academic Press [Google Scholar]
- 31. Pavel J., Harter C., Wieland F. T. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 33. Lenzen C., Cool R. H., Wittinghofer A. (1995) Methods Enzymol. 255, 95–109 [DOI] [PubMed] [Google Scholar]
- 34. Helms J. B., Palmer D. J., Rothman J. E. (1993) J. Cell Biol. 121, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bremser M., Nickel W., Schweikert M., Ravazzola M., Amherdt M., Hughes C. A., Söllner T. H., Rothman J. E., Wieland F. T. (1999) Cell 96, 495–506 [DOI] [PubMed] [Google Scholar]
- 36. Duijsings D., Lanke K. H., van Dooren S. H., van Dommelen M. M., Wetzels R., de Mattia F., Wessels E., van Kuppeveld F. J. (2009) Traffic 10, 316–323 [DOI] [PubMed] [Google Scholar]
- 37. Gaschet J., Hsu V. W. (1999) J. Biol. Chem. 274, 20040–20045 [DOI] [PubMed] [Google Scholar]
- 38. D'Souza-Schorey C., van Donselaar E., Hsu V. W., Yang C., Stahl P. D., Peters P. J. (1998) J. Cell Biol. 140, 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ooi C. E., Dell'Angelica E. C., Bonifacino J. S. (1998) J. Cell Biol. 142, 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takatsu H., Yoshino K., Toda K., Nakayama K. (2002) Biochem. J. 365, 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robinson M. S. (2004) Trends Cell Biol. 14, 167–174 [DOI] [PubMed] [Google Scholar]
- 42. Hoffman G. R., Rahl P. B., Collins R. N., Cerione R. A. (2003) Mol. Cell 12, 615–625 [DOI] [PubMed] [Google Scholar]
- 43. Watson P. J., Frigerio G., Collins B. M., Duden R., Owen D. J. (2004) Traffic 5, 79–88 [DOI] [PubMed] [Google Scholar]
- 44. Hsia K. C., Hoelz A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11271–11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu W., Lin J., Jin C., Xia B. (2009) J. Mol. Biol. 386, 903–912 [DOI] [PubMed] [Google Scholar]
- 46. Lee C., Goldberg J. (2010) Cell 142, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Drake M. T., Zhu Y., Kornfeld S. (2000) Mol. Biol. Cell 11, 3723–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Austin C., Boehm M., Tooze S. A. (2002) Biochemistry 41, 4669–4677 [DOI] [PubMed] [Google Scholar]
- 49. Wegmann D., Hess P., Baier C., Wieland F. T., Reinhard C. (2004) Mol. Biol. Cell 24, 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun Z., Anderl F., Fröhlich K., Zhao L., Hanke S., Brügger B., Wieland F., Béthune J. (2007) Traffic 8, 582–593 [DOI] [PubMed] [Google Scholar]
- 51. Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K., Sinning I., Hurt E., Brügger B., Béthune J., Wieland F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11731–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manolea F., Chun J., Chen D. W., Clarke I., Summerfeldt N., Dacks J. B., Melançon P. (2010) Mol. Biol. Cell 21, 1836–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stamnes M. A., Craighead M. W., Hoe M. H., Lampen N., Geromanos S., Tempst P., Rothman J. E. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8011–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Amor J. C., Harrison D. H., Kahn R. A., Ringe D. (1994) Nature 372, 704–708 [DOI] [PubMed] [Google Scholar]
- 55. Pasqualato S., Ménétrey J., Franco M., Cherfils J. (2001) EMBO Rep. 2, 234–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y., Kahn R. A., Prestegard J. H. (2010) Nat. Struct. Mol. Biol. 17, 876–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gommel D., Orci L., Emig E. M., Hannah M. J., Ravazzola M., Nickel W., Helms J. B., Wieland F. T., Sohn K. (1999) FEBS Lett. 447, 179–185 [DOI] [PubMed] [Google Scholar]
- 58. Renault L., Guibert B., Cherfils J. (2003) Nature 426, 525–530 [DOI] [PubMed] [Google Scholar]
- 59. Tsai S. C., Adamik R., Tsuchiya M., Chang P. P., Moss J., Vaughan M. (1991) J. Biol. Chem. 266, 8213–8219 [PubMed] [Google Scholar]
- 60. Tsuchiya M., Price S. R., Tsai S. C., Moss J., Vaughan M. (1991) J. Biol. Chem. 266, 2772–2777 [PubMed] [Google Scholar]
- 61. Tsuchiya M., Price S. R., Nightingale M. S., Moss J., Vaughan M. (1989) Biochemistry 28, 9668–9673 [DOI] [PubMed] [Google Scholar]
- 62. Vizcaíno J. A., Côté R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) Proteomics 9, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.