Abstract
Niemann-Pick C1-like 1 (NPC1L1) is a multi-transmembrane protein that mediates the absorption of dietary and biliary cholesterol through vesicular endocytosis. The subcellular localization of NPC1L1 is regulated by cholesterol. Cholesterol depletion induces the transport of NPC1L1 to plasma membrane (PM) from endocytic recycling compartment that requires MyoVb·Rab11a·Rab11-FIP2 triple complex, and cholesterol-replenishment renders the internalization of NPC1L1 together with cholesterol. Here, we find that GTP-bound Cdc42 interacts with NPC1L1. Cholesterol depletion regulates the activation of Cdc42 and enhances NPC1L1-Cdc42 interaction. Overexpression of constitutive GTP-bound Cdc42 mutant form or knockdown of Cdc42 inhibits the transport of NPC1L1 to the PM and disturbs the cholesterol-regulated binding of NPC1L1 to Rab11a, MyoVb, and actin. Knockdown of Cdc42 downstream effectors N-WASP or Arp3 also leads to the similar results. In liver-specific Cdc42 knock-out (Cdc42 LKO) mice, NPC1L1 fails to localize to bile canaliculi, and the biliary cholesterol cannot be efficiently reabsorbed. These results indicate that Cdc42 controls the cholesterol-regulated transport and localization of NPC1L1, and plays a role in cholesterol absorption.
Keywords: Cdc42, Cholesterol Regulation, Plasma Membrane, Trafficking, Vesicles
Introduction
Intestinal absorption is a major way for mammals to obtain the exogenous cholesterol, which is mainly mediated by a multi-transmembrane protein Niemann-Pick C1-like 1 (NPC1L1)2 (1, 2). Human NPC1L1 is highly expressed in small intestine and liver, and rodent NPC1L1 is selectively expressed in intestine (1, 2). The NPC1L1 protein localizes on the brush border membrane of small intestine and canalicular membrane of liver. It is critical for dietary cholesterol absorption and biliary cholesterol reabsorption (1, 3). In humans the nonsynonymous variants of NPC1L1 are associated with cholesterol absorption variations (4–7).
NPC1L1 forms a complex with lipid raft proteins Flotillin-1 and -2 (8). Cholesterol depletion induces them to transport to PM from ERC, which depends on microfilaments and MyoVb·Rab11a·Rab11-FIP2 complex (9, 10). On PM, NPC1L1 binds exogenous cholesterol and form cholesterol-enriched membrane domains (8, 11). The internalization of these NPC1L1·Flotillin·cholesterol microdomains ensures the efficient uptake of cholesterol (8).
Cdc42, a member of the Rho family of small GTPases, has been ascribed to numerous functions both in membrane trafficking and cell polarity (12). Cdc42 switches between GDP-bound inactive state and GTP-bound active state, a process regulated by guanine nucleotide exchange factors (GEFs), guanine nucleotide dissociation inhibitors (GDIs), and GTPase-activating proteins (GAPs). Once bound to GTP, Cdc42 interacts with N-WASP and releases its autoinhibitory conformation, which then binds and activates Arp2/3 complex to initiate branched actin polymerization (13, 14). N-WASP and Arp2/3 complex are also closely related to the movement of vesicles and actin polymerization, which is essential for vesicle budding and trafficking (15, 16).
Here we aim to understand the role of Cdc42 in cholesterol-regulated transport of NPC1L1. We find that Cdc42 associates with NPC1L1 and cholesterol depletion activates Cdc42. Overexpression and knockdown of Cdc42 blocks the transport of NPC1L1 from ERC to PM upon cholesterol depletion. Co-immunoprecipitation (co-IP) assays also demonstrate that Cdc42 together with N-WASP and Arp2/3 are required for cholesterol regulated binding of NPC1L1 to Rab11a, MyoVb, and actin. In addition, in vivo analyses in Cdc42 LKO mice indicate that depletion of Cdc42 disrupts canalicular localization of NPC1L1 and decreases the reabsorption of biliary cholesterol.
EXPERIMENTAL PROCEDURES
Materials
Mouse monoclonal anti-Cdc42 antibodies were from Millipore and Cytoskeleton, rabbit polyclonal anti-Cdc42 antibody was from Santa Cruz Biotechnology; mouse monoclonal anti-Rac1 antibody was from Millipore, mouse monoclonal anti-Arp3 antibody was from BD Biosciences, and rabbit anti-Mrp2 antibody was described previously (17). Mouse monoclonal anti-T7, anti-Myc IgG-9E10, rabbit polyclonal anti-Myc, anti-EGFP, and secondary antibodies were described previously (18). Cholesterol, methyl-β-cyclodextrin (CDX), and other materials were described previously (9). Cdc42flox/flox mice were as described (19).
Cell Culture
CRL1601 (McArdle RH7777 rat hepatoma cell) and CRL1601-NPC1L1-EGFP cells that express NPC1L1-EGFP were grown in monolayer at 37 °C in 5% CO2 (9). Cells were maintained in medium A (Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 10% fetal bovine serum (FBS). For CRL1601-NPC1L1-EGFP cells, 200 μg/ml G418 was supplemented. Cholesterol-depleting medium is medium A supplemented with 5% lipoprotein-deficient serum (LPDS), 10 μm compactin, 50 μm mevalonate, and 1.5% (w/v) CDX. Transfection of cells was performed with Fugene HD (Roche) according to the manufacturer's manual.
Plasmids
NPC1L1-EGFP, NPC1L1-T7, EGFP-Rab11a, and EGFP-MyoVb expression plasmids were described previously (10). Full-length Cdc42 coding sequences were amplified from human cDNA and inserted in-frame into modified vectors with N-terminal RFP or 5×Myc tags. Cdc42(G12V) and Cdc42(T17N) mutants were generated by QuickChange mutagenesis.
Co-immunoprecipitation
24 h after transfection, cells were treated as indicated, and were immediately washed twice with ice-cold PBS before harvested in ice-cold lysis buffer (PBS containing 0.5% digitonin, 5 mm EGTA, 5 mm EDTA, protease, and phosphatase inhibitors). Whole cell lysates were incubated with anti-EGFP or anti-T7 beads and rotated at 4 °C for 2 h. The beads were washed five times with lysis buffer, resuspended in an appropriate amount of SDS-PAGE loading buffer, and incubated at 37 °C for 30 min before loading onto an SDS-PAGE gel.
GST Pull-down/Cdc42 Activity Assay
GST-PAK1-PBD (amino acids 69–150 of human PAK1) was constructed and expressed as described (20). 107 CRL1601 cells for each sample were treated with 1% CDX for indicated times. Cells were washed twice with ice-cold PBS and then lysed in Mg2+ lysis buffer (MLB) as described by the Millipore user manual. The supernatant was incubated with 20 μg of GST-PBD-agarose at 4 °C for 2 h. The beads were washed three times by MLB and then resuspended in appropriate amounts of SDS-PAGE-loading buffer. Proteins were separated via 15% SDS-PAGE gel and analyzed via Western blotting.
RNA Interference
Oligo duplexes of siRNA were synthesized by Genepharma (Shanghai, China). The sequences of the siRNAs were as follows: AGACTCCTTTCTTGCTTGT (Cdc42-1), TGACAGACTACGACCGTTA (Cdc42-2), CGACAAAGGAAATCTGAAA (N-WASP-1), GTTCCGAAAAGCAGTTACA (N-WASP-2), GGTTTATGGAGCAAGTGAT (Arp3-1), and GAGACCGAGAAGTAGGAAT (Arp3-2). Transfection of siRNA was carried out as previously described (9).
Adenovirus-mediated Gene Expression
The AdEasyTM Adenovirial vector system was utilized to construct the adenovirus expression vectors (21). Adenovirus expressing Cre recombinase was a kind gift from Dr. Hongbin Ji (Shanghai Institute of Biochemistry and Cell Biology). For EGFP and NPC1L1-EGFP expression, the coding sequences were subcloned into pShuttle-CMV vector and recombined with pAdEasy vector. The adenoviruses were packaged in HEK293A cells and purified with CsCl ultracentrifugation. The viruses were titered and administrated via tail vein injection (5 × 108 pfu viruses per mouse for each gene expression). 4 days later, mouse tissues and bile were collected following 12-h overnight fasting.
Immunofluorescence
CRL1601 cells were transfected as described. Cells on coverslips were fixed in 4% formaldehyde (PFA) for 20 min, then washed with PBS, and mounted. Mouse livers infected by adenovirus were perfused with PBS followed by 4% PFA, and further fixed in 4% PFA at 4 °C for 4 h, dehydrated in 20% sucrose in PBS overnight, and then were embedded in OCT (Leica Microsystem) and frozen at −80 °C before preparation of 8-μm-thick frozen section. Sections were permeabilized and blocked with 0.2% Triton X-100 in PBS containing 10% FBS for 1 h at room temperature. They were incubated with anti-Mrp2 antibody (1:200 dilution) overnight at 4 °C and then Alexa Fluor 555-conjugated second antibody (1:500 dilution) at room temperature for 1 h. Slides were mounted with FluorSave (Merk), and images were acquired on a Leica TCS SP5 imaging microscopy.
Measurements of Hepatic and Biliary Total Cholesterol
Male mice 8–10 weeks of age were fed chow diet. Gallbladder bile was collected after mice were fasted overnight (12 h). For hepatic cholesterol extraction, 50 mg of liver tissue were snap-frozen in liquid nitrogen and homogenized in 1 ml of chloroform/methanol (2:1) mixed with 200 μl of MilliQ water and centrifuged at 2000 × g for 10 min; then 20 μl of organic phase were freeze-dried and applied to measure liver total cholesterol. To determine biliary lipid concentrations, 5 μl of bile was solubilized in 45 μl of MilliQ water and extracted with 200 μl of chloroform/methanol (2:1). The organic phase and aqueous phase were both freeze-dried and dissolved in ethanol and MilliQ water, respectively. The organic phase was analyzed for total cholesterol by enzymatic kits (Wako Cholesterol E).
RESULTS
NPC1L1 Preferentially Interacts with GTP-bound Form of Cdc42
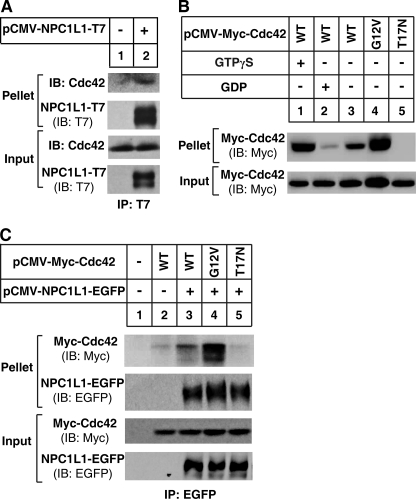
To identify proteins involved in NPC1L1-mediated cholesterol absorption, we performed a large-scale co-IP followed by SDS-PAGE and tandem mass spectrometry using the NPC1L1-EGFP cell line (9). Among the NPC1L1-associated proteins, Cdc42, a small GTPase of the Rho family, was identified in one specific band, indicating that Cdc42 plays a role in NPC1L1 trafficking. The interaction between NPC1L1 and Cdc42 was confirmed by co-IP analysis followed by Western blotting from CRL1601 cells transiently transfected with control vector or plasmid encoding NPC1L1-T7 (Fig. 1A).
FIGURE 1.
Cdc42 interacts with NPC1L1. A, interaction of NPC1L1 with endogenous Cdc42. The plasmids encoding NPC1L1-T7 or vector pcDNA3 were transfected into CRL1601 cells. 24 h later, the cells were harvested in IP buffer and immunoprecipitation was performed by pulling down T7-tagged NPC1L1 with anti-T7 agarose. B, activity of wild type Cdc42, Cdc42-G12V, and Cdc42-T17N. The plasmids encoding Myc-tagged wild type or mutants of Cdc42 were transfected into CRL1601 cells. 24 h later, the cells were harvested in Mg2+ lysis buffer, and GST pull-down was performed with GST-PAK1-PBD-agarose. C, preferential interaction of NPC1L1 with GTP-bound Cdc42. The plasmids encoding NPC1L1-EGFP and Myc-tagged wild type or mutants of Cdc42 were co-transfected into CRL1601 cells. 24 h later, the cells were harvested in IP buffer, and immunoprecipitation was performed by pulling down EGFP-tagged NPC1L1 with anti-EGFP-agarose.
Cdc42 switches between GDP-bound inactive state and GTP-bound active state in response to a variety of stimuli. To further investigate whether the interaction of NPC1L1 and Cdc42 depends on the activity of Cdc42, we constructed the constitutively active (GTP-locked form, G12V) and inactive (GDP-locked form, T17N) Cdc42 mutants, and performed GST pull-down assays with GST-PAK1-PBD-agarose (20, 22–24). As shown in Fig. 1B, GST-PAK1-PBD could effectively interact with both GTPγS-bound Cdc42 and Cdc42(G12V), while there is no obvious interaction with GDP-bound Cdc42 and Cdc42(T17N). Furthermore, NPC1L1 co-precipitated with the constitutively active mutant Cdc42(G12V) but not Cdc42(T17N), and only weakly bound to wild type Cdc42 (Fig. 1C). These data indicate that NPC1L1 preferentially interacts with GTP-bound form of Cdc42.
Activity of Cdc42 and Its Interaction with NPC1L1 Are Both Regulated by Cholesterol Depletion
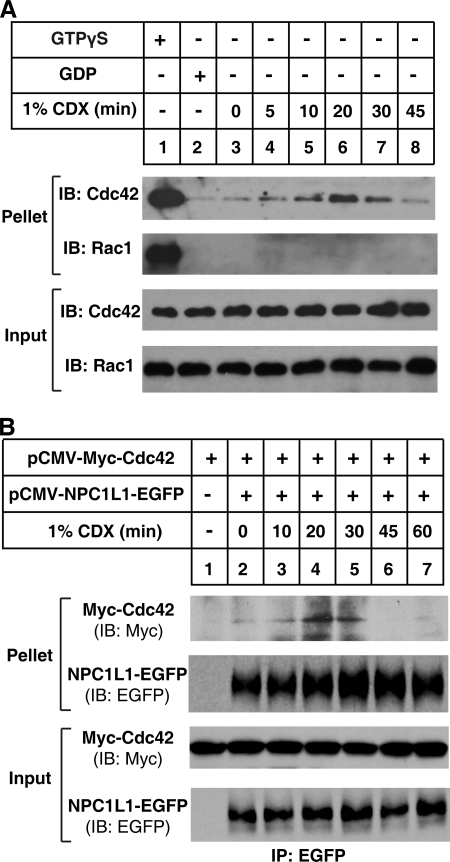
Previous studies revealed that cholesterol depletion induced the transport of NPC1L1 toward PM (9, 10). And Cdc42 was reported to be activated by cholesterol depletion with CDX (25, 26). To investigate the activation of Cdc42 during NPC1L1-translocation to PM upon cholesterol depletion, we performed GST-PAK1-PBD pull-down assays when the cholesterol was depleted. As shown in Fig. 2A, Cdc42 was gradually activated while the cholesterol was depleted by CDX. After depletion of cholesterol for 20 min, GTP-bound form of Cdc42 reached the peak level. Then, Cdc42 started to lose its activity even though cholesterol depletion is continuing. During the cholesterol depletion process, Rac1, another small GTPase with homology to Cdc42, was not activated, indicating that the activation of Cdc42 induced by cholesterol depletion was specific.
FIGURE 2.
Cdc42 activity and its interaction with NPC1L1 are both regulated by cholesterol depletion. A, activation of endogenous Cdc42 by cholesterol depletion. CRL1601 cells were depleted of cholesterol with 1% CDX for various time durations. Then the cells were harvested in Mg2+ lysis buffer, and GST pull-down was performed with GST-PAK1-PBD-agarose. B, regulation of the interaction between NPC1L1 and Cdc42 by cholesterol depletion. The plasmids encoding NPC1L1-EGFP and wild type Myc-Cdc42 were co-transfected into CRL1601 cells. 24 h later, the cells were depleted of cholesterol with 1% CDX for various time durations. Then cells were harvested in IP buffer and immunoprecipitation was performed by pulling down EGFP-tagged NPC1L1 with anti-EGFP-agarose.
Subsequent co-IP of Myc-tagged wild type Cdc42 and NPC1L1 during cholesterol depletion showed that at steady state (time 0 min) the interaction of Cdc42 with NPC1L1 was very weak (Fig. 2B, lane 1), but was strengthened to the peak level after cholesterol depletion for 20 min (Fig. 2B, lane 3). Similar with the dynamics of Cdc42 activity, the interaction between Cdc42 and NPC1L1 was gradually reduced by longer depletion of cholesterol (Fig. 2B). These results demonstrate that the activity of Cdc42 and its interaction with NPC1L1 are both regulated by cholesterol depletion at a similar time scale, suggesting that Cdc42 may play a role in NPC1L1 translocation.
Both Overexpression and Down-regulation of Cdc42 Inhibit the Transport of NPC1L1 to PM
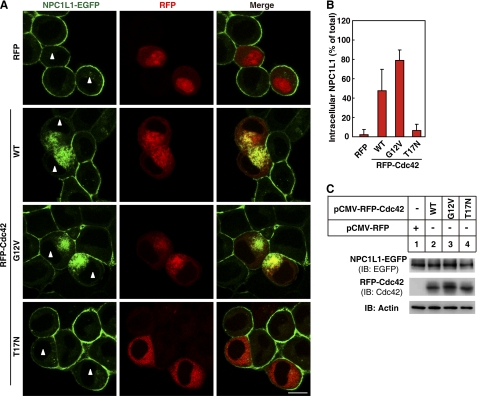
To directly test whether Cdc42 is involved in the transport of NPC1L1 to PM, we expressed RFP-fused wild type (WT), GTP-locked form (G12V), and GDP-locked form (T17N) of Cdc42, respectively in CRL1601 cells stably expressing NPC1L1-EGFP (CRL1601-NPC1L1-EGFP). At steady state, overexpression of Cdc42(WT), Cdc42(G12V), or Cdc42(T17N) did not affect the perinuclear localization of NPC1L1 (results not shown). When cholesterol was depleted by CDX, NPC1L1 was transported to PM in control and Cdc42(T17N)-expressing cells (Fig. 3A, white triangle-labeled cells in the first and fourth panels). In contrast, the transport of NPC1L1 to PM was inhibited in Cdc42(WT)- and Cdc42(G12V)-expressing cells (Fig. 3A, white triangle-labeled cells in the second and third panels). Quantification of intracellular localized NPC1L1 illustrated that after cholesterol depletion, less than 10% of the total NPC1L1 resided within the control or Cdc42(T17N)-expressing cells (Fig. 3B). However, more than 50% of the total NPC1L1 was restrained within the cells expressing Cdc42(WT) or Cdc42(G12V) (Fig. 3B). Meanwhile, the indistinguishable expression level of Cdc42 variants examined by immunoblotting indicated that the different effects caused by these Cdc42 variants were not ascribed to the differences of expression levels (Fig. 3C). Therefore, we conclude that excessive GTP-bound Cdc42 inhibits the transport of NPC1L1 to PM.
FIGURE 3.
Overexpression of Cdc42 inhibits the transport of NPC1L1 to PM. A, plasmids encoding RFP (for control of Cdc42 variants) or RFP-fused wild type or mutants of Cdc42 were transfected into CRL1601 cells stably expressing NPC1L1-EGFP. 24 h later, the cells were depleted of cholesterol by 1.5% CDX for 60 min. Scale bar represents 10 μm. B, quantification of ratio of the intracellular NPC1L1-EGFP shown in A. EGFP fluorescence intensity was measured by Image-Pro Plus 5.02 as described under “Experimental Procedures.” Error bars represent standard deviations. C, expression level of NPC1L1-EGFP, RFP-fused Cdc42 variants. Transfection and cholesterol depletion were performed as in A, then cells were harvested and subjected to Western blot.
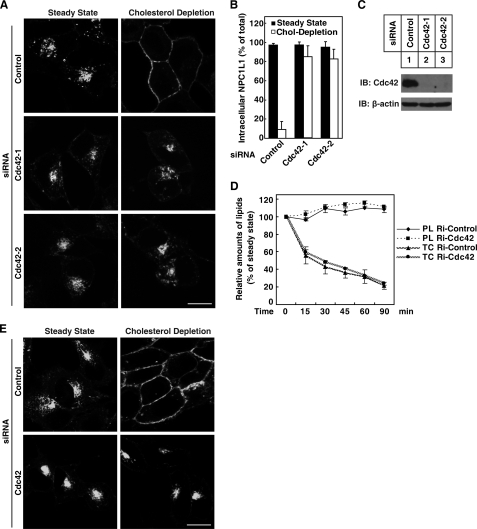
Because overexpression of GTP-bound Cdc42 showed strong inhibition of NPC1L1 translocation, we wondered whether knockdown of endogenous Cdc42 would also affect the transport of NPC1L1. To answer this question, RNAi of endogenous Cdc42 in CRL1601-NPC1L1-EGFP cells was performed. Western blot analysis showed that Cdc42 protein levels were substantially reduced by both siRNAs (Fig. 4C). Knockdown of Cdc42 had no effect on NPC1L1 localization at steady state (Fig. 4A), whereas, after cholesterol depletion for 60 min following Cdc42 RNAi, around 80% of total NPC1L1 was retained in perinuclear regions (Fig. 4, A and B). So, lower Cdc42 level inhibits NPC1L1 translocation to PM.
FIGURE 4.
Knockdown of Cdc42 inhibits the translocation of NPC1L1 to PM. A, CRL1601 cells stably expressing NPC1L1-EGFP were transfected with control siRNAs or siRNAs against rat Cdc42. 48 h later, siRNAs were transfected again. And 24 h later, the cells were depleted of cholesterol by 1.5% CDX for 60 min. B, quantification of ratio of the intracellular NPC1L1-EGFP shown in A. Error bars represent standard deviations. C, knockdown efficiencies of siRNAs against Cdc42. siRNAs were transfected two times as in A. Then cells were harvested and subjected to Western blot. D, knockdown of Cdc42 does not interfere with cholesterol depletion by CDX. siRNAs were transfected two times as in A. The siRNA Cdc42–1 was applied to knockdown Cdc42 in this experiment. Then cells were treated with 1% CDX for various durations before harvested and lysed in chloroform/methanol (2:1) to extract lipids as described under “Experimental Procedures.” Total cholesterol (TC) and phospholipids (PL) were measured enzymatically. E, knockdown of Cdc42 inhibits the transport of NPC1L1 to PM upon cholesterol depletion by LPDS, lovastatin, and 25-hydroxycholesterol. siRNA were transfected as in D. Cells were then incubated in DMEM medium containing 10% FBS (steady state) or DMEM medium containing 5% LPDS, 1 μm Lovastatin and 1 μg/ml 25-hydroxycholesterol (cholesterol depletion) for 16 h. Scale bar represents 10 μm.
One possibility was that the ability of CDX to deplete cholesterol was diminished in Cdc42-deficient cells, thus NPC1L1 remained in ERC as cholesterol depletion was not efficient. To rule out this possibility, cholesterol concentration was measured in both control and Cdc42 knockdown cells treated by CDX for different time durations. Phospholipids were also measured as a control. CDX decreased cholesterol level in control and Cdc42 RNAi cells, while phospholipids remained constant (Fig. 4D). This indicates that cholesterol depletion by CDX is specific. The indistinguishable curves of cholesterol level in both control and Cdc42 RNAi cells indicate that Cdc42 deficiency does not interfere with cholesterol depletion by CDX.
Although CDX is widely used to rapidly deplete cholesterol in a variety of cells (27–29), another cholesterol depletion approach was also applied to confirm the results. Cells were incubated in medium containing LPDS plus 1 μm Lovastatin and 1 μg/ml 25-hydroxycholesterol for 16 h to deplete cellular cholesterol (27). Substitution of LPDS with FBS prevents the cells from obtaining cholesterol from lipoproteins. Lovastatin and 25-hydroxycholesterol inhibit cholesterol biosynthesis pathway (27, 30). NPC1L1 in control cells was transported to PM after cholesterol depletion, while NPC1L1 in Cdc42 RNAi cells still remained in the perinuclear region (Fig. 4E), consistent with the results under CDX treatment (Fig. 4A). Taken together, Cdc42 is required for the transport of NPC1L1 to PM upon cholesterol depletion.
Cdc42 Downstream Effectors N-WASP and Arp2/3 Complex Are Required for Transport of NPC1L1 to PM
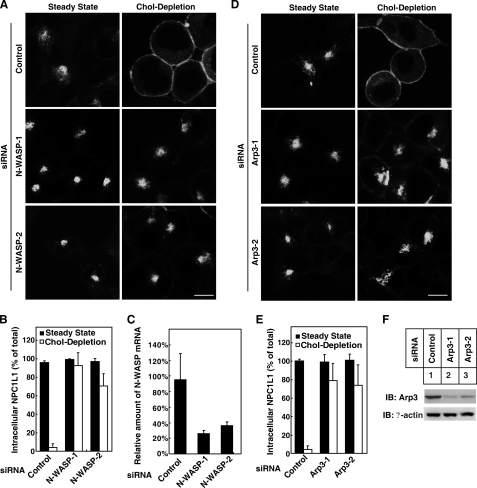
Previous studies have shown that the transport of NPC1L1 depends on microfilaments (9, 10). As Cdc42 and its downstream effectors N-WASP and Arp2/3 complex regulate actin polymerization, which is involved in many intracellular membrane traffic events (14–16), we next investigated whether they played a role in the transport of NPC1L1 to the PM using siRNAs-mediated silencing. Real-time PCR analysis showed that N-WASP expression was reduced by ∼72% and ∼62%, respectively, by siRNAs (Fig. 5C). After cholesterol depletion following RNA interference, more than 65% of the NPC1L1 was restrained in intracellular perinuclear region in N-WASP knockdown cells, while almost all NPC1L1 were transported to the PM in the control cells (Fig. 5, A and B). Similarly, in Arp3 knockdown cells, NPC1L1 localized in perinuclear region and the cholesterol depletion-induced transport of NPC1L1 to PM was inhibited (Fig. 5, D–F). These results indicate that Cdc42 downstream effectors N-WASP and Arp2/3 play important role in the transport of NPC1L1 to PM.
FIGURE 5.
Cdc42 downstream effectors N-WASP and Arp2/3 complex are required for the transport of NPC1L1 to PM. A, knockdown of N-WASP inhibits the transport of NPC1L1 to PM. CRL1601 cells stably expressing NPC1L1-EGFP were transfected with control siRNAs or siRNAs against rat N-WASP. siRNAs were transfected 2 times as in Fig. 4A, then the cells were depleted of cholesterol by 1.5% CDX for 60 min. B, quantification of ratio of the intracellular NPC1L1-EGFP shown in A. Error bars represent standard deviations. C, knockdown efficiencies of siRNAs against N-WASP. siRNAs were transfected the same with A, cells were harvested in Tri reagent and total RNA was extracted to perform RT Real-time RCR. The amounts of mRNA of N-WASP in siRNA-transfected cells were normalized with that of cyclophilin. Values with indicated siRNAs represent the normalized amount of mRNA relative to that of cells transfected with control siRNA. D, knockdown of Arp3 inhibits the transport of NPC1L1 to PM. Cells were treated as in A except that the siRNAs were against Arp3. E, quantification of ratio of the intracellular NPC1L1-EGFP shown in D. Error bars represent standard deviations. F, knockdown efficiencies of siRNAs against Arp3. siRNAs were transfected as in D, and cells were harvested and subjected to Western blot. Scale bar represents 10 μm.
Cdc42, N-WASP, and Arp2/3 Complex Is Involved in the Cholesterol-regulated Binding of NPC1L1 to Rab11a, MyoVb, and Actin
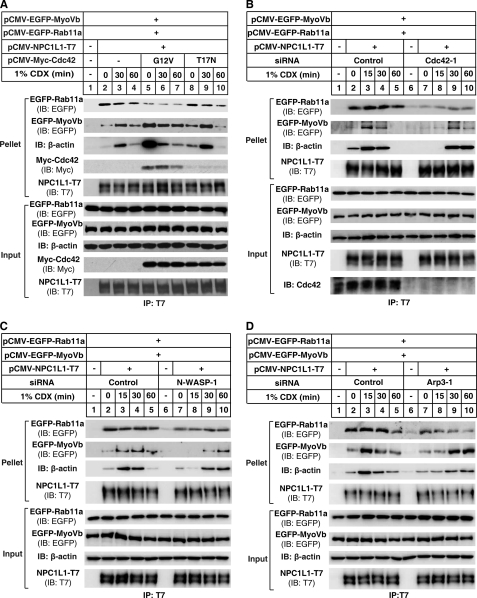
Our studies have demonstrated that MyoVb·Rab11a·Rab11-FIP2 was required for the translocation of NPC1L1 toward the PM, and Rab11a and MyoVb dissociated sequentially from NPC1L1 in this process (10). Because Cdc42, N-WASP and Arp2/3 were also important for the transport of NPC1L1 from ERC to PM, we then tested whether they affected the interaction between NPC1L1 and Rab11a·MyoVb complex upon cholesterol depletion. In control cells (without exogenous expression of Cdc42), NPC1L1 associated with Rab11a at steady state (Fig. 6A, lane 2), and the interaction gradually decreased (Fig. 6A, lanes 2–4) during cholesterol depletion. On the other hand, the binding of NPC1L1 to MyoVb and actin was weak at steady state, then increased to the peak level after cholesterol depletion for 30 min, but was drastically reduced at 60-min time point (Fig. 6A, lanes 2–4), which is similar with the binding pattern of NPC1L1 and wild type Cdc42 (Fig. 2B). When Cdc42(G12V) (GTP-locked form) was co-expressed, the interaction between NPC1L1 and Rab11a was reduced to a low level in the whole process. Meanwhile, NPC1L1 bound to high level of MyoVb and actin at steady state and gradually decreased after cholesterol depletion. In contrast, overexpression of Cdc42(T17N) (GDP-locked form) did not affect the association pattern of NPC1L1 upon cholesterol depletion as compared with control cells (Fig. 6A, lanes 8–10). These results indicated that the binding of GTP-bound Cdc42 to NPC1L1 complex promoted the association of MyoVb and actin with NPC1L1 complex while dissociated Rab11a.
FIGURE 6.
Cdc42 and its downstream effectors N-WASP and Arp2/3 complex are required for the cholesterol depletion regulated binding of NPC1L1 to Rab11a, MyoVb and actin. A, overexpression of Cdc42(G12V) but not Cdc42(T17N) reduced the binding of Rab11a to NPC1L1 and changed the binding pattern of MyoVb and actin with NPC1L1. CRL1601 cells were transfected with plasmids encoding NPC1L1-T7, EGFP-Rab11a, EGFP-MyoVb, and Myc-Cdc42(G12V) or Myc-Cdc42(T17N). 24 h after transfection, the cells were depleted of cholesterol with 1% CDX for various durations. Then cells were harvested in IP buffer, and immunoprecipitation was performed by pulling down T7-tagged NPC1L1 with anti-T7-agarose. B—D, knocking down the expression of Cdc42 (B), N-WASP (C), or Arp3 (D) reduced the binding of Rab11a to NPC1L1 and changed the binding pattern of MyoVb and actin with NPC1L1. CRL1601 cells were transfected with control siRNAs or siRNAs against rat Cdc42 (B), N-WASP (C), or Arp3 (D). 24 h later, plasmids encoding NPC1L1-T7, EGFP-Rab11a, and EGFP-MyoVb were co-transfected into the cells. 24 h later, cells were transfected with siRNAs for a second time. 24 h later, the cells were depleted of cholesterol with 1% CDX for various durations. Then cells were harvested in IP buffer, and immunoprecipitation was performed by pulling down T7-tagged NPC1L1 with anti-T7 agarose.
Then the binding of NPC1L1 to Rab11a, MyoVb and actin was examined in Cdc42 knockdown cells after cholesterol depletion. Disruption of Cdc42 dramatically reduced the interaction between NPC1L1 and Rab11a, and delayed the association with actin and MyoVb (Fig. 6B, compare lanes 7–10 with lanes 2–5), as compared with the control siRNA transfected cells. These results showed that the decreased Cdc42 protein resulted in delayed recruitment of MyoVb and actin to NPC1L1, while excessive GTP-bound Cdc42(G12V) accelerated this process, indicating the role of Cdc42 is to promote association of actin and MyoVb with NPC1L1.
Furthermore, similar results were also observed in cells with N-WASP or Arp3 knockdown (Fig. 6C-D, respectively), indicating that Cdc42·N-WASP·Arp2/3 complex-mediated actin polymerization may be required for cholesterol-regulated binding of NPC1L1 to Rab11a and MyoVb.
NPC1L1 Failed to Localize to Bile Canalicular Membrane in Cdc42 LKO Mouse Liver, and Biliary Cholesterol Could Not Be Reabsorbed
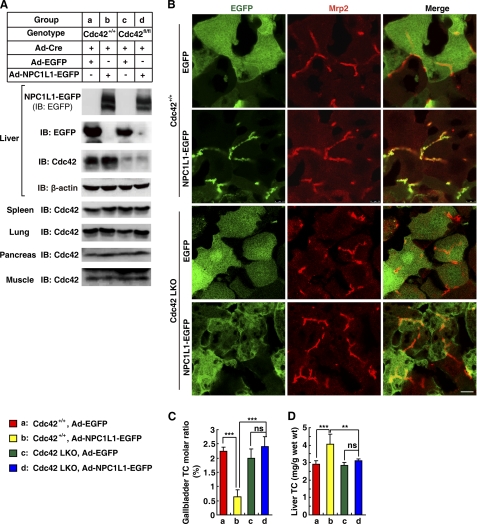
NPC1L1 is mainly expressed in the small intestine in mice, whereas it's highly expressed in both liver and small intestine in humans (1, 2). Previous studies revealed that exogenously expressed NPC1L1 in mouse liver localizes to bile canaliculi and facilitates biliary cholesterol reabsorption (3, 8). To investigate the physiological role of Cdc42 on the localization and function of NPC1L1 in vivo, liver-specific Cdc42 knock-out mice were generated using adenoviral delivery of Cre recombinase (Ad-Cre). We performed tail vein injection of Ad-Cre along with Ad-NPC1L1-EGFP or Ad-EGFP (as a control) into Cdc42+/+ and Cdc42flox/flox mice. In livers of Cdc42flox/flox mice but not Cdc42+/+ mice, 90% deletion of Cdc42 was detected after Cre recombinase expression, and there was no visible loss of Cdc42 in spleen, lung, pancreas, and muscle measured by immunoblotting (Fig. 7A). The obtained hepatic Cdc42-deleted mice were referred to as Cdc42 LKO mice henceforth. Next, we examined the localization of NPC1L1 in livers of Cdc42+/+ and Cdc42 LKO mice. Immunofluorescence studies showed that NPC1L1 localized to the canalicular membrane in Cdc42+/+ mice as it co-localized with the multidrug resistance protein 2 (Mrp2), a canalicular transporter for bile salts (31). In sharp contrast, in liver samples of Cdc42 LKO mice NPC1L1 lost its apically polarized distribution and was dispersed in the whole cell (Fig. 7B). These results demonstrated that Cdc42 was required for the apical membrane localization of hepatic NPC1L1, which is consistent with our data in cultured cells (Fig. 4A).
FIGURE 7.
NPC1L1 failed to localize to canalicular membrane and reabsorb biliary cholesterol in Cdc42 LKO mice. 8-week-old wild type C57B/L6 mice and Cdc42flox/flox (abbreviated as Cdc42fl/fl in a) mice fed on chow diet were tail vein injected with adenovirus expressing Cre recombinase (abbreviated as Ad-cre) and adenovirus expressing NPC1L1-EGFP (Ad-NPC1L1-EGFP) or EGFP (Ad-EGFP) (109 pfu viruses per mouse, four mice for each group). 4 days later, the mice were fasted overnight before sacrifice. A, knock-out efficiency and specificity of liver Cdc42 in Cdc42flox/flox mice. The liver Cdc42 level from each mouse was examined (data not shown here). Liver, pancreas, spleen, kidney, and muscle samples from each group were homogenized and mixed together before subjected to SDS-PAGE. Cdc42 were recognized by anti-Cdc42 polyclonal antibody (Santa Cruz Biotechnology). Liver-specific Cdc42 deleted Cdc42flox/flox mice by Ad-Cre were designated as Cdc42 LKO mice henceforth. B, 8-μm-thick frozen section sample was subjected to immunostaining with rabbit anti-Mrp2 antiserum and Alexa Fluor 555-conjugated goat anti-rabbit secondary antibody. NPC1L1-EGFP and EGFP were detected by its intrinsic fluorescence excited at 488 nm. Scale bar represents 10 μm. C and D, total cholesterol (TC) in gallbladder and liver were measured. (***: p < 0.001; **: 0.001<p < 0.01; *: 0.01<p < 0.05; ns: p > 0.05 (two-way ANOVA). Error bars represent standard deviations.
The concentrations of cholesterol in bile and liver were subsequently analyzed to investigate whether the function of hepatic NPC1L1 was dependent on Cdc42. Expression of NPC1L1 in Cdc42+/+ mouse liver significantly reduced the molar ratio of biliary cholesterol, but increased the liver cholesterol level (Fig. 7, C and D, compare columns a with b). This indicates that the hepatic NPC1L1 mediates biliary cholesterol reabsorption into liver. However, these effects were blunted in Cdc42 LKO mice (Fig. 7, C and D, compare columns c and d). In conclusion, the function of NPC1L1 in reabsorbing biliary cholesterol was impaired when hepatic Cdc42 was deleted.
DISCUSSION
In the current study, we elucidate that Cdc42 plays a key role in the transport of NPC1L1 toward PM upon cholesterol depletion, and provide a potential link between Cdc42 and its effectors, microfilament system and translocation of NPC1L1.
We find that Cdc42 is activated by cholesterol depletion in a time-dependent manner, which is consistent with previous studies that CDX-induced cholesterol depletion from lipid raft can activate Cdc42 (25, 26). Because the activation of Cdc42 is generally mediated by specific GEFs (32), cholesterol depletion may change the structural components of membranes and cause spatial rearrangement of membrane associated proteins, which may lead to the contact of GEFs with Cdc42 and the following activation of Cdc42 (33, 34). This speculation is also supported by Jaksits et al. report that disruption of lipid rafts, which bear GEF-like activity, can induce Cdc42 activation (25).
Our studies show that Cdc42 activation is required for the transport of NPC1L1. However, overexpression of GTP-locked Cdc42 can retain NPC1L1 in ERC when cholesterol is depleted (Fig. 3). This implies that the cycling of Cdc42 may be vital for NPC1L1 movement. At steady state, Cdc42 is mainly maintained in GDP-bound form and almost no GTP-bound Cdc42 exists (Fig. 2A). When cholesterol is depleted, NPC1L1 associates with GTP-bound Cdc42, but not GDP-bound form (Figs. 1C and 6A). Overexpression of GTP-locked Cdc42(G12V) leads to enhanced binding between NPC1L1 and MyoVb and actin, and blocks the transport of NPC1L1 to PM. However, this is not the case for the over-expression of GDP-locked Cdc42(T17N), as it cannot bind to NPC1L1 and show no impact on the translocation of NPC1L1. GTP-locked Cdc42(G12V) may disrupt the balance of Cdc42 cycling. It may constitutively bind and activate N-WASP and Arp2/3 complex to initiate actin polymerization around NPC1L1 vesicles even at the steady state, which makes the signaling not properly turned off as in the control cells (Fig. 6A).
Previous studies have demonstrated that the transport of NPC1L1 to PM depends on MyoVb·Rab11a·Rab11-FIP2 triple complex (10). The co-IP experiments have shown that Rab11a binds to NPC1L1 at the steady state (Fig. 6). When the cellular cholesterol level drops, NPC1L1 translocates to PM, during which it dissociates from Rab11a and binds MyoVb and microfilaments (Fig. 4). After arriving at the PM, MyoVb and actin are separated from NPC1L1. The current study demonstrates that Cdc42 plays an essential role in this process. Cholesterol depletion activates Cdc42 and the binding of GTP-bound Cdc42 to NPC1L1 promotes Rab11a dissociation and MyoVb and actin association. Meanwhile, GTP-bound Cdc42 may activate N-WASP·Arp2/3 complex to regulate the actin polymerization near NPC1L1 vesicles, which may drive vesicle budding, scission, and movement.
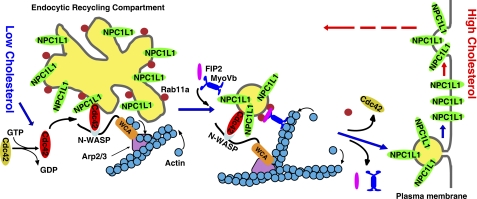
In summary, we propose a Cdc42 working model illustrating its role in cholesterol-induced NPC1L1 translocation to the PM (Fig. 8). At the steady state, Rab11a binds to NPC1L1 and bridges the interaction between NPC1L1 and MyoVb. When the cellular cholesterol level drops, Rab11a dissociated from NPC1L1·MyoVb complex and Cdc42 is activated to bind this complex. The GTP-bound Cdc42 further activates its downstream effectors N-WASP and Arp2/3, which promotes actin assembly. Then the vesicle budding is initiated, which further allows export of NPC1L1·MyoVb complex out of ERC. With the dynamics of Cdc42, NPC1L1 vesicles are transported to PM.
FIGURE 8.
Schematic model of Cdc42 regulated transport of NPC1L1 toward PM. At steady state, NPC1L1 associates with Rab11a and localizes in ERC. When cholesterol level drops, Cdc42 is activated and binds to NPC1L1 complex. The GTP-bound Cdc42 further activates N-WASP and Arp2/3 complex, which promotes the branched actin assembly. Then Rab11a gradually dissociates from NPC1L1 complex and the vesicles are transported from ERC to PM by MyoVb. When the cholesterol level is high, cholesterol will induce the internalization of NPC1L1 together with cholesterol from PM (8, 9).
In summary, our study demonstrates that Cdc42, which is activated by cholesterol depletion, interacts with NPC1L1 and facilitates its transport to the PM upon cholesterol depletion. The actin polymerization mediated by Cdc42 may contribute to the translocation of NPC1L1 to the PM. In vivo study demonstrates that the localization and cholesterol absorption function of NPC1L1 is dependent on Cdc42.
Acknowledgments
We thank Hong-Hua Miao, Yu-Xiu Qu, Qin Li, Jie Xu, Shu-Hua Zhou, and Li-Juan Wang for technical assistance, and Dr. Wei Qi for critical reading of the manuscript.
This work was supported by the grants from the Ministry of Science and Technology of China (2009CB919000 and 2011CB910900), National Natural Science Foundation of China (30925012), and Shanghai Science and Technology Committee (10QH1402900).
- NPC1L1
- Niemann-Pick C1-like 1
- Cdc42 LKO
- liver-specific Cdc42 knockout
- CDX
- methyl-β-cyclodextrin
- co-IP
- co-immunoprecipitation
- EGFP
- enhanced green fluorescent protein
- ERC
- endocytic recycling compartment
- MyoVb
- myosin Vb
- PM
- plasma membrane
- RFP
- red fluorescent protein.
REFERENCES
- 1. Altmann S. W., Davis H. R., Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., Wang L., Murgolo N., Graziano M. P. (2004) Science 303, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 2. Davis H. R., Jr., Zhu L. J., Hoos L. M., Tetzloff G., Maguire M., Liu J., Yao X., Iyer S. P., Lam M. H., Lund E. G., Detmers P. A., Graziano M. P., Altmann S. W. (2004) J. Biol. Chem. 279, 33586–33592 [DOI] [PubMed] [Google Scholar]
- 3. Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A., Davies J. P., Nilsson L. M., Yu L. (2007) J. Clin. Invest. 117, 1968–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen J. C., Pertsemlidis A., Fahmi S., Esmail S., Vega G. L., Grundy S. M., Hobbs H. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1810–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fahmi S., Yang C., Esmail S., Hobbs H. H., Cohen J. C. (2008) Hum. Mol. Genet. 17, 2101–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamanashi Y., Takada T., Suzuki H. (2009) Pharmacogenet. Genomics 19, 884–892 [DOI] [PubMed] [Google Scholar]
- 7. Wang L. J., Wang J., Li N., Ge L., Li B. L., Song B. L. (2011) J. Biol. Chem. 286, 7397–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ge L., Qi W., Wang L. J., Miao H. H., Qu Y. X., Li B. L., Song B. L. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ge L., Wang J., Qi W., Miao H. H., Cao J., Qu Y. X., Li B. L., Song B. L. (2008) Cell Metab. 7, 508–519 [DOI] [PubMed] [Google Scholar]
- 10. Chu B. B., Ge L., Xie C., Zhao Y., Miao H. H., Wang J., Li B. L., Song B. L. (2009) J. Biol. Chem. 284, 22481–22490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J. H., Ge L., Qi W., Zhang L., Miao H. H., Li B. L., Yang M., Song B. L. (2011) J. Biol. Chem. 286, 25088–25097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris K. P., Tepass U. (2010) Traffic. 11, 1272–1279 [DOI] [PubMed] [Google Scholar]
- 13. Abdul-Manan N., Aghazadeh B., Liu G. A., Majumdar A., Ouerfelli O., Siminovitch K. A., Rosen M. K. (1999) Nature 399, 379–383 [DOI] [PubMed] [Google Scholar]
- 14. Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. (1999) Cell 97, 221–231 [DOI] [PubMed] [Google Scholar]
- 15. Gasman S., Chasserot-Golaz S., Malacombe M., Way M., Bader M. F. (2004) Mol. Biol. Cell 15, 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matas O. B., Martínez-Menárguez J. A., Egea G. (2004) Traffic. 5, 838–846 [DOI] [PubMed] [Google Scholar]
- 17. Yuan H., Zhang H., Wu X., Zhang Z., Du D., Zhou W., Zhou S., Brakebusch C., Chen Z. (2009) Hepatology 49, 240–249 [DOI] [PubMed] [Google Scholar]
- 18. Cao J., Wang J., Qi W., Miao H. H., Wang J., Ge L., DeBose-Boyd R. A., Tang J. J., Li B. L., Song B. L. (2007) Cell Metab. 6, 115–128 [DOI] [PubMed] [Google Scholar]
- 19. Wu X., Quondamatteo F., Lefever T., Czuchra A., Meyer H., Chrostek A., Paus R., Langbein L., Brakebusch C. (2006) Genes Dev. 20, 571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benard V., Bohl B. P., Bokoch G. M. (1999) J. Biol. Chem. 274, 13198–13204 [DOI] [PubMed] [Google Scholar]
- 21. Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., He T. C. (2007) Nat. Protoc. 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 22. Miller P. J., Johnson D. I. (1994) Mol. Cell. Biol. 14, 1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weber M., Salo V., Uuskallio M., Raudaskoski M. (2005) Fungal. Genet. Biol. 42, 624–637 [DOI] [PubMed] [Google Scholar]
- 24. Erickson J. W., Cerione R. A., Hart M. J. (1997) J. Biol. Chem. 272, 24443–24447 [DOI] [PubMed] [Google Scholar]
- 25. Jaksits S., Bauer W., Kriehuber E., Zeyda M., Stulnig T. M., Stingl G., Fiebiger E., Maurer D. (2004) J. Immunol. 173, 1628–1639 [DOI] [PubMed] [Google Scholar]
- 26. Fessler M. B., Arndt P. G., Frasch S. C., Lieber J. G., Johnson C. A., Murphy R. C., Nick J. A., Bratton D. L., Malcolm K. C., Worthen G. S. (2004) J. Biol. Chem. 279, 39989–39998 [DOI] [PubMed] [Google Scholar]
- 27. Gong Y., Lee J. N., Lee P. C., Goldstein J. L., Brown M. S., Ye J. (2006) Cell Metab. 3, 15–24 [DOI] [PubMed] [Google Scholar]
- 28. Liu B., Ramirez C. M., Miller A. M., Repa J. J., Turley S. D., Dietschy J. M. (2010) J. Lipid Res. 51, 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugii S., Reid P. C., Ohgami N., Du H., Chang T. Y. (2003) J. Biol. Chem. 278, 27180–27189 [DOI] [PubMed] [Google Scholar]
- 30. Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. (2002) Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 31. Kullak-Ublick G. A., Stieger B., Meier P. J. (2004) Gastroenterology 126, 322–342 [DOI] [PubMed] [Google Scholar]
- 32. Pan J. Y., Wessling-Resnick M. (1998) Bioessays 20, 516–521 [DOI] [PubMed] [Google Scholar]
- 33. Johnson D. I. (1999) Microbiol. Mol. Biol. Rev. 63, 54–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meller N., Merlot S., Guda C. (2005) J. Cell Sci. 118, 4937–4946 [DOI] [PubMed] [Google Scholar]