Background: The androgen receptor (AR) and its splice variants are critical for castration resistance of prostate cancer.
Results: We have cloned a novel membrane-bound AR splice variant AR8 that is required for optimal activation of AR.
Conclusion: AR8 potentiates AR transcriptional activity and promotes castration resistance.
Significance: Our findings provide new insights into mechanisms by which AR and its variants promote castration resistance.
Keywords: Nuclear Receptors, Phosphorylation, Prostate Cancer, Src, Steroid Hormone Receptor, Alternative Splicing
Abstract
Progression from the androgen-sensitive to androgen-insensitive (or castration-resistant) stage is the major obstacle for sustained effectiveness of hormonal therapy for prostate cancer. The androgen receptor (AR) and its splice variants play important roles in regulating the transcription program essential for castration resistance. Here, we report the identification of a novel AR splice variant, designated as AR8, which is up-regulated in castration-resistant prostate cancer cells. AR8 is structurally different from other known AR splice variants because it lacks a DNA binding domain and therefore, unlikely functions as a transcription factor on its own. Immunofluorescence staining revealed that AR8 was primarily localized on the plasma membrane, possibly through palmitoylation of two cysteine residues within its unique C-terminal sequence. Mutation of these putative palmitoylation sites in AR8 led to loss of its plasma membrane localization. In addition, we demonstrated that overexpression of AR8 in prostate cancer cells promoted association of Src and AR with the EGF receptor in response to EGF treatment and enhanced tyrosine phosphorylation of AR. Conversely, specific knockdown of AR8 expression in prostate cancer cells compromised EGF-induced Src activation and AR phosphorylation. This effect was accompanied with attenuation of proliferation and increased apoptosis in prostate cancer cells cultured in androgen-depleted medium. We also showed that AR8 was required for optimal transcriptional activity of AR in response to treatment of both androgen and EGF. Taken together, our results demonstrate that the membrane-associated AR8 isoform may contribute to castration resistance by potentiating AR-mediated proliferative and survival responses to hormones and growth factors.
Introduction
Androgen ablation therapy is the most common treatment for patients with metastatic prostate cancer. Despite an initially beneficial response to hormonal therapy, a majority of patients eventually develop recurrent castration-resistant tumors which are the major cause of mortality related to prostate cancer. Therefore, understanding mechanisms underlying progression to a castration-resistant state is critical for developing more effective treatments for advanced prostate cancer (1, 2).
Development of castration resistance in prostate cancer is a complicated process due to the nature of its heterogeneity. The androgen receptor (AR)2 is a key signaling molecule that exerts physiological effects of androgens in the prostate. It is well established that AR regulates transcription of androgen-responsive genes by binding to specific DNA sequences, known as androgen response elements (1). The human AR gene is composed of at least eight exons. As other members of steroid hormone receptor family, AR contains several functional domains, including an N-terminal transactivation domain (encoded by exon 1), a DNA binding domain (encoded by exons 2 and 3) and a ligand binding domain (encoded by exons 5–8). Mounting evidence suggests that AR signaling remains active in castration-resistant tumors (2). Multiple mechanisms have been proposed for reactivation of AR signaling under androgen-deprived conditions, including deregulation of the AR gene, steroid metabolism enzymes, steroid hormone receptor co-activators, as well as growth factors and cytokines (3–5). Amplification of the AR locus was shown to associate with anti-androgen resistance (6). Autocrine/paracrine factor-induced tyrosine phosphorylation of AR via Src, ETK/BMX, or Ack-1 kinases may contribute to androgen-independent activation of AR or sensitize it to respond to low levels of hormone (7–10). The detailed mechanisms by which AR is phosphorylated by these kinases remain elusive. It is also unclear how membrane-bound active Src kinase phosphorylates cytosolic AR. On the other hand, several independent studies showed that a series of AR splice variants lacking the ligand binding domain are up-regulated in hormone-resistant prostate cancer cell lines and tissues and promote castration-resistant growth (11–15). It was shown that some of these truncated AR isoforms, including AR3/AR-V7, and ARv567es, play an indispensable role in androgen-insensitive growth by regulating a unique set of target genes (12, 14). Expression of one of the major AR splice variants, AR3, predicts the risk of tumor recurrence after radical prostatectomy (12). Therefore, deregulation of AR splice variants may contribute to prostate cancer progression.
In this study, we identified a novel membrane-bound AR splice variant AR8, which is up-regulated in castration-resistant prostate cancer cell lines. Our data show that AR8 co-operates with the prototype AR to potentiate androgen and growth factor response in prostate cancer cells by promoting AR association with EGFR on the plasma membrane and enhancing AR tyrosine phosphorylation. Our study provides new insights into understanding the role of AR splice variants in development of castration resistance.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
LNCaP, 22Rv1 and COS-1 cells were purchased from American Type Culture Collection (Manassas, VA). Prostate cancer cell lines C4-2, C4-2B, and CWR-R1 were kindly provided by Drs. D. Tindall and E. Wilson, respectively. Cells were maintained in RPMI 1640 medium with 10% FBS. Transfection experiments were carried out using FuGENE 6 or FuGENE HD (Roche Applied Science) following the manufacturer's instructions.
Cloning and Constructs
The AR8 transcript was amplified using the 5′/3′-Rapid Amplification of cDNA Ends (RACE) kit (Roche Applied Science) as described previously (12). The PCR products were cloned and sequenced. The full-length AR8 cDNA sequence has been deposited in GenBank (accession number HM055487). The AR8 coding sequence was subcloned into a lentiviral expression vector as described previously (8). AR8 mutation was generated by a PCR-based method (QuikChange kit; Stratagene) and confirmed by sequencing. The short hairpin RNAs specific for human AR8 (shAR8) were constructed based on the Addgene protocol for pLKO.1 using the following oligonucleotide sequences: shAR8-1, 5′-ccggctcattatcaggtctatcactcgagtgatagacctgataatgagtttttg-3′ and 5′-aattcaaaaactcattatcaggtctatcactcgagtgatagacctgataatgag-3′; shAR8-2, 5′-ccgggactctgga aactcattatctcgagataatgagtttccagagtctttttg-3′ and 5′-aattcaaaaagactctggaaactcattatctcgagataatgagtttccagagtc-3′.
Antibodies
The antibodies used in this study include mouse monoclonal anti-FLAG M2 (Sigma), anti-tubulin (Abm), anti-EGFR Ab12 (NeoMarkers), anti-EGFR Ab13 (NeoMarkers), anti-AR (H280), anti-AR (N-20), anti-AR (C-19), anti-AR (441) (Santa Cruz Biotechnology), and anti-phospho SrcY416 (anti-pSrc) (Cell Signaling). The phospho-specific antibody for AR Tyr-534 (anti-pAR) has been described previously (8). The polyclonal antibody against AR8 was generated by immunizing two rabbits with the purified GST fusion protein containing the last 33 unique amino acids at the C terminus of AR8 following the standard protocol (Cocalico Biologicals). The antiserum was then partially purified by depleting anti-GST through a conjugated GST column.
Quantitative Real-time PCR
Quantitative real-time PCR was performed as described previously (12). The sequences of primers used for AR isoforms are: AR sense, 5′-ctactccggaccttacggggacatgcg-3′ and antisense, 5′-gggctgacattcatagccttcaatgtgtgac-3′; AR8 sense, 5′-cgacttcaccgcacctgatg-3′ and antisense 5′-ctctttcttcgggtatttcgcatg-3′; and 18 S rRNA sense, 5′-ttgacggaagggcaccaccag-3′ and antisense 5′-gcaccaccacccacggaatcg-3′. Primers for human POV1 and PSA are as described previously (8). Relative abundance of each target transcript to 18 S rRNA was quantified using the comparative ΔΔCt. The ratio of AR8 to AR was calculated by using the Pfaffl method (16).
Immunofluorescence
Cells were grown on coverslips and treated as indicated in figure legends and fixed in Lana's fix buffer (paraformaldehyde-picric acid fixative) 4% paraformaldehyde, 14% (v/v) saturated picric acid, 0.16 m phosphate buffer, pH 6.9 for 1 h and washed four times with PBS. The coverslides were blocked in PBS containing 0.3% Triton X-100, 1% BSA, and 1% normal donkey serum for 1 h at room temperature. Primary antibody was added and incubated for 1 h at room temperature. After washing with PBS, the coverslides were incubated with rhodamine-conjugated anti-mouse and fluorescein isothiocyanate-conjugated anti-rabbit secondary antibodies for 1 h at room temperature. Finally, cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei before mounting. The coverslides were examined by using a Nikon Eclipse TE2000-U microscope system.
Luciferase Reporter Assays
Luciferase assays were carried out as described previously (8). Briefly, at 24 h after transfection, cells were incubated with fresh phenol red-free serum-free medium for the experiments with growth factors or with phenol red-free medium containing 5% charcoal-stripped FBS for the experiments using dihydrotestosterone (DHT). The Dual-Luciferase assays were carried out according to the manufacturer's protocol (Promega). The transfection efficiency was normalized using a promoter-less Renilla internal control. The results are presented as the mean relative light units ± S.D. of triplicate samples.
Immunoprecipitation, Biotinylated Peptide Pulldown
Cells were washed twice with ice-cold PBS and lysed using lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm Na3VO4, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm PMSF) at 4 °C for 30 min. Cell lysates were centrifuged to remove cell debris before incubation with the antibody at 4 °C for 1 h. The immunocomplexes were collected using protein A/G-Sepharose beads. The biotin-GDMRNTRRKRLWKLIIRSINSCICSPREAEVPVRQQK-OH peptide and a biotinylated control were synthesized by NEO Peptide (Cambridge, MA) and incubated with CWR-R1 cell extracts. Associated proteins captured by NeutrAvidin-agarose beads were resolved by SDS-PAGE and visualized using Coomassie Blue staining. Selected protein bands were excised and subjected to mass spectrometry (MS) analysis as described previously (17).
In Vitro Cell Proliferation and Apoptosis Assays
Cell growth was determined using a Cell Proliferation Reagent WST-1 Kit (Roche Applied Science). Briefly, LNCaP or CWR-R1 cells were grown in 100 μl of culture medium into 96-well plates at a concentration of 3 × 103 cells/well. The A450 nm and A690 nm were measured using a plate reader. Experiments were repeated at least three times. Cell proliferation assays were conducted using Click-iT™ EdU Imaging kits (Invitrogen). Briefly, cells were plated on coverslips and incubated with 10 μm EdU solution for 16 h. Cells were then fixed with 3.7% formaldehyde and incubated with 0.5 ml Click-iT™ reaction mixture for 30 min at room temperature. The coverslides were examined using a Nikon Eclipse TE2000-U microscope system. Apoptosis was determined by the TUNEL assays using In Situ Cell Death Detection kit (Roche Applied Science). Briefly, cells were grown on coverslips and fixed with a freshly prepared fixation (4% paraformaldehyde in PBS, pH 7.4) and then incubated in permeabilization solution (0.1% Triton X-100 in 0.1% sodium citrate) for 2 min on ice. TUNEL reaction mixture (50 μl) was added and the sample incubated for 60 min at 37 °C. Finally, cells were counterstained with DAPI to visualize nuclei before mounting. The coverslides were examined using a Nikon Eclipse TE2000-U microscope system.
RESULTS
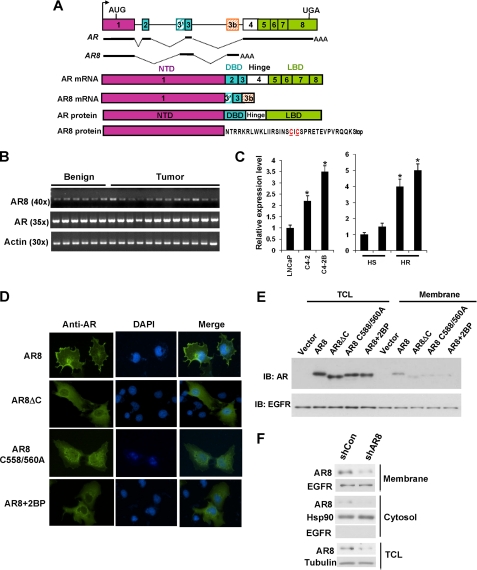
In our previous study, more than twenty alternatively spliced AR transcripts were identified in CWR-R1 cells using 3′-Rapid Amplification of cDNA Ends (12). In addition to the variants reported previously, we have identified a novel AR splice variant composed of exons 1, 3, and 3b (Fig. 1A). Due to usage of an alternative splice acceptor site in exon 3 (marked as 3′), this transcript is deduced to encode a protein containing the N-terminal transactivation domain and a 33-amino acid unique sequence at the C terminus (Fig. 1A and supplemental Fig. 1). RT-PCR analysis using a pair of primers exclusively recognizing the unique junction of exon 1 and exon 3′ revealed that the AR8 transcript was detectable in a panel of human prostate benign and tumor tissues (Fig. 1B). Interestingly, the level of AR8 transcript is elevated in castration-resistant LNCaP derivatives (C4-2, C4-2B) and CWR22 xenograft tumors compared with their hormone-sensitive counterparts (Fig. 1C and supplemental Fig. 2B). In addition, the level of AR8 transcript relative to AR appeared to be increased in CWR-R1 cells in response to treatment with androgen or EGF (supplemental Fig. 2C), suggesting that the relative ratio of AR8 to AR is higher when cell proliferation is enhanced. AR8 does not contain a DNA binding domain, and therefore, it is unlikely to function as a transcription factor. The lack of transcription activity for AR8 was confirmed by testing a series of reporters driven by androgen response element-containing promoters (supplemental Fig. 3). Surprisingly, when AR8 was overexpressed in COS-1 cells, it mainly localized to the plasma membrane and only sparsely in some perinuclear compartments (Fig. 1D), whereas AR or another AR splice variant AR3 was either localized in the cytosol or the nucleus of COS-1 cells under the same conditions (supplemental Fig. 4). Similar results were obtained when AR8 was overexpressed in LNCaP and CWR-R1 cells (supplemental Fig. 5), suggesting that this protein is preferentially associated with the plasma membrane. To test whether the unique C-terminal sequence is involved in AR8 membrane targeting, we deleted this region. Fig. 1D shows that, in the absence of its unique C terminus, AR8 failed to localize to the plasma membrane. Because post-translational modifications such as myristoylation and palmitoylation can regulate the steady-state localization and function of various peripheral membrane proteins, such as Ras family small GTPases and Src family kinases (18, 19), we examined whether any amino acid residue in AR8 could be lipidated. A bioinformatic search (20) predicted two cysteine residues, Cys-558 and Cys-560, located in the AR8 C-terminal region as putative palmitoylation sites. Substitution of both cysteine residues led to the loss of membrane localization of AR8. In addition, treatment of 2-bromopalmitic acid, a known palmitoylation inhibitor (21), blocked AR8 membrane association. Similar results were obtained when we examined the subcellular localization of AR8 and its mutants in membrane fractionation experiments (Fig. 1E). The endogenous AR8 was also found to be enriched in membrane fraction in CWR-R1 cells (Fig. 1F). Therefore, AR8 may target the plasma membrane through palmitoylation of its unique C terminus.
FIGURE 1.
Cloning and expression of membrane-bound AR8. A, schematic structure of human AR splice variants. The hatched cassettes indicate the cryptic exons. Solid thick lines represent the transcribed exon sequences. The AR8 unique amino acid sequence is shown, and putative palmitoylation sites are in red and underlined. B, expression of AR8 in human prostate tissues. Total RNA was isolated from 6 benign and 12 malignant human prostate tissues and subjected to reverse transcription-PCR. The primer sets used to amplify AR- or AR8-specific transcripts were described under “Experimental Procedures.” C, relative expression levels of AR8 in LNCaP, C4-2, and C4-2B were quantified using real-time PCR (left). Their expression in two pairs of CW22R xenograft tumors derived from intact (HS) and castrated (HR) male mice were also quantified (right). *, p < 0.05. Error bars, S.D. D, plasma membrane localization of AR8. COS-1 cells were transfected with AR8 or AR8 mutants. Some cells were treated with 25 μm 2-bromopalmitic acid (2BP) for 16 h as indicated. Cells were then subjected to immunofluorescence staining with anti-AR (N20) antibody. Nucleus was visualized using DAPI staining. E, effects of AR8 mutation on its membrane localization. COS-1 cells were transfected with the indicated constructs. Membrane fractionation was carried out using the Eukaryotic Membrane Protein Extraction kit (Pierce). AR protein present in membrane fractions and total lysates were detected by Western blotting (IB) with an anti-AR antibody. EGFR served as a loading control. F, CWR-R1 cells were infected with lentivirus encoding the shRNA for AR8 (shAR8) or the scrambled control (shCon). At 48 h after infection membrane fractionation was carried out as above. The level of AR8 protein in the membrane or cytosol fraction as well as in the total lysates was detected by Western blotting with the anti-AR8 antibody. Tubulin serves as a loading control and anti-Hsp90, anti-EGFR to assign the majorities of the marker proteins to their expected cytoplasmic, membrane fractions respectively.
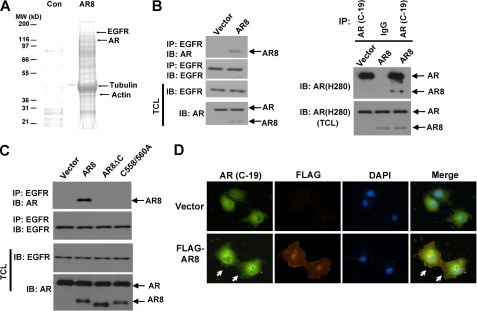
To investigate further the role of AR8 in prostate cancer cells, a biotin-conjugated peptide containing the AR8 unique C-terminal sequence was synthesized and used as bait to identify potential interacting proteins with AR8 in CWR-R1 cells. Our MS analysis revealed that multiple proteins were associated with AR8 C-terminal unique sequence (Fig. 2A). Tubulins and actins are among the most abundant proteins interacting with the biotinylated peptide, suggesting that AR8 may be associated with cytoskeletons in prostate cancer cells. This is consistent with our observation that overexpressed AR8 is primarily localized to the plasma membrane where these cytoskeletal proteins are enriched. Interestingly, we also found that the AR8 C-terminal peptide was associated with EGFR and AR. We then confirmed the association of AR8 with EGFR and AR in LNCaP cells overexpressing AR8 by performing co-immunoprecipitation experiments. As shown in Fig. 2B (left), AR8 co-immunoprecipitated with EGFR whereas endogenous AR failed to do so under our experimental conditions. In addition, AR8 appeared to form a complex with endogenous AR as it was co-immunoprecipitated by an antibody specific for the C terminus of AR (Fig. 2B, right). We also examined whether membrane targeting of AR8 was required for its association with EGFR. As shown in Fig. 2C, deletion of the C-terminal sequence or mutation of the palmitoylated cysteine residues completely abolished its association with EGFR. We further demonstrated that overexpression of AR8 in COS-1 cells enhanced membrane association of AR detected using the antibody specific for its C terminus (Fig. 2D).
FIGURE 2.
AR8 promotes AR association with EGFR. A, proteins associated with a biotinylated AR8 C-terminal peptide (AR8) or control peptide (Con) were identified as described under “Experimental Procedures.” Some of the identified proteins are indicated. B, LNCaP cells were infected with lentivirus encoding AR8 or control vector. At 48 h after infection, cells lysates were immunoprecipitated (IP) with anti-EGFR (left), AR (C-19), or IgG control (right), followed by Western blot (IB) analysis using the indicated antibodies. C, LNCaP cells were infected with lentivirus encoding AR8, AR8 mutants, or a vector control. At 48 h after infection, cells lysates were immunoprecipitated with anti-EGFR, followed by Western blot analysis using the indicated antibodies. D, COS-1 cells were transfected with the plasmids encoding AR with or without FLAG-tagged AR8. Cells were then subjected to immunofluorescence staining using anti-AR (C-19) or anti-FLAG antibodies. The nucleus was visualized using DAPI staining.
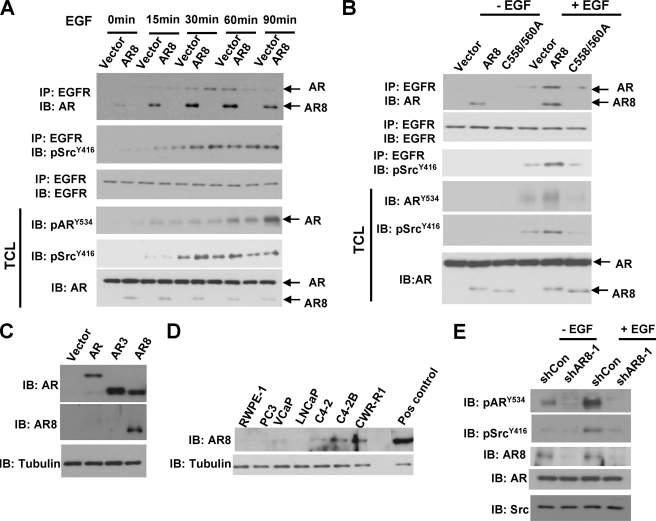
We showed previously that EGF can induce AR tyrosine phosphorylation via activating Src kinase (8). Because activated Src kinase is believed to localize to the plasma membrane whereas AR is largely present in the cytosol or nucleus, we examined whether the membrane-anchored AR8 could be involved in this process and serve as a mediator in Src-induced AR activation. As shown in Fig. 3A, consistent with previous studies, EGF treatment promoted Src association with EGFR and induced Src kinase activity, which were further boosted by AR8. This interaction was accompanied by an increase in association of AR8 and AR with EGFR in response to EGF stimulation. EGF-induced AR association with EGFR was further enhanced when AR8-expressing cells were treated with EGF for 15 and 30 min. However, 60–90 min after EGF treatment, dissociation of AR from EGFR appeared to be coupled with a high level of AR Y534 phosphorylation, which was accelerated in AR8-expressing cells. In addition, the AR8 C558/C560 mutant failed to enhance AR tyrosine phosphorylation (Fig. 3B), suggesting that AR8 membrane targeting is required for its activity. To study further the function of endogenous AR8, we developed a polyclonal antibody specific for the unique C terminus of AR8 protein. This antibody could detect the overexpressed AR8 but not AR or AR3 (Fig. 3C). This antibody could also detect endogenous AR8 protein in a panel of prostate cancer cell lines including CWR-R1, VcaP, and LNCaP derivatives (Fig. 3D). Consistent with our RT-PCR data, AR8 expression is elevated in castration-resistant C4-2 and C4-2B cells compared with the parental LNCaP cells. Furthermore, knocking down endogenous AR8 significantly diminished both basal and EGF-induced AR Y534 phosphorylation (Fig. 3E). Taken together, these data suggested that AR8 sensitized prostate cancer cells to respond to EGF stimulation and enhanced EGF-induced Src activation and AR Y534 phosphorylation.
FIGURE 3.
AR8 enhanced AR tyrosine phosphorylation in response to EGF treatment. A, LNCaP cells infected with lentivirus encoding AR8 or a control vector. At 48 h after infection, cells were treated with EGF for the indicated period of time. Cell lysates were immunoprecipitated (IP) with anti-EGFR followed by Western blot (IB) analysis using the indicated antibodies. B, LNCaP cells infected with lentivirus encoding AR8, the C558A/C560A mutant, or a control vector as described in A. The effects of C558A/C560A mutation on AR activity were determined as above. C, specificity of anti-AR8 antibody. COS-1 were transfected with AR, AR3, AR8 or vector. Total protein lysates were immunoblotted with anti-AR8 and anti-AR, respectively. D, expression of AR8 protein in prostate cells. Total cell lysates from various prostate cell lines were immunoblotted with anti-AR8. Overexpressed AR8 in LNCaP cells was used as a positive control (Pos control). Tubulin was used as a loading control. E, CWR-R1 cells infected with lentivirus encoding the shRNA for AR8 (shAR8-1) or a scrambled control (shCon). At 48 h after infection, cells were serum-starved overnight followed by treatment with vehicle or 100 ng/ml EGF for 30 min. Cell lysates were immunoblotted using the indicated antibodies.
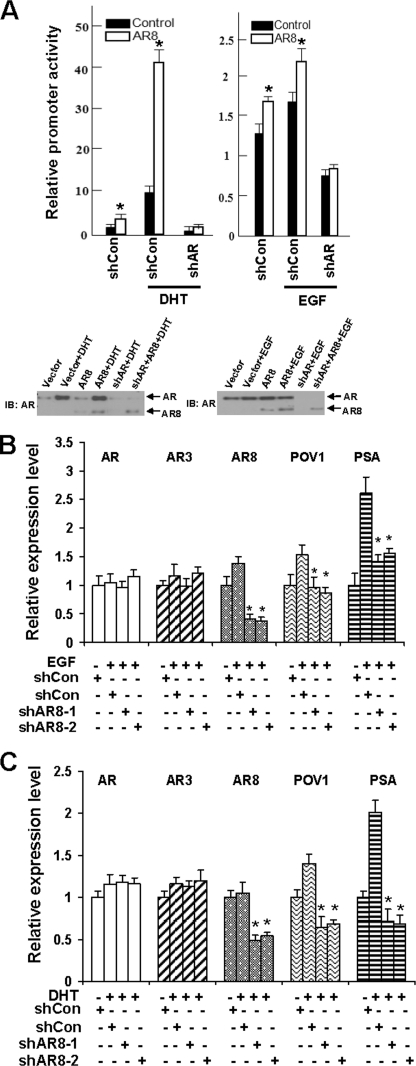
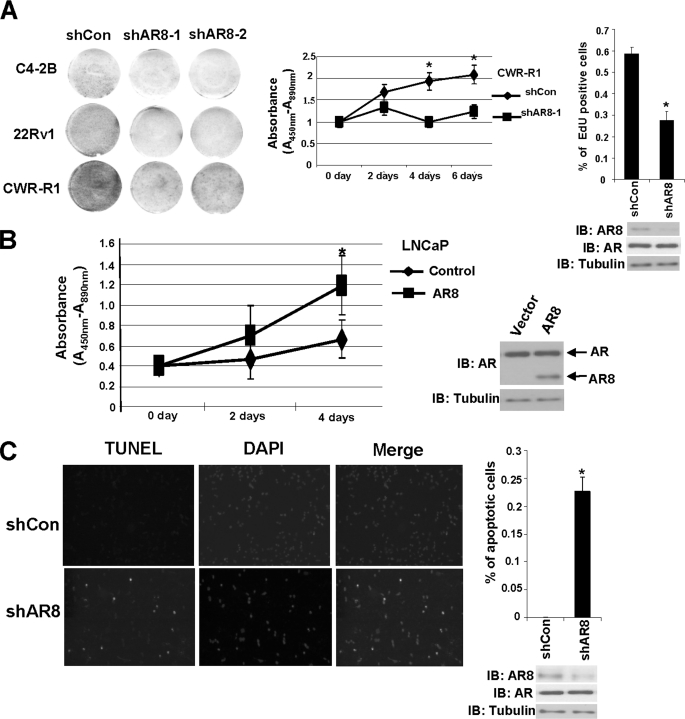
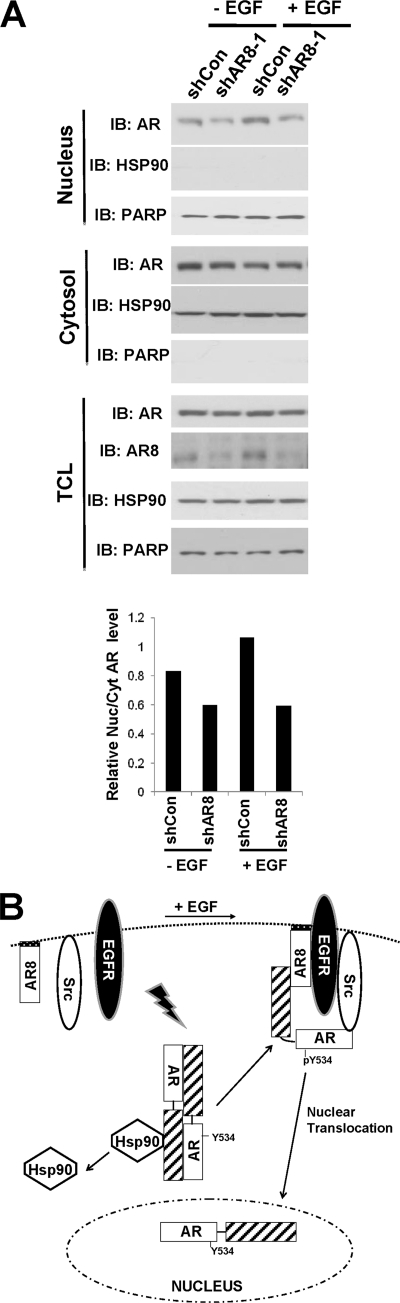
Because we showed previously that Y534 phosphorylation can enhance AR activity in response to growth factors and low levels of androgen, we examined whether AR8 could promote AR transcriptional activity. When AR8 was overexpressed in LNCaP cells, we detected elevated ARR2-driven reporter activities in response to both androgen and EGF treatment (Fig. 4A). Conversely, when we knocked down endogenous AR8 expression in CWR-R1 cells by two independent shRNAs, expression of AR-regulated genes, including PSA and POV1, was significantly diminished in response to both EGF and androgen treatment (Fig. 4, B and C). Furthermore, knockdown of AR8 expression in several hormone-insensitive prostate cancer lines, including C4–2B, 22Rv1, and CWR-R1, attenuated their proliferation whereas overexpression of AR8 in hormone-sensitive LNCaP cells promoted cell proliferation under androgen-depleted conditions (Fig. 5, A and B). AR8 appears to be required for survival of CWR-R1 cells because inhibition of AR8 expression by a specific shRNA led to a dramatic increase in the number of apoptotic cells (Fig. 5C). We showed previously that AR Y534 phosphorylation plays a role in regulation of AR nuclear translocation in response to EGF treatment. We therefore examined whether AR8 is required for this process. As shown in Fig. 6A, knocking down of AR8 expression in CWR-R1 cells significantly reduced the level of nuclear AR protein. Taken together, these data suggested that AR8 plays a critical role in both survival and proliferation of prostate cancer cells under androgen-depleted conditions by modulating Src-mediated AR tyrosine phosphorylation.
FIGURE 4.
The effects of AR8 on EGF- or DHT-induced AR transcription activity. A, LNCaP cells were infected with lentivirus encoding AR8 or a control vector. At 16 h after infection, cells were transfected with ARR2-luciferase reporter. At 24 h after transfection, cells were treated with 1 nm DHT (left) or 10 ng/ml EGF (right) for 12 h before measuring luciferase activity. B, CWR-R1 cells were infected with lentivirus encoding an shRNA for AR8 (shAR8-1 or shAR8-2) or a scrambled control. Cells were treated with vehicle or 10 ng/ml EGF for 12 h, and total RNAs were isolated. The relative change of AR, AR3, AR8, POV1, or PSA transcripts was quantified by real-time PCR. The value of the control sample without stimulation was set as 1. C, CWR-R1 cells were treated as in B, except that EGF was replaced with 0.1 nm DHT. The relative change gene expression was monitored as in B. *, p < 0.05 compared with the control. Error bars, S.D.
FIGURE 5.
Regulation of prostate cancer cell proliferation and survival by AR8 under androgen-depleted conditions. A, C4-2B, 22Rv1, and CWR-R1 cells were infected with lentivirus encoding an shRNAs for AR8 (shAR8-1 or shAR8-2) or a control shRNA (shCon). After a 2-week culture in androgen-depleted (CS) medium, cells were visualized using Coomassie Blue staining (left). CWR-R1 cells were infected with lentivirus encoding the shAR8-1 or a scrambled control (shCon), followed by measurement of cell proliferation using WST-1 assays at indicated times (center) or the Click-iT™ EdU assay at 48 h after infection (right). B, LNCaP cells were infected with lentivirus encoding AR8 or a control vector. Cell proliferation was measured using WST-1 assays at the indicated times. C, CWR-R1 cells were plated on coverslips and infected with lentivirus encoding shAR8-1 or a scrambled control. At 48 h after infection, apoptosis was detected by TUNEL assays. Apoptotic cells were quantified by counting TUNEL-positive cells in 1000 cells from three independent random fields. *, p < 0.05 compared with the control. Error bars, S.D.
FIGURE 6.
A, regulation of AR nuclear translocation by AR8. CWR-R1 cells were infected with lentivirus encoding the shRNA for AR8 (shAR8-1) or the scrambled control (shCon). Cells were treated with vehicle or 100 ng/ml EGF for 1 h. The cytoplasmic and nuclear fractions were isolated using the Nuclear and Cytoplasmic Extraction kit (Pierce). The level of AR protein in each fraction was determined by Western blotting (IB) using a monoclonal AR antibody (441). Hsp90 and Poly(ADP-ribose)polymerase were used as markers for the cytoplasmic and nuclear fractions, respectively. The intensity of AR in each fraction was quantified by using the software Quantity One. The ratio of nuclear versus cytoplasmic AR was determined. The value of the EGF-treated control was set as 1. B, postulated role of AR8 in EGF-induced activation of AR. AR8 may function as a lipid-anchor protein to bring AR to the proximity of the plasma membrane and promote formation of a dynamic signaling complex containing EGFR, Src, AR, and AR8 in response to EGF. The level of AR8 could modulate kinetics of the assembly and dissociation of this complex, allowing sequential phosphorylation and subsequent nuclear translocation of AR.
DISCUSSION
Development of castration resistance in prostate cancer is a complicated process that may involve multiple genetic and epigenetic alterations. Aberrant expression of AR splice variants may be involved (22). In this report, we showed that deregulation of a novel membrane-associated AR splice variant (AR8) may play a role in prostate cancer progression by potentiating proliferative and survival responses to hormones and growth factors through modulating the activity of AR and possibly other splice variants. In contrast to AR and other known AR splice variants, AR8 lacks the DNA binding domain required for transcription activity and is primarily localized to the plasma membrane. Therefore, AR8 most likely functions through a nongenomic mechanism. We showed that AR8 could rapidly associate with EGFR in prostate cancer cells in response to EGF as well as enhanced Src activation and AR tyrosine phosphorylation. In addition, in the presence of high levels of AR8, the kinetics of association of phosphorylated AR with EGFR was also altered. Within the first 30 min of EGF stimulation, AR8 appeared to enhance AR association with EGFR and allowed membrane-localized Src kinase to phosphorylate AR. These events were followed by AR8 promoting dissociation of AR from EGFR when the level of phosphorylated AR reached a high level at 60–90 min after EGF treatment. We showed previously that AR could be phosphorylated at multiple tyrosine residues in Src-overexpressing cells (8). It is possible that subsequent phosphorylation of different tyrosine residues by Src or other kinases (e.g. ETK/BMX, Ack-1) may attribute to dissociation of AR from EGFR. Our data suggested that EGFR, Src, AR, and AR8 form a dynamic signaling complex in response to EGF, and the level of AR8 could modulate kinetics of the assembly and dissociation of this complex, allowing sequential phosphorylation and its subsequent nuclear translocation (Fig. 6B). Therefore, AR8 may augment androgen and growth factor responses in prostate cancer cells by bringing AR to the proximity of the plasma membrane and enhancing AR tyrosine phosphorylation. Future study using antibodies specific for other tyrosine phosphorylation sites or quantitative MS analysis would be helpful to dissect further the role of AR8 in promoting transient membrane association and phosphorylation of AR in response to growth factors.
Although AR8 is less abundant than AR and its known splice variants such as AR3, it may play an important role in regulating a rate-limiting step of AR activation induced by growth factors, possibly androgens as well. Bringing AR to the proximity of the plasma membrane may enhance its ligand binding efficiency, especially under androgen-deprived conditions. This possibility is supported by our observation that knockdown of AR8 expression in prostate cancer cells desensitized androgen response. In addition to modulating AR activity by promoting Tyr-534 phosphorylation, AR8 may also regulate the activity of other AR splice variants. We also detected a reduction of AR3 protein level in AR8 knocked-down CWR-R1 cells.3 However, such effect does not likely result from nonspecific knockdown of AR3 transcript because the transcript level of neither AR3 nor AR was altered in AR8 knocked-down CWR-R1 cells (Fig. 4, B and C). It is possible that a downstream effector activated by AR8, such as Src kinases, may mediate this effect by modulating the stability of AR3 protein through phosphorylation. This is currently under investigation. It would be interesting to find out how AR8 could modulate both AR and AR3 activity and potential interplays among AR and its various splice variants.
Taken together, our findings provide new insights into mechanisms by which AR and its splice variants promote castration resistance in prostate cancer cells. Given that inhibition of AR8 activity diminished AR tyrosine phosphorylation and attenuated tumor cell growth under androgen-depleted conditions, targeting the AR8-AR-Src-EGFR complex could potentially serve as a basis for developing new intervention regimen.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA106504 (to Y. Q.) and Grant HHSN261200800001E through the NCI. This work was also supported by Department of Defense Grant W81XWH-08-1-0174 (to Y. Q.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) HM055487.
X. Yang and Y. Qiu, unpublished data.
- AR
- androgen receptor
- DHT
- dihydrotestosterone
- EGFR
- EGF receptor
- PSA
- prostate-specific antigen.
REFERENCES
- 1. Heinlein C. A., Chang C. (2004) Endocr. Rev. 25, 276–308 [DOI] [PubMed] [Google Scholar]
- 2. Culig Z., Klocker H., Bartsch G., Hobisch A. (2002) Endocr. Relat. Cancer 9, 155–170 [DOI] [PubMed] [Google Scholar]
- 3. Scher H. I., Buchanan G., Gerald W., Butler L. M., Tilley W. D. (2004) Endocr. Relat. Cancer 11, 459–476 [DOI] [PubMed] [Google Scholar]
- 4. Scher H. I., Sawyers C. L. (2005) J. Clin. Oncol. 23, 8253–8261 [DOI] [PubMed] [Google Scholar]
- 5. Shen M. M., Abate-Shen C. (2010) Genes Dev. 24, 1967–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. (2004) Nat. Med. 10, 33–39 [DOI] [PubMed] [Google Scholar]
- 7. Dai B., Chen H., Guo S., Yang X., Linn D. E., Sun F., Li W., Guo Z., Xu K., Kim O., Kong X., Melamed J., Qiu S., Chen H., Qiu Y. (2010) Cancer Res. 70, 5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo Z., Dai B., Jiang T., Xu K., Xie Y., Kim O., Nesheiwat I., Kong X., Melamed J., Handratta V. D., Njar V. C., Brodie A. M., Yu L. R., Veenstra T. D., Chen H., Qiu Y. (2006) Cancer Cell 10, 309–319 [DOI] [PubMed] [Google Scholar]
- 9. Mahajan N. P., Liu Y., Majumder S., Warren M. R., Parker C. E., Mohler J. L., Earp H. S., Whang Y. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8438–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DaSilva J., Gioeli D., Weber M. J., Parsons S. J. (2009) Cancer Res. 69, 7402–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dehm S. M., Schmidt L. J., Heemers H. V., Vessella R. L., Tindall D. J. (2008) Cancer Res. 68, 5469–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo Z., Yang X., Sun F., Jiang R., Linn D. E., Chen H., Chen H., Kong X., Melamed J., Tepper C. G., Kung H. J., Brodie A. M., Edwards J., Qiu Y. (2009) Cancer Res. 69, 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu R., Dunn T. A., Wei S., Isharwal S., Veltri R. W., Humphreys E., Han M., Partin A. W., Vessella R. L., Isaacs W. B., Bova G. S., Luo J. (2009) Cancer Res. 69, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun S., Sprenger C. C., Vessella R. L., Haugk K., Soriano K., Mostaghel E. A., Page S. T., Coleman I. M., Nguyen H. M., Sun H., Nelson P. S., Plymate S. R. (2010) J. Clin. Invest. 120, 2715–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watson P. A., Chen Y. F., Balbas M. D., Wongvipat J., Socci N. D., Viale A., Kim K., Sawyers C. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16759–16765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu K., Shimelis H., Linn D. E., Jiang R., Yang X., Sun F., Guo Z., Chen H., Li W., Chen H., Kong X., Melamed J., Fang S., Xiao Z., Veenstra T. D., Qiu Y. (2009) Cancer Cell 15, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baekkeskov S., Kanaani J. (2009) Mol. Membr. Biol. 26, 42–54 [DOI] [PubMed] [Google Scholar]
- 19. Berthiaume L., Resh M. D. (1995) J. Biol. Chem. 270, 22399–22405 [DOI] [PubMed] [Google Scholar]
- 20. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Webb Y., Hermida-Matsumoto L., Resh M. D. (2000) J. Biol. Chem. 275, 261–270 [DOI] [PubMed] [Google Scholar]
- 22. Guo Z., Qiu Y. (2011) Int. J. Biol. Sci. 7, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.