Background: S. aureus is an important cause of severe pneumonia acquired following upper airway colonization.
Results: S. aureus-expressing SpA induces RhoA, EGFR, and calpain activity resulting in disruption of tight junctions.
Conclusion: SpA is necessary but not sufficient to mediate invasive infection across the airway epithelium.
Significance: S. aureus activates multiple epithelial signaling cascades to initiate invasive pulmonary infection.
Keywords: Bacterial Pathogenesis, Calpain, Epithelial Cell, Rho, Staphylococcus aureus, Tight Junctions
Abstract
Staphyococcus aureus and especially the epidemic methicillin-resistant S. aureus strains cause severe necrotizing pneumonia. The mechanisms whereby these organisms invade across the mucosal epithelial barrier to initiate invasive infection are not well understood. Protein A (SpA), a highly conserved and abundant surface protein of S. aureus, activates TNF receptor 1 and EGF receptor (EGFR) signaling cascades that can perturb the cytoskeleton. We demonstrate that wild-type S. aureus, but not spa mutants, invade across polarized airway epithelial cell monolayers via the paracellular junctions. SpA stimulated a RhoA/ROCK/MLC cascade, resulting in the contraction of the cytoskeleton. SpA+ but not SpA− mutants stimulated activation of EGFR and along with subsequent calpain activity cleaved the membrane-spanning junctional proteins occludin and E-cadherin, facilitating staphylococcal transmigration through the cell-cell junctions. Treatment of polarized human airway epithelial monolayers with inhibitors of ROCK, EGFR, MAPKs, or calpain prevented staphylococcal penetration through the monolayers. In vivo, blocking calpain activity impeded bacterial invasion into the lung parenchyma. Thus, S. aureus exploits multiple receptors available on the airway mucosal surface to facilitate invasion across epithelial barriers.
Introduction
Staphylococcus aureus and particularly the epidemic community-associated USA300 methicillin-resistant S. aureus (MRSA)2 strains are an increasingly prevalent cause of invasive infection including pneumonia in the context of antecedent influenza (1). The initial stages of S. aureus pulmonary infection follow aspiration of the organisms from the upper airways (2). Yet, despite their ubiquity, exactly how staphylococci progress from innocuous colonization of the respiratory tract to invasive pneumonia is not well understood. Several staphylococcal components have been shown to contribute to virulence in models of pneumonia. However, the general lack of susceptibility of mice to S. aureus infection (3) has raised doubts about the applicability of the mouse data to human infection (4). Nonetheless, the Panton Valentin Leukocidin (5, 6), phenol-soluble modulins (7), and the α-hemolysin (8–10) may all contribute to the staphylococcal virulence in humans. However, staphylococcal toxins are generally expressed during the stationary phase of bacterial growth and may not be present in vivo in sufficient concentration during the early stages of pulmonary infection to participate in staphylococcal invasion (11).
In contrast, protein A (SpA) is a conserved surface protein of all S. aureus strains, highly expressed during the early stages of bacterial growth and abundantly shed from the cell surface (12). SpA has numerous interactions with host immune effectors binding TNF receptor 1 (TNFR1) (13), EGF receptor (EGFR) (14), IgG (15), and von Willebrand factor (16) as well as activating B cell clonal expansion (17). SpA also has a role in the pathogenesis of murine pneumonia because spa mutants are unable to establish pulmonary infection in a mouse model, and tnfr1-null mice, lacking the SpA receptor, are highly resistant to S. aureus pneumonia (18). In this model system, it was noted that despite the high intranasal inoculum, tnfr1-null mice, in contrast to the wild-type control animals, did not become bacteremic. These observations suggested that the ability of staphylococci to activate TNF signaling may be important in invasion across the tight junctions of the mucosal barrier.
It has been well established that TNF signaling has substantial effects on the cytoskeleton of many different cell types (19). The involvement of Rho family GTPases in cytoskeletal responses to TNF has been described in diverse cell types including macrophages, endothelial, and epithelial cells (20, 21). Ridley and co-workers noted that TNF treatment of human endothelial cells activated Rho GTPases leading to actomyosin contraction and the formation of intercellular gaps (13, 19). Changes in the distribution of actin, stress fiber formation, and actomyosin contraction are mediated by ROCK, the RhoA associated coiled-coil kinase that targets MLC (22). Myosin light chain kinase activation by itself is sufficient to induce changes in the epithelial barrier through actomyosin contraction and subsequent effects on tight junctions (23). Exactly how TNFR1 activates changes in the small GTPases is not well established but has been suggested to involve JNK and/or TRAF4 signaling (20, 24).
S. aureus SpA also activates EGFR which can contribute to actomyosin contraction (25, 26) as well as stimulating ERK1/2 and ADAM17, a metalloproteinase with sheddase function (14). The ERK MAPKs, also activated by TLR2 signaling, induce m-calpains in epithelial cells, proteases that cleave the transmembrane portion of the junctional proteins occludin and E-cadherin (27) and facilitate the transmigration of polymorphonuclear leukocytes (PMNs) to the airway (28). Thus, S. aureus has several epithelial targets that could potentially affect barrier function. In the experiments detailed in this report we used polarized human airway epithelial monolayers as well as mouse models of pneumonia and bacteremia to demonstrate that S. aureus protein A activates a RhoA/ROCK/MLC cascade (22) and that SpA+ organisms stimulate proteolytic activity to facilitate contraction of the epithelial cytoskeleton and translocation through paracellular junctions of the mucosal epithelium.
EXPERIMENTAL PROCEDURES
Cell Lines and Bacteria
16HBE cells (D. Gruenert, California Pacific Medical Center Research Institute, San Francisco, CA), were grown as previously detailed (28). S. aureus strain Newman wild type, spa mutant, and sortase mutants, or LAC USA300 MRSA were resuspended in 16HBE media without antibiotics (Cellgro MEM with 10% FCS) at a density of 108 cfu/ml. Escherichia coli BL21 (DE3) (Invitrogen) was used for expression of recombinant SpA proteins.
Bacterial Transmigration
16HBE cells were grown on 3-μm pore size Transwell-Clear filters (Corning-Costar) with an air-liquid interface to form polarized monolayers. 108 cfu/ml of Newman wild type, spa mutant, or sortase mutant was added to the apical compartment of the monolayer with or without exogenous recombinant full-length SpA (2.5 μm) or TNF (100 ng/ml) or TGFα (10 ng/ml). For inhibitor studies, monolayers were pretreated with EGFR inhibitor, BPDQ (50 μm), ERK1/2 inhibitor, U0126 (50 μm), JNK inhibitor, SP600125 (50 μm), p38 inhibitor, SB202190 (12 μm), calpain inhibitor, calpeptin (20 μm), TNFα-converting enzyme inhibitor, TAPI (50 μm), general protease inhibitor, GM6001 (20 μm) and ROCK inhibitor, Y-27632 (1 μm and 10 μm) for 1 h prior to addition of bacteria resuspended in the same concentration of inhibitor. 24 h after stimulation, bacterial counts (cfu/ml) were determined from the basolateral compartment.
Rho Activation Assay
Rho activation was determined using a G-LISA small G-protein activation assay (Cytoskeleton) and Rho activation assay kit (Millipore) according to the manufacturer's instructions. Briefly, for the Millipore assay, 16HBE cells were grown on 6-well plates to 80% confluence and stimulated with S. aureus. Cells were lysed, and active Rho GTP was isolated using Rhotekin RBD-agarose beads and immunoblotted using a monoclonal Rho antibody.
Western Blotting
16HBE cells were grown on 6-well plates to 50–60% confluence, changed to medium with 0.1% FBS overnight, and stimulated at various time points. Immunodetection of occludin and E-cadherin cleavage products was performed as previously described (28), and phospho-MLC detection was performed using a polyclonal phosphoserine (19) MLC antibody (Abcam) and a polyclonal MLC antibody (Cell Signaling) or monoclonal β-actin antibody (Sigma) as loading control.
Confocal Microscopy
16HBE cells were grown on 3-μm pore size Transwell-Clear filters with an air-liquid interface to form polarized monolayers. For confocal imaging cells were fixed with 4% paraformaldehyde, blocked with 5% normal donkey serum, and stained with phalloidin-rhodamine or phalloidin-Alexa Fluor 647 (Molecular Probes), monoclonal anti-S. aureus antibody (Abcam), and monoclonal anti-ZO-1 antibody (Zymed Laboratories Inc.) with secondary anti-mouse IgG Alexa Fluor 488 (Molecular Probes). After washing, filters were removed from Transwells using a scalpel, mounted with Vectashield Hard Set (Vector Laboratories) onto glass slides, and imaged using a Zeiss LSM 510 Meta scanning confocal microscope with a Plan-Neofluar 100×/1.3 oil objective at room temperature. Image acquisition and presentation were performed using Zeiss LSM Image Browser version 4.2. For biotin labeling experiments, 1 mg/ml EZ-Link Sulfo-NHS-LC-Biotin (Pierce) was added apically for 30 min at 4°C before fixing the cells. Alexa Fluor 555-conjugated streptavidin (Molecular Probes) was added for detection of biotin.
Dextran Permeability
Confluent monolayers of 16HBE cells, polarized at an air-liquid interface, were grown on 3-μm pore size Transwell-Clear filters. After apical stimulation with purified SpA from Sigma for 24 h, dextran-fluorescein (molecular weight 3,000) was added apically for 1 h and fluorescence in the basal compartment monitored at ex 485, em 535 on a SpectraFluor Plus fluorometer (Tecan).
Mouse Model of Infection
8-week-old female C57BL/6 wild-type mice (Jackson Laboratories) were anesthetized with ketamine/xylazine and inoculated intranasally with 50 μl of S. aureus (108 cfu) or PBS. Mice were pretreated with 15 mg/kg Y-27632 (in 20% dimethyl sulfoxide in PBS; Enzo Life Sciences) by intraperitoneal injection, 100 mg/kg gefitinib (Iressa; in 1% Tween 80 in PBS; Tocris Bioscience) by oral gavage, or 20 mg/kg calpeptin (in 25% dimethyl sulfoxide in PBS; EMD Bioscience) by intraperitoneal injection 24 and 1 h prior to intranasal inoculation. Bronchoalveolar lavage fluid and lungs were harvested 4–24 h after infection and used for PMN detection and determination of bacterial cfu/ml. Animal experiments were performed in accordance with the guidelines of the IACUC at Columbia University (protocol number AAAA7681).
PMN Detection
Lung cell suspensions were double stained with phycoerythrin-labeled anti-CD45 (to detect leukocytes) and fluorescein isothiocyanate (FITC)-labeled anti-Ly6C/Ly6G antibodies (to detect neutrophils) (BD Pharmingen) and analyzed by flow cytometry with a FACSCalibur using Cell Quest software (Becton Dickinson) (18). Irrelevant, isotype-matched antibodies were used as a control. Cells were gated on the basis of their forward scatter and side scatter profile and analyzed for double expression of CD45 and Ly6C/Ly6G.
Statistical Analysis
Samples with normal distribution were analyzed by Student's t test. For multiple comparisons an ANOVA followed by post hoc Dunnett's test was used to compare with the control group. Mouse samples were compared using the nonparametric Mann-Whitney test. Differences between groups were considered significant at p < 0.05. Statistical analysis was determined using GraphPad Instat version 3.0.
RESULTS
S. aureus Induces Changes in the Distribution of Actin and ZO-1 in Airway Cells
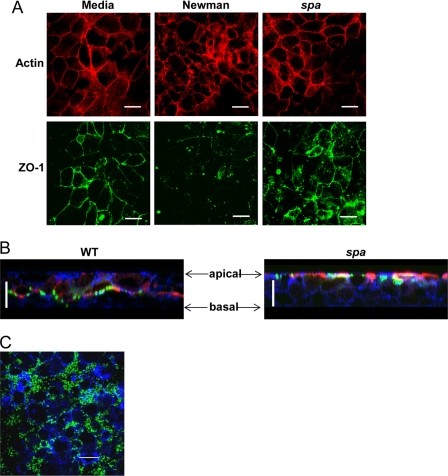
The effects of S. aureus on the cytoskeleton of polarized human airway epithelial cells grown at an air-liquid interface were examined using phalloidin staining. Under control conditions, these cells exhibit relatively uniform phalloidin staining, neatly outlining the borders of the epithelial cells in a typical “chicken wire” distribution. After 24 h of exposure to wild-type S. aureus strain Newman, the distribution of actin was much more heterogeneous, with clumping, regions of discontinuous borders, and overall a much more disorganized pattern of phalloidin staining (Fig. 1A). The distribution of the tight junction protein ZO-1 was substantially disrupted in response to the wild-type but not spa mutant staphylococci. Epithelial cells exposed to a Newman spa mutant were indistinguishable from the media-treated control. Although S. aureus Newman is a pathogenic strain, it lacks several virulence factors commonly expressed in the epidemic CA-MRSA USA300 clones. The effects of the LAC CA-MRSA S. aureus USA300 were visualized in z-sections of the same polarized 16HBE human airway epithelial cells (Fig. 1B). Using biotin as a marker for exposed proteins, MRSA are observed to penetrate through the paracellular junctions and traverse the monolayers in contrast to the spa mutant that remained at the apical surface. Biotin remains apical in the spa-exposed monolayers throughout the course of the experiment, whereas it is detected at the basal portions of the monolayers exposed to the wild-type organisms, suggesting that the perturbation of the cytoskeleton was enabling these nonmotile organisms to breach the tight junctions of the epithelial barrier. The MRSA primarily accumulated in the paracellular junctions and were not observed within the epithelial cells (Fig. 1C).
FIGURE 1.
S. aureus induces SpA-dependent changes in actin organization and tight junctions. A, 16HBE cells grown polarized on Transwells were stimulated with media, S. aureus strain Newman wild type (WT) or spa mutant. Cells were stained with phalloidin (red) or anti-ZO-1 (green), and x-y sections at comparable apical levels are shown. Scale bars, 20 μm. B, biotin (red) was added to polarized monolayers following stimulation, and cells were stained for phalloidin (blue) and S. aureus (green). z-sections are shown. Scale bars, 20 μm. C, cells were stained with anti-S. aureus (green) and phalloidin (blue). x-y sections from an apical layer are shown.
SpA Alters the Barrier Properties of Epithelial Monolayers
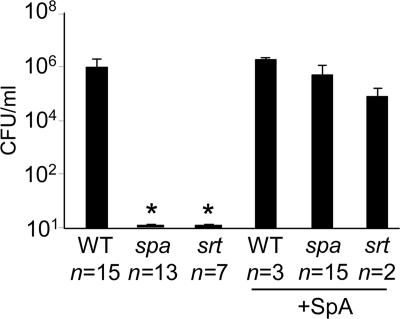
To determine whether the SpA-associated changes in the cytoskeleton were sufficient to enable bacterial passage across polarized airway epithelial cells we quantified transmigration of S. aureus strain Newman, a spa mutant, or a sortase mutant (lacking secreted surface proteins including protein A). The numbers of organisms recovered from the basal compartment of Transwells were quantified 24 h after an inoculum was added to the apical surface of human airway epithelial cells grown in a polarized fashion on Transwells (Fig. 2). Approximately 1 × 106 cfu/ml of a 1 × 109 cfu/ml inoculum of strain Newman were quantified in the basal compartment; whereas neither the spa nor sortase mutants crossed the monolayer during this period of time. Complementation of the spa or sortase mutants with exogenous purified SpA restored bacterial transmigration, virtually equivalent to that observed with the wild-type strain. Addition of LPS to account for any contamination of the recombinant SpA proteins produced in E. coli did not enhance transmigration (data not shown). Involvement of TRAF4, which has been reported to link TNFR1 and RhoA (29), was not found to be involved in the responses to SpA (data not shown).
FIGURE 2.
Staphylococcal transmigration across 16HBE monolayers. The number of organisms in the basal compartment of Transwells 24 h following apical inoculation with strain Newman wild type (WT), spa or sortase (srt) mutants or strains complemented with purified SpA are shown. *, p < 0.05. Error bars, S.D. n for each group is shown on the figure. Statistical analysis performed on data from three individual experiments.
SpA Signaling Activates Rho, ROCK, and MLC Activities
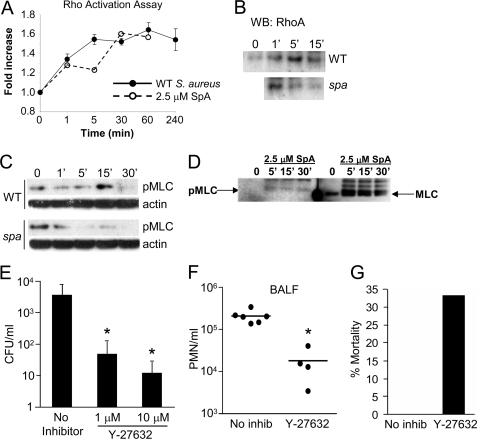
The IgG binding domain of SpA recognizes and initiates signaling through TNFR1 (13), which could be associated with effects on the cytoskeleton and alter epithelial permeability (30–32). As TNF activation induces actomyosin contraction through RhoA/ROCK/MLC-mediated signaling, we predicted that SpA signaling would similarly activate RhoA. In response to short term incubation with either wild-type S. aureus or purified SpA, we observed RhoA activation using a G-LISA assay (Fig. 3A). Wild-type S. aureus, but not a spa mutant, induced active Rho GTP detected by immunoblotting (Fig. 3B). Similarly, MLC phosphorylation was stimulated in the epithelial cells exposed to wild-type S. aureus but not the spa mutant (Fig. 3C), and SpA alone also induced MLC phosphorylation (Fig. 3D). To determine whether the induction of RhoA-MLC activity was required to facilitate staphylococcal translocation, we interrupted signaling with the ROCK inhibitor Y-27632 (33) and monitored effects on S. aureus translocation. Treatment of the monolayers with the ROCK inhibitor Y-27632 for 24 h resulted in a dose-dependent decrease in staphylococcal transmigration (Fig. 3E), confirming the participation of this pathway in the biologically relevant outcome. We also tested whether ROCK inhibition in vivo might prevent S. aureus invasive infection in a murine model of S. aureus pneumonia, but unfortunately systemic treatment of mice with Y-27632 also blocked PMN RhoA signaling in PMNs, inhibiting their mobility (Fig. 3F) and causing significantly increased morbidity and mortality (Fig. 3G). There were no untoward effects of the inhibitor on the mice, as determined by histopathology, nor did it affect the growth rate of the bacteria (data not shown).
FIGURE 3.
S. aureus activates the RhoA/ROCK/MLC cascade in airway epithelial cells. A, RhoA activation (-fold increase over media alone control) by S. aureus Newman wild type (WT) or 2.5 μm SpA (Sigma). B, immunoblot of activated RhoA complex in 16HBE cells exposed to WT or spa mutant bacteria. C, kinetics of induction (in minutes) of MLC phosphorylation induced by WT and spa mutant organisms. D, 2.5 μm SpA added to 16HBE cells under control conditions. E, staphylococcal transmigration across monolayers pretreated with the ROCK inhibitor Y-27632. *, p < 0.05. Data shown are n = 4, representative of two individual experiments. Error bars, S.D. F, C57BL/6 mice treated with the ROCK inhibitor Y-27632 18 h prior to infection with 108 CFU S. aureus strain Newman. At 18 h after inoculation PMNs in the lung were quantified by flow cytometry. G, mortality assessed compared with PBS-treated controls. *, p = 0.0095. Each dot represents one mouse, and lines represent the median for each group.
Neither Cytokines Alone nor Purified SpA Activates Changes in Tight Junctions
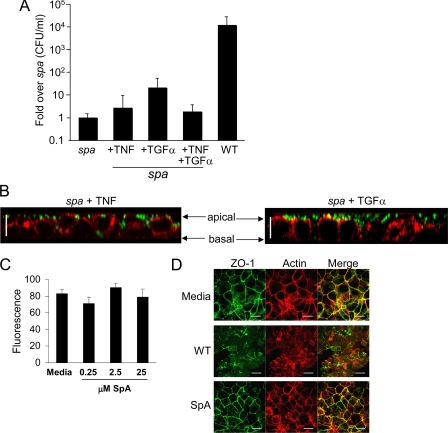
The IgG binding domain of SpA recognizes and activates both TNFR1 and EGFR (14), raising the possibility that the endogenous ligands for these receptors, namely TNF and TGFα, might have an effect on the barrier properties of the human airway epithelium. However, the addition of TNF did not complement the transmigration defect of the spa mutant, nor did the addition of TGFα, an EGFR agonist, or both cytokines together (Fig. 4A). The EGFR ligand EGF did not activate junctional changes either (data not shown). Treatment of the monolayers with a spa mutant plus either TNF or TGFα alone did not induce the changes in the cytoskeleton seen with wild-type organisms (Fig. 4B). Of note, treatment of the monolayers with purified SpA alone did not alter the junctions as detected by monitoring the transmigration of fluorescent dextran across monolayers (Fig. 4C) or by visualizing the distribution of ZO-1 and actin (Fig. 4D). These observations indicate that SpA is necessary but not sufficient to induce changes in the epithelial tight junctions.
FIGURE 4.
Effects of TNF, TGFα, and SpA on epithelial junctions. A, staphylococcal transmigration across monolayers stimulated with Newman spa complemented with TNF, TGFα, or TNF + TGFα compared with the wild-type (WT) strain. Data are shown in n = 8, representative of two individual experiments. Error bars, S.D. B, z-sections of polarized monolayers following stimulation for 20 h. Green, S. aureus; red, phalloidin. Scale bars, 20 μm. C, epithelial permeability across 16HBE monolayers measured by quantifying fluorescein-conjugated molecular weight 3,000 dextran translocation following stimulation with purified SpA at concentrations shown. D, cells stained with phalloidin (red) or anti-ZO-1 (green) with co-localization of actin and ZO-1 appearing yellow (Merge). x-y sections at comparable apical levels are shown. Scale bars, 20 μm.
SpA-EGFR Signaling Contributes to Staphylococcal Transmigration
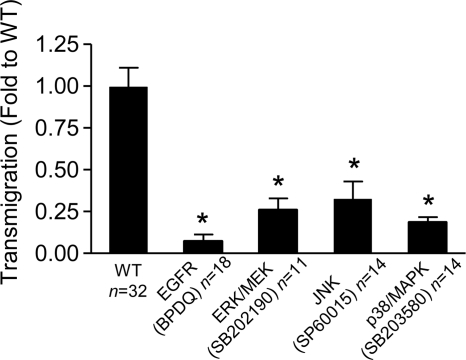
The SpA-EGFR interaction could activate a number of downstream pathways that affect the integrity of the epithelial barrier. EGFR signaling also activates RhoA/MLC activity (25), as well as stimulating protease expression. The EGFR-RhoA interaction is mediated by ERK1/2 MAPK, and guanine nucleotide exchange factor-H1 and may itself be activated by TNF (34). MAPKs are involved in actin remodeling and changes in barrier function (35). We tested the consequences of either EGFR or MAPK inhibition (comparing ERK1/2, JNK, and p38) inhibition on staphylococcal transmigration (Fig. 5). Treatment of the monolayers with BPDQ to block EGFR phosphorylation significantly inhibited transmigration. Inhibitors of the MAPKs (ERK1/2, p38, and JNK) also significantly blocked S. aureus translocation. These results are consistent with the involvement of MAPKs in the induction of RhoA activation. However, there are additional consequences of MAPK activation that also could alter the epithelial barrier.
FIGURE 5.
Staphylococcal transmigration is blocked by EGFR and MAPK inhibition. S. aureus strain Newman (WT) transmigration 24 h after inoculation across monolayers pretreated with EGFR inhibitor BPDQ, ERK1/2/MEK inhibitor UO126, JNK inhibitor SP600125, or p38/MAPK inhibitor SB202190 is shown. *, p < 0.05 Student's t test. n for each group is shown on figure, representative of at least two individual experiments. Error bars, S.D.
EGFR-induced Proteolytic Activity Modulates the Paracellular Junctions
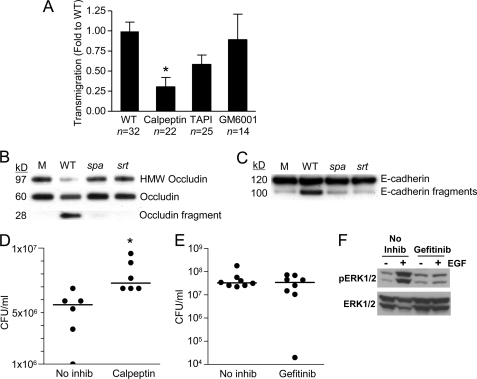
Immediately downstream of EGFR, ERK1/2 phosphorylation activates both TNFα-converting enzyme (ADAM17) proteolytic activity and the m-calpains (27). For staphylococci to invade through the paracellular junctions they must negotiate a web of membrane-spanning proteins involved in homotypic interactions between adjacent epithelial cells. At least two of these junctional proteins, occludin and E-cadherin, are substrates of the calpains (28). Treatment of the monolayers with the general calpain inhibitor calpeptin significantly blocked staphylococcal penetration through the epithelial monolayers (Fig. 6A), whereas a general protease inhibitor (GM6001), had no effect, and TAPI, which is specific for ADAM17, had only a modest effect on S. aureus transmigration. To confirm that S. aureus activates epithelial proteolytic activity that target these proteins, we screened lysates of airway epithelial cells following exposure to wild-type S. aureus, spa mutant, or sortase mutant, for the generation of the expected occludin and E-cadherin cleavage products, which would indicate the activation of calpains. Fragments corresponding to the extracellular domains of each of these proteins were detected in the epithelial cells exposed to the bacteria with SpA secretion, but not those exposed to the spa or sortase mutant (Fig. 6, B and C), suggesting that S. aureus-induced epithelial proteolytic activity contributes to bacterial translocation.
FIGURE 6.
S. aureus activates epithelial proteases. A, transmigration of S. aureus across monolayers treated with calpain inhibitor calpeptin, ADAM17 inhibitor TAPI, or a general protease inhibitor GM6001, 24 h after inoculation.*, p < 0.05 Student's t test. n for each group is shown on the figure, representative of at least two individual experiments. Error bars, S.D. B and C, generation of high molecular weight (HMW) occludin (B) and E-cadherin cleavage (C) products in Triton X-100-soluble lysates of polarized airway epithelial cells exposed to S. aureus Newman wild type (WT), spa or sortase (srt) mutants identified by immunoprecipitation and immunoblotting. D and E, CFU/ml recovered from bronchoalveolar lavage fluid of C57BL/6 mice pretreated with calpeptin (D) (*, p = 0.0043) or gefitinib (EGFR inhibitor) (E) 4 h after infection intranasally with 108 cfu S. aureus. Each dot represents one mouse, and lines represent the median for each group. F, gefitinib inhibits EGFR-dependent ERK1/2 phosphorylation in vivo. Mice were pretreated with gefitinib or vehicle (No inhib), then received an intraperitoneal injection of 1 μg of EGF or PBS, and 15 min later lungs were collected and prepared for immunoblotting. Phospho- and total ERK1/2 were detected by immunoblotting. Each lane represents one mouse; one independent experiment.
To confirm the participation of calpain activation in the pathogenesis of S. aureus pneumonia, mice were pretreated with calpeptin or a PBS control prior to intranasal inoculation with S. aureus USA300. Pretreatment with calpeptin resulted in significantly increased numbers of staphylococci in the bronchoalveolar lavage fluid compared with vehicle-treated controls (Fig. 6D), suggesting that inhibition of calpains interfered with staphylococcal invasion across the airway epithelium. Blocking EGFR signaling in vivo by pretreating mice with gefitinib (36) did not inhibit the invasive ability of USA300 (Fig. 6E), although we documented EGFR inhibition by demonstrating decreased induction of ERK1/2 phosphorylation (Fig. 6F). Thus, the ability of wild-type staphylococci to invade through the epithelial junction appears to require SpA-mediated induction of both TNF and EGFR signaling and activation of epithelial proteolytic activity.
DISCUSSION
S. aureus are tremendously successful human pathogens due at least in part, to their ability to adapt to immune responses in the host (15, 37). In the studies presented we found that S. aureus also exploit the epithelial machinery that regulates actomyosin dynamics and maintains the integrity of the epithelial barrier. Production of the abundant surface protein SpA that occurs during periods of active bacterial growth and before the production of secreted toxins, activates RhoA GTPase, ROCK, and MLC to perturb the epithelial cytoskeleton, a consequence of both TNF and EGFR activation. The Rho family of GTPases is affected by many bacterial pathogens that subvert the actomyosin machinery for their own motility and cellular invasion (30, 32). The Gram-negative mucosal pathogens, Salmonella, Shigella, Clostridia, Vibrio, Pseudomonas, pathogenic E. coli and others express a large number of genes devoted to the type III secretion system and effectors that target GTPases to enable bacteria to modify and exploit the host cell cytoskeleton (32, 38). This appears to be a widely conserved mechanism of pathogenesis as we now demonstrate that extracellular Gram-positive staphylococci also use host GTPases to promote mucosal invasion.
The SpA-TNF interaction by itself was not sufficient to allow staphylococcal invasion. TNF did not activate significant changes in the cytoskeleton of human respiratory epithelial cells, nor did TNF complement a spa mutant. This is reassuring, as it seems counterintuitive that the major proinflammatory cytokine in the airway would impair the integrity of the mucosal barrier possibly to enhance bacterial invasion. This is quite different from what has been observed in the gut, where TNF induces changes in tight junction permeability (39). In the respiratory epithelium, however, a more complex interaction appears to be necessary to change the barrier function of the epithelium. Our data suggest that TNF/EGFR/MAPK and Rho GTPase signaling are all likely to be involved in the response to S. aureus infection. Once S. aureus penetrates across the epithelial barrier, its ability to stimulate TNF signaling may be sufficient to perturb cell-cell junctions as has been demonstrated in other model systems, particularly in the gut (22, 23).
The ability of S. aureus to activate EGFR signaling is also important in facilitating transepithelial invasion. Not only does EGFR also stimulate the RhoA cascade (25), EGFR-mediated activation of epithelial proteases, particularly m-calpain through MAPK activity, contributed substantially to bacterial invasion across the tight junctions of the airway epithelium (27), demonstrated both in vitro and in vivo. However, although SpA alone was sufficient to activate RhoA signaling, by itself, it did not activate the proteolytic changes in the paracellular junctions unless expressed within the context of intact bacteria. The requirement for intact bacteria is likely to reflect the contribution of staphylococcal lipoproteins and TLR2 signaling that also activate calpain activity in airway epithelial cells airway (28). This is consistent with previous studies of SpA activation of eukaryotic cascades in which B cell activation also requires SpA to be expressed within the context of intact organisms (40). Our observations that S. aureus actively modify the paracellular junctions suggest that the potential spaces between adjacent epithelial cells are a preferred route for invasive infection, particularly early in pathogenesis. There are other routes for staphylococcal invasion. Staphylococcal endocytosis by pulmonary endothelial cells in vitro has been reported and would provide another route of bacterial entry into the lung parenchyma (41). In addition, organisms endocytosed by macrophages or neutrophils but not killed can be a source of infection disseminated to distant sites (42). However, even at other locations, SpA recognition of TNFR1 and EGFR in the endothelium or vasculature could similarly initiate changes in the cell-cell junctions that would facilitate staphylococcal penetration and spread across anatomical barriers.
It is increasingly apparent that much of the virulence of S. aureus is due to its sophisticated interactions with the host. Whereas secreted bacterial toxins, especially α-hemolysin, contribute to leukocyte damage (10), bacterial manipulation of host immune signaling; the TNF cascade (18), the IFN-β cascade (43), or the EGFR-MAPK cascade (14), and interference with B cell activation (17), all have roles in the pathogenesis of S. aureus pneumonia. Once the epithelial barrier is breached and bacteria reach the subepithelial matrix, there are abundant receptors for the many staphylococcal surface adhesins offering a nidus for continuing bacterial replication. Tissue damage from host proteolytic enzymes and reactive oxygen intermediates released by recruited PMNs expose matrix components to provide further binding sites and liberate iron, amino acids, and carbohydrates to foster bacterial growth. Staphylococcal invasion into the bloodstream, possibly facilitated by RhoA-activated cytoskeletal contraction and cleavage of junctional proteins, is a major complication of S. aureus pneumonia and an important cause of sepsis and multiorgan failure. Strategies to prevent the initial stages of staphylococcal invasion, particularly in patients identified to be at increased risk; those colonized with S. aureus, especially patients on a ventilator or with an antecedent significant viral infection, might be useful in preventing this potentially devastating infection.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL079395 (to A. P.).
- MRSA
- methicillin-resistant S. aureus
- BPDQ
- 4-[(3-bromophenyl)amino]-6,7-diaminoquinazoline
- EGFR
- EGF receptor
- MLC
- myosin light chain
- PMN
- polymorphonuclear leukocyte
- ROCK
- RhoA-associated coiled-coil kinase
- SpA
- protein A
- TNFR1
- TNF receptor 1.
REFERENCES
- 1. Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., Townes J. M., Craig A. S., Zell E. R., Fosheim G. E., McDougal L. K., Carey R. B., Fridkin S. K., and for the Active Bacterial Core surveillance, M. I. (2007) JAMA 298, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 2. Safdar N., Bradley E. A. (2008) Am. J. Med. 121, 310–315 [DOI] [PubMed] [Google Scholar]
- 3. Pishchany G., McCoy A. L., Torres V. J., Krause J. C., Crowe J. E., Jr., Fabry M. E., Skaar E. P. (2010) Cell Host Microbe 8, 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Otto M. (2011) Nat. Med. 17, 169–170 [DOI] [PubMed] [Google Scholar]
- 5. Labandeira-Rey M., Couzon F., Boisset S., Brown E. L., Bes M., Benito Y., Barbu E. M., Vazquez V., Höök M., Etienne J., Vandenesch F., Bowden M. G. (2007) Science 315, 1130–1133 [DOI] [PubMed] [Google Scholar]
- 6. Diep B. A., Chan L., Tattevin P., Kajikawa O., Martin T. R., Basuino L., Mai T. T., Marbach H., Braughton K. R., Whitney A. R., Gardner D. J., Fan X., Tseng C. W., Liu G. Y., Badiou C., Etienne J., Lina G., Matthay M. A., DeLeo F. R., Chambers H. F. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5587–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clarke S. R. (2010) Cell Host Microbe 7, 423–424 [DOI] [PubMed] [Google Scholar]
- 8. Bhakdi S., Tranum-Jensen J. (1991) Microbiol. Rev. 55, 733–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bubeck Wardenburg J., Schneewind O. (2008) J. Exp. Med. 205, 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bubeck Wardenburg J., Bae T., Otto M., Deleo F. R., Schneewind O. (2007) Nat. Med. 13, 1405–1406 [DOI] [PubMed] [Google Scholar]
- 11. Schmidt K. A., Manna A. C., Gill S., Cheung A. L. (2001) Infect. Immun. 69, 4749–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheung A. L., Eberhardt K., Heinrichs J. H. (1997) Infect. Immun. 65, 2243–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gómez M. I., O'Seaghdha M., Magargee M., Foster T. J., Prince A. S. (2006) J. Biol. Chem. 281, 20190–20196 [DOI] [PubMed] [Google Scholar]
- 14. Gómez M. I., Seaghdha M. O., Prince A. S. (2007) EMBO J. 26, 701–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Foster T. J. (2005) Nat. Rev. Microbiol. 3, 948–958 [DOI] [PubMed] [Google Scholar]
- 16. Hartleib J., Köhler N., Dickinson R. B., Chhatwal G. S., Sixma J. J., Hartford O. M., Foster T. J., Peters G., Kehrel B. E., Herrmann M. (2000) Blood 96, 2149–2156 [PubMed] [Google Scholar]
- 17. Goodyear C. S., Silverman G. J. (2003) J. Exp. Med. 197, 1125–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gómez M. I., Lee A., Reddy B., Muir A., Soong G., Pitt A., Cheung A., Prince A. (2004) Nat. Med. 10, 842–848 [DOI] [PubMed] [Google Scholar]
- 19. Wójciak-Stothard B., Entwistle A., Garg R., Ridley A. J. (1998) J. Cell. Physiol. 176, 150–165 [DOI] [PubMed] [Google Scholar]
- 20. Mathew S. J., Haubert D., Krönke M., Leptin M. (2009) J. Cell Sci. 122, 1939–1946 [DOI] [PubMed] [Google Scholar]
- 21. Mong P. Y., Petrulio C., Kaufman H. L., Wang Q. (2008) J. Immunol. 180, 550–558 [DOI] [PubMed] [Google Scholar]
- 22. McKenzie J. A., Ridley A. J. (2007) J. Cell. Physiol. 213, 221–228 [DOI] [PubMed] [Google Scholar]
- 23. Shen L., Black E. D., Witkowski E. D., Lencer W. I., Guerriero V., Schneeberger E. E., Turner J. R. (2006) J. Cell Sci. 119, 2095–2106 [DOI] [PubMed] [Google Scholar]
- 24. Sax J. K., El-Deiry W. S. (2003) J. Biol. Chem. 278, 36435–36444 [DOI] [PubMed] [Google Scholar]
- 25. Mateus A. R., Seruca R., Machado J. C., Keller G., Oliveira M. J., Suriano G., Luber B. (2007) Hum. Mol. Genet. 16, 1639–1647 [DOI] [PubMed] [Google Scholar]
- 26. den Hartigh J. C., van Bergen en Henegouwen P. M., Verkleij A. J., Boonstra J. (1992) J. Cell Biol. 119, 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glading A., Bodnar R. J., Reynolds I. J., Shiraha H., Satish L., Potter D. A., Blair H. C., Wells A. (2004) Mol. Cell. Biol. 24, 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chun J., Prince A. (2009) Cell Host Microbe 5, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kédinger V., Alpy F., Baguet A., Polette M., Stoll I., Chenard M. P., Tomasetto C., Rio M. C. (2008) PLoS One 3, e3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gruenheid S., Finlay B. B. (2003) Nature 422, 775–781 [DOI] [PubMed] [Google Scholar]
- 31. Clayburgh D. R., Barrett T. A., Tang Y., Meddings J. B., Van Eldik L. J., Watterson D. M., Clarke L. L., Mrsny R. J., Turner J. R. (2005) J. Clin. Invest. 115, 2702–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Finlay B. B. (2005) Curr. Top. Microbiol. Immunol. 291, 1–10 [DOI] [PubMed] [Google Scholar]
- 33. Liao J. K., Seto M., Noma K. (2007) J. Cardiovasc. Pharmacol. 50, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kakiashvili E., Dan Q., Vandermeer M., Zhang Y., Waheed F., Pham M., Szászi K. (2011) J. Biol. Chem. 286, 9268–9279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Usatyuk P. V., Natarajan V. (2004) J. Biol. Chem. 279, 11789–11797 [DOI] [PubMed] [Google Scholar]
- 36. Harari P. M., Allen G. W., Bonner J. A. (2007) J. Clin. Oncol. 25, 4057–4065 [DOI] [PubMed] [Google Scholar]
- 37. Fournier B., Philpott D. J. (2005) Clin. Microbiol. Rev. 18, 521–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soong G., Parker D., Magargee M., Prince A. S. (2008) J. Bacteriol. 190, 2814–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marchiando A. M., Shen L., Graham W. V., Weber C. R., Schwarz B. T., Austin J. R., 2nd, Raleigh D. R., Guan Y., Watson A. J., Montrose M. H., Turner J. R. (2010) J. Cell Biol. 189, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bekeredjian-Ding I., Inamura S., Giese T., Moll H., Endres S., Sing A., Zähringer U., Hartmann G. (2007) J. Immunol. 178, 2803–2812 [DOI] [PubMed] [Google Scholar]
- 41. Jarry T. M., Memmi G., Cheung A. L. (2008) Cell Microbiol. 10, 1801–1814 [DOI] [PubMed] [Google Scholar]
- 42. Thwaites G. E., Gant V. (2011) Nat. Rev. Microbiol. 9, 215–222 [DOI] [PubMed] [Google Scholar]
- 43. Martin F. J., Gomez M. I., Wetzel D. M., Memmi G., O'Seaghdha M., Soong G., Schindler C., Prince A. (2009) J. Clin. Invest. 119, 1931–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]