Abstract
The specific inhibition of the biosynthesis of target proteins is a relatively novel strategy in pharmacology and is based mainly on antisense approaches (e.g. antisense oligonucleotides or RNA interference). Recently, a novel class of substances was described acting at a later step of protein biosynthesis. The cyclic heptadepsipeptides CAM741 and cotransin were shown to inhibit selectively the biosynthesis of a small subset of secretory proteins by preventing stable insertion of the nascent chains into the Sec61 translocon complex at the endoplasmic reticulum membrane (Besemer, J., Harant, H., Wang, S., Oberhauser, B., Marquardt, K., Foster, C. A., Schreiner, E. P., de Vries, J. E., Dascher-Nadel, C., and Lindley, I. J. (2005) Nature 436, 290–293; Garrison, J. L., Kunkel, E. J., Hegde, R. S., and Taunton, J. (2005) Nature 436, 285–289). These peptides act in a signal sequence-discriminatory manner, which explains their selectivity. Here, we have analyzed the cotransin sensitivity of various G protein-coupled receptors in transfected HEK 293 cells. We show that the biosynthesis of the human endothelin B receptor (ETBR) is highly sensitive to cotransin, in contrast to that of the other G protein-coupled receptors analyzed. Using a novel biosynthesis assay based on fusions with the photoconvertible Kaede protein, we show that the IC50 value of cotransin action on ETBR biosynthesis is 5.4 μm and that ETBR signaling could be completely blocked by treating cells with 30 μm cotransin. Taken together, our data add an integral membrane protein, namely the ETBR, to the small group of cotransin-sensitive proteins.
Keywords: G Protein-coupled Receptors (GPCR), Protein Export, Protein Translocation, Receptor Regulation, Receptors
Introduction
Signal sequences play an important role during biogenesis and transport of secretory and membrane proteins (1–3). They bind the signal recognition particle and consequently mediate targeting of the ribosome-nascent chain-signal recognition particle complex to the translocon machinery of the ER2 membrane. After engaging the Sec61 translocon complex, signal sequences are also involved in translocon gating. In the case of secretory proteins, the signal sequences are located at the N terminus of the proteins (signal peptides) and cleaved off following ER translocation by the signal peptidases of the ER membrane. Integral membrane proteins may also possess N-terminal cleavable signal peptides. The majority of the proteins, however, possess signal sequences that are part of the mature protein. These “signal anchor sequences” are usually formed by the first transmembrane domain, but the other transmembrane domains may also have such a potential. In the case of opsin, for example, it was shown that five of the six transmembrane segments studied could function as a signal anchor sequence (4).
The translocon machinery must handle a broad range of secretory and membrane proteins, and due to this lack in specificity, its components were not considered to represent suitable drug targets. However, the recently described first inhibitors of the Sec61 translocon complex act in a surprisingly selective and signal sequence-discriminatory manner (5, 6). On the basis of the natural fungal substance HUN-7293 (7, 8), the derivative CAM741 was synthesized. CAM741 was shown to prevent selectively cotranslational translocation of the vascular cell adhesion molecule 1 (VCAM1) through the Sec61 translocon complex (5). More recently, it was shown that the vascular endothelial growth factor (VEGF) is also sensitive (9). Cotransin represents a simplified derivative of HUN-7293, which also blocks cotranslational VCAM1 translocation (6). In the same study, four other proteins were described to be cotransin-sensitive at low micromolar concentrations as follows: P-selectin, angiotensinogen, β-lactamase, and one integral membrane protein, namely the CRF1R (6).
The detailed mechanism of action of these cyclodepsipeptides is not fully understood. Although targeting of the nascent chain to the Sec61 translocon complex is not affected, productive interaction of the target signal sequences with the translocon is prevented, and the channel gating process and subsequent cotranslational translocation of the target protein are prohibited (6). Cross-linking studies indicate that the cyclodepsipeptides interact with the Sec61α core component (protein-conducting channel) of the Sec61 translocon complex (10) and enhance cross-linking of the nascent chains to the Sec61β component (5, 11). These data point toward a mechanism where the cyclodepsipeptides compete with the signal sequences for binding to a specific site within the protein-conducting channel (9).
Strikingly, selectivity of the substances is conferred by the signal sequences of the target proteins. Selectivity seems to be based on the fact that signal sequences do not have any sequence homologies (1, 2), although their conformation is conserved. However, it was not possible to define a consensus within the sensitive signal sequences as yet, although some critical residues have been described (11). It is also unknown whether the cyclodepsipeptides bind to the signal sequences directly.
The selective mechanism of action of CAM741 and cotransin raised the question whether specific derivatives interfering with biosynthesis of only a single protein may be found. A proof of principle was recently published for the CAM741 derivative NFI028; although CAM741 inhibits both the VCAM1 and VEGF biosynthesis, NFI028 is specific for VCAM1 (9).
Here, we have analyzed the effect of cotransin on the biosynthesis of various G protein-coupled receptors (GPCRs) possessing signal peptides or signal anchor sequences. Moreover, we have established a biosynthesis assay based on fusions with the Kaede protein from the stony coral Trachyphyllia geoffroy (12, 13). We show that the human endothelin B receptor (ETBR), a GPCR with a cleavable signal peptide, also belongs to the family of cotransin-sensitive proteins.
EXPERIMENTAL PROCEDURES
Materials
Cotransin was synthesized in our group using our previously described solid phase protocol (14) and dissolved in DMSO. [125I]ET-1 (2000 Ci/mmol) was purchased from Amersham Biosciences. The vector plasmids pEGFP-N1 (encoding the red-shifted variant of GFP) and the Tet-On gene expression system (containing the doxycycline-inducible vector plasmid pTRE-Tight and the HEK 293 Tet-On® advanced cell line) were from Clontech. The vector CoralHueTM pKaede-MN1 was purchased from MBL International (Woburn, MA). Primary-cultured mouse astrocytes were a gift from H. Kettenmann (Max-Delbrück-Zentrum für Molekulare Medizin, Berlin, Germany). The transfection reagent polyethyleneimine (PEI) was from Polysciences Inc. (Eppelheim, Germany). DNA-modifying enzymes were from New England Biolabs (Frankfurt am Main, Germany). Oligonucleotides were purchased from Biotez (Berlin, Germany). The RNeasy mini kit was from Qiagen (Hamburg, Germany). TRIzol, SuperScript II reverse transcriptase, and the oligo(dT)12–18 primer, and the AlamarBlue kit were purchased from Invitrogen. The TaqMan universal master mix and the TaqMan gene expression assay mix were from Applied Biosystems (Darmstadt, Germany). Trypan blue was purchased from Seromed (Berlin, Germany). The RotiLoad sample buffer was from Carl Roth (Karlsruhe, Germany). The polyclonal rabbit anti-GFP antiserum 02 (raised against a GST-GFP fusion protein) has been described previously (15). The monoclonal mouse anti-GFP antibody was purchased from Clontech. Horseradish peroxidase-conjugated anti-mouse IgG was purchased from Dianova (Hamburg, Germany). The 35-mm diameter ibidi® dishes were obtained from ibidi LLC (Martinsried, Germany). All other reagents were from Sigma.
DNA Manipulations
Standard DNA manipulations were carried out according to the handbooks of Sambrook and Russel (16). Nucleotide sequences of the plasmid constructs were verified using the FS Dye Terminator kit from PerkinElmer Life Sciences.
Plasmid Constructs
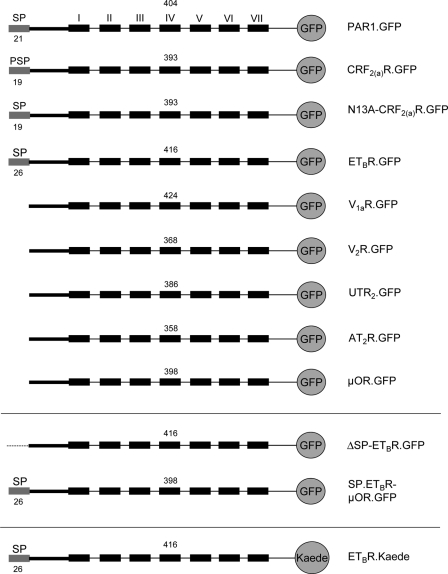
A GFP tag was C-terminally fused to various GPCRs with and without cleavable signal peptide by cloning the cDNAs of the receptors into vector plasmid pEGFP-N1 (thereby replacing the stop codons of the receptors; Fig. 1, details of the cloning procedure on request). The resulting constructs were PAR1.GFP, CRF2(a)R.GFP, N13A-CRF2(a)R.GFP, ETBR.GFP, V1aR.GFP, V2R.GFP, UTR2.GFP, AT2R.GFP, and μOR.GFP. A signal peptide deletion mutant for ETBR.GFP was constructed by deleting the sequence encoding the N-terminal 26 amino acid residues. The resulting construct was ΔSP-ETBR.GFP. In addition, the sequence encoding the signal peptide of the ETBR was fused to the μOR.GFP yielding construct SP.ETBR-μOR.GFP. The GFP tag of the ETBR was also exchanged for the photoconvertible Kaede protein by cloning the receptor cDNA into vector plasmid CoralHueTM pKaede-MN1.
FIGURE 1.
Schematic representation of the constructs used in this study (see the text for details). The indicated GPCRs were fused C-terminally with GFP or Kaede to quantify their expression by confocal LSM or by flow cytometry measurements. N-terminal signal peptides and their predicted size (program “SignalP 3.0”) (27, 28) are indicated by gray boxes. Roman numerals indicate transmembrane domains (only shown for the first construct); arabic numerals above each construct refer to the number of receptor amino acid residues present (without signal peptide).
Cell Culture and Transfection
HEK 293 cells were cultured at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% (v/v) fetal calf serum (FCS), penicillin (100 units/ml), and streptomycin (100 μg/ml). Transfection of the cells with plasmids and PEI was carried out according to the supplier's recommendations. Equal amounts of plasmid were transfected in each experiment allowing comparison of the receptor expression levels.
Quantitative Real Time PCR (qRT-PCR)
HEK 293 cells (3 × 105) grown on 6-well plates for 24 h were transiently transfected with 0.8 μg of plasmid DNA and PEI according to the supplier's recommendations. Cells were cultured for 5 h and afterward treated with cotransin (30 μm) or DMSO for another 17 h. Cells were cultured and washed with PBS (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.76 mm KH2PO4, pH 7.4), and total RNA was extracted with 500 μl of TRIzol reagent and 100 μl of chloroform and precipitated with 250 μl of isopropyl alcohol. The total RNA was washed with 500 μl of ethanol (70% v/v) and cleaned using the RNeasy mini kit. The cDNA was synthesized using 1 μg of total RNA, the SuperScript II reverse transcriptase, and the oligo(dT)12–18 primer according to the supplier's recommendations. The qRT-PCR was performed in an ABI7300 sequence detection system (Applied Biosystems) using 2 μl of cDNA, 12.5 μl of TaqMan 2× universal master mix, 1.25 μl of 20× TaqMan gene expression assay mix, and 9.25 μl of water (total volume = 25 μl/well). The thermocycler program was as follows: 2 min, 50 °C; 10 min, 95 °C; 40 cycles, 15 s, 95 °C; 1 min, 60 °C. Each assay was carried out in triplicate. GAPDH expression was used as internal control for normalization.
Cell Viability Assay
HEK 293 cells (1.3 × 104) were grown on 96-well plates in 100 μl of DMEM for 48 h. Cells were supplemented either with cotransin (0.1–100 μm) or puromycin (10−3–10−1 μg/ml) and incubated for another 17 h. The AlamarBlue dye (10 μl/well) was added, and cells were incubated for another 2 h at 37 °C. The absorbance of the reduced (metabolized) AlamarBlue dye was measured photometrically at 570 and 600 nm, and the A570/A600 difference was calculated.
[125I]ET-1 Binding and Displacement Assay
HEK 293 cells (8.7 × 105) grown for 24 h on 60-mm diameter dishes were transiently transfected with 1.6 μg of plasmid DNA and PEI according to the supplier's recommendations. 24 h after transfection, cells were treated with cotransin (30 μm) or DMSO and cultured for another 17 h. Cells were washed twice with PBS and resuspended in 1 ml of Tris-BAME buffer (50 mm Tris-HCl, 2 mm EGTA, 10 mm MgCl2, 1.4 μg/ml aprotinin, 0.2 mg/ml bacitracin, 3.2 μg/ml trypsin inhibitor, 0.5 mm PMSF, pH 7.2). Cells were disrupted using a Potter homogenizer and centrifuged at 26,000 × g for 35 min at 4 °C. Crude membranes were resuspended in 150 μl of Tris-BAME buffer, and [125I]ET-1 (diluted in 100 μl of the same buffer) was added to achieve the indicated concentrations. Nonspecific binding was determined in the presence of 1 μm unlabeled ET-1. Cells were incubated with the ligand for 2 h on ice (to avoid receptor internalization), washed quickly two times with ice-cold PBS, and lysed with 100 μl of 0.1 m NaOH. The lysate was transferred to tubes, and radioactivity was measured using a γ-counter. When using primary-cultured mouse astrocytes for the binding assay, ETBR-specific binding was guaranteed by subtracting values obtained in the presence of the ETBR-specific agonist IRL 1620 (Sigma) (1 μm) instead of unlabeled ET-1. The [125I]ET-1 displacement assay was carried out accordingly, but cotransin or unlabeled ET-1 (1 μm each) was added to compete for [125I]ET-1 (50 pm) in ligand binding.
Inositol Phosphate Accumulation Assay
HEK 293 cells (7.5 × 104) grown for 24 h on 24-well plates were transiently transfected with 0.5 μg of plasmid DNA and PEI according to the supplier's recommendations. Cells were cultured for 5 h and treated with cotransin (30 μm) or DMSO. Cells were cultured for another 17 h, and the inositol phosphate accumulation assay was carried out as described previously (17). All data were analyzed using the GraphPad Prism software (release 3.02; La Jolla, CA).
Immunoprecipitation of ETBR.GFP and Immunoblotting
HEK 293 cells (3 × 105) grown for 24 h on 6-well plates were transiently transfected with 0.8 μg of plasmid DNA and PEI according to the supplier's recommendations. Cells were cultured for 5 h and treated with cotransin (30 μm) or DMSO. Cells were cultured for another 17 h, washed twice with PBS, pH 7.4, and lysed for 1 h with 1 ml of lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 10 mm EGTA, 10 mm EDTA, 0.25% (w/v) SDS, 1% Igepal, 0.5 mm benzamidine, 1.4 μg/ml aprotinin, 3.2 μg/ml trypsin inhibitor, pH 8.0). Insoluble debris was removed by centrifugation (20 min, 20,000 × g). The supernatant was supplemented with polyclonal rabbit anti-GFP antiserum 02 coupled to protein A-Sepharose Cl-4B beads, and the sample was incubated overnight (beads were prepared by equilibrating 3.5 mg of the beads with lysis buffer and subsequent overnight incubation with 1 μl of polyclonal rabbit anti-GFP antiserum 02). GFP-tagged receptors were precipitated, and the beads were washed twice with 1 ml of washing buffer (50 mm Tris-HCl, 500 mm NaCl, 10 mm EDTA, 10 mm EGTA, 0.25% (w/v) SDS, 1% Igepal, 0.5% (v/v), pH 8.0) and once with 1 ml of PBS. EndoH and PNGaseF digestions were performed according to the supplier's recommendations. Samples were supplemented with RotiLoad sample buffer, incubated for 5 min at 95 °C, and analyzed by SDS-PAGE/immunoblotting (8% acrylamide) using a monoclonal mouse anti-GFP antibody and horseradish peroxidase-conjugated anti-mouse IgG. Immunoblotting was carried out as described previously (18).
Quantification of the GFP-tagged GPCRs by Flow Cytometry Measurements
HEK 293 cells or COS-7 cells (1.8 × 105) grown for 24 h on 12-well plates were transiently transfected with 1.2 μg of plasmid DNA and PEI according to the supplier's recommendations. Cells were cultured for 5 h and treated with cotransin (GPCR cotransin sensitivity assay, 10 μm; signal peptide specificity assay, 30 μm) or DMSO. Cells were cultured for another 17 h and washed twice with PBS. The GFP signals of the receptors were analyzed by flow cytometry analysis using a FACSCalibur apparatus (BD Biosciences). For each sample, total fluorescence intensity of 1 × 104 cells was analyzed using the BD CellQuest Pro software (release 6.0; BD Biosciences). The fluorescence background of vector-transfected cells was subtracted. Data of the cotransin-treated cells were expressed in % of the DMSO control.
Modulation of Receptor Expression Using the Tet-On System
We used the Tet-On gene expression system from Clontech for our study. Construct ETBR.GFP was cloned into the vector plasmid pTRE-Tight and the HEK 293 Tet-On advanced cell line was transiently transfected with the resulting construct. Receptor expression was induced by doxycycline according to the supplier's recommendations. The GFP fluorescence signals of the receptors were quantified by flow cytometry measurements as described above.
Localization of ETBR.GFP Using Confocal LSM
HEK 293 cells (2.5 × 105) grown for 24 h in a 35-mm diameter dish were transfected with 0.8 μg of plasmid DNA and PEI according to the supplier's recommendations. Cells were cultured for 5 h and treated with cotransin (30 μm) or DMSO. Cells were cultured for another 17 h, washed twice with PBS, and transferred immediately, between two coverslips, into a self-made chamber (details on request). For colocalization of the receptor's GFP signals with the plasma membrane trypan blue, live cells were covered with PBS, and trypan blue was added in a final concentration of 0.05% (v/v). After 1 min of staining, GFP and trypan blue signals were visualized at room temperature using a Zeiss LSM510-META invert confocal laser scanning microscope (objective lens, ×100/1.3 oil; optical section, <0.9 μm; multitrack mode; GFP, λexc, 488 nm, argon laser; BP filter, 500–530 nm; trypan blue, λexc, 543 nm, HeNe laser, LP filter, 560 nm). The overlay of both signals was computed using the Zeiss LSM510 software (release 3.2; Carl Zeiss AG, Jena, Germany). Images were imported into Photoshop software (release 10.0; Adobe Systems Inc., San Jose, CA), and contrast was adjusted to approximate the original image. To quantify GFP signals, the signal intensities at the plasma membrane and in the cell's interior were measured using the 8-bit gray scale (ranging from 0 to 255) provided by the LSM510 software (n = 16–27 cells).
Quantification of ETBR.Kaede Biosynthesis by Kaede-based Assays
Microscopic Single Cell Assay
HEK 293 cells (2.5 × 105) grown for 24 h in a 35-mm diameter ibidi dish were transfected with 0.8 μg of plasmid DNA and PEI according to the supplier's recommendations. After 24 h of incubation, cells were incubated with cotransin (30 μm, 3 h), and dishes were transferred to a 37 °C microscope chamber. Cells were covered in phenol red-free DMEM with FCS. The gKaede and rKaede signals of the receptors were analyzed on a Zeiss LSM510-META inverted confocal LSM in a time series using different channels (multitrack mode; objective lens, ×100/1.3 oil; optical section, <2.0 μm; λexc gKaede, 488 nm, argon laser, BP filter, 505–530 nm; λexc rKaede,= 543 nm, argon laser, LP filter, 560 nm). Images were imported into Photoshop 10.0 software (Adobe Systems Inc, San Diego), and the contrast was adjusted to approximate the original image. To measure biosynthesis, the gKaede fluorescence signals of the receptors were photoconverted to rKaede by irradiation of the cells by a conventional UV lamp for 10 s. The gKaede and rKaede signals were recorded in a time series (180 min total; one picture every 15 min) using different channels as described above. The reappearance of the gKaede signals at the plasma membrane was also analyzed quantitatively for single cells as described (15).
Flow Cytometry Assay
HEK 293 cells (1.8 × 105) grown for 24 h on 12-well plates were transiently transfected with 1.2 μg of plasmid DNA and PEI according to the supplier's recommendations. Cells were cultured for 24 h; the gKaede signals were photoconverted to rKaede for 4 min using a 50-watt mercury UV lamp, and cells were supplemented with different cotransin concentrations (0.1–100 μm) or DMSO. Cells were cultured for another 17 h, washed twice with PBS, and the gKaede signals of 1 × 104 cells were analyzed by flow cytometry analysis using a FACSCalibur apparatus and the CellQuest Pro software (release 6.0; BD Biosciences). The fluorescence background of vector-transfected cells and residual gKaede signals present immediately after photoconversion were subtracted. Data of the cotransin-treated cells were expressed in % of the respective untreated sample.
RESULTS
Biosynthesis of the ETBR Is Cotransin-sensitive
GPCRs follow one of two different ER insertion pathways. Some GPCRs possess cleavable N-terminal signal peptides like secretory proteins that are cleaved off following signal peptide-mediated integration of the receptors into the ER membrane. The majority of GPCRs do not possess signal peptides, and a transmembrane domain of the mature receptor (usually TM1) must take over signaling functions as a so-called signal anchor sequence. Here, we have assessed the influence of cotransin on the biosynthesis of various GPCRs with and without cleavable signal peptides.
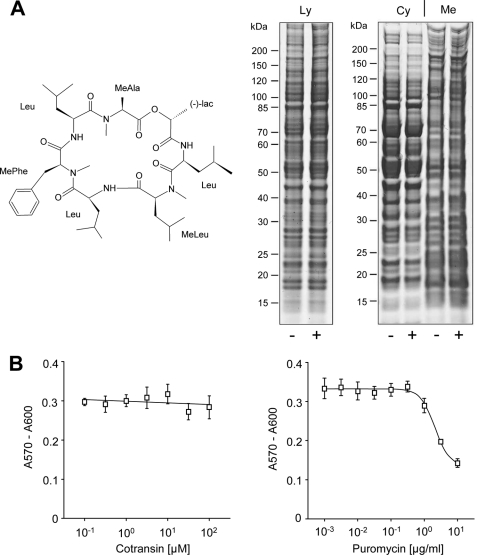
We first studied the toxicity of cotransin in our HEK 293 cell model. Untransfected cells were treated with 30 μm cotransin for 17 h, and total lysates as well as fractionated cytosolic and membrane proteins were separated and analyzed by SDS-PAGE (Fig. 2A, right panel). In neither case was the pattern of detectable proteins significantly affected by cotransin treatment confirming the previously described selective action of this peptide on protein synthesis (6). Cytotoxicity was analyzed by treating HEK 293 cells for 17 h with increasing concentrations of cotransin (0.1-100 μm) and by measuring metabolic reduction of the preloaded AlamarBlue REDOX indicator. No cytotoxicity was observed with cotransin concentrations up to 100 μm (Fig. 2B, left panel). As a control for the functionality of the cell viability assay, the drug puromycin was used and a toxic effect was observed starting at a concentration of 10 μg/ml (Fig. 2B, right panel).
FIGURE 2.
Cotransin toxicity assays. A, chemical structure (left panel) and SDS-PAGE analysis (right panel) of HEK 293 cell proteins. Cells were treated with 30 μm cotransin (+) or DMSO (−). Total lysates (Ly) or cytosolic (Cy) and crude membrane proteins (Me) were loaded on a SDS gradient gel (4–12%). B, cell viability assay. Following treatment of cells with increasing concentrations of cotransin (left panel) or puromycin (right panel), HEK 293 cells were preloaded with AlamarBlue redox indicator, and its metabolic reduction was analyzed (measurement wavelength λexc, 570 nm; reference wavelength λem, 600 nm). Data points indicate cell viability and represent mean values of the A570/A600 difference of three independent experiments each performed in triplicate (±S.D.).
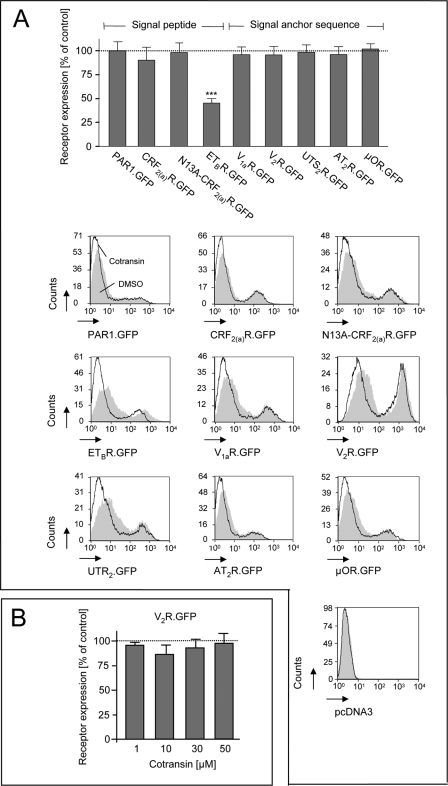
For a pilot study analyzing cotransin sensitivity of GPCRs, we used nine receptors possessing various types of GPCR signal sequences (see Fig. 1 for the constructs). Cleavable signal peptides are present in the case of the PAR13 and the ETBR (19). The CRF2(a)R contains an N-terminal pseudo signal peptide that is unable to mediate ER targeting, remains uncleaved, and forms part of the mature receptor (20). The N13A mutant of the CRF2(a)R converts the pseudo signal peptide into a conventional and cleaved signal peptide (20). The V1aR, the V2R, the UTR2, the AT2R, and the μOR do not possess cleavable signal peptides and must possess instead putative internal signal anchor sequences. To analyze cotransin sensitivity, all receptors were C-terminally tagged with GFP (thereby deleting their stop codons) to facilitate their detection and to compare their expression levels. Transiently transfected HEK 293 cells expressing the receptor constructs were treated for 17 h with 10 μm cotransin, and receptor expression was quantified by flow cytometry measurements of the GFP fluorescence signals (Fig. 3A). In the case of the ETBR, the amount of detectable GFP signals was decreased substantially to 45% of the DMSO control, indicating a significant inhibition of receptor biosynthesis following cotransin treatment. Expression of all the other GPCRs studied was unaffected. Even when using higher cotransin concentrations up to 50 μm, receptor expression was not influenced significantly in the case of construct V2R.GFP (Fig. 3B).
FIGURE 3.
A, GPCR cotransin sensitivity assays using transiently transfected HEK 293 cells. Upper panel, flow cytometry measurement of the GFP fluorescence signals of various GPCRs following cotransin treatment (10 μm, 17 h; see the text and Fig. 1 for the details of the constructs). Columns represent mean values of the GFP fluorescence signals of 1 × 104 cells of three independent experiments (±S.D.) and are expressed in % of the DMSO control (***, p < 0.001; one-sample two-tailed t test). Lower panel, flow cytometry histograms of a single representative experiment for each GPCR. The cell counts were plotted against the GFP fluorescence intensities. Gray areas represent the DMSO-treated control cells; white areas below the black lines represent the cotransin-treated cells. Signal intensities below 10 result from nontransfected or weakly transfected cells. Vector transfected cells were used as a control (pcDNA3). B, flow cytometry measurement of the GFP fluorescence signals of V2R.GFP following treatment with increasing cotransin concentrations (10–50 μm, 17 h). Columns represent mean values of the GFP fluorescence signals of 1 × 104 cells of three independent experiments (±S.D.) and are expressed in % of the DMSO control (differences were not significant in one-sample two-tailed t and ANOVA tests).
In the flow cytometry diagrams of the individual experiments shown in Fig. 3A, two peaks were visible for each receptor construct. The second peak represents the fluorescence of transfected cells and was substantially influenced by cotransin treatment only in the case of the ETBR.GFP (see above). The first peak consists of a cotransin-insensitive autofluorescence signal (fluorescence intensities <10; see the vector control) and signals of transfected cells expressing the receptors at a very low level. These weak receptor fluorescence signals were decreased in the case of all studied receptors following cotransin treatment. Thus, cotransin may also have a (albeit very weak) nonselective effect on protein biosynthesis, which is readily superimposed once a sensitive signal sequence is present.
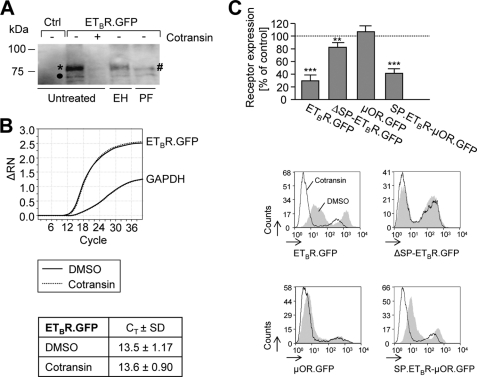
To confirm the results that the ETBR is a highly cotransin-sensitive protein, construct ETBR.GFP was precipitated from cotransin-treated transiently transfected HEK 293 cells (30 μm, 17 h) and detected by SDS-PAGE/immunoblotting (Fig. 4A). Two immunoreactive protein bands were detectable. The upper stronger protein band represents the mature ETBR.GFP (* in Fig. 4A, apparent molecular mass ∼75 kDa) (21); the lower faint protein band (● in Fig. 4A) represents a degradation product resulting from N-terminal receptor cleavage by an endogenous metalloprotease that could not be completely removed even upon EGTA/EDTA treatment (10 mm each) (21). Cotransin treatment leads to a substantial decrease of the ETBR receptor expression consistent with the above results. Treatment with EndoH did not influence the apparent molecular mass of the upper protein band, whereas treatment with PNGaseF reduces the upper band to the nonglycosylated form (# in Fig. 4A) demonstrating that the receptor possesses the previously described N-glycosylation in the N-tail increasing the apparent molecular mass only to a minor extent (∼3 kDa) (21). The fact the lower faint band (● in Fig. 4A) is not sensitive to either EndoH or PNGaseF demonstrates that this band does not represent the high mannose form of the receptor but is indeed the cleavage product.
FIGURE 4.
Cotransin action on protein and mRNA expression levels of ETBR.GFP in transiently transfected HEK 293 cells and signal peptide specificity of cotransin. A, SDS-PAGE/immunoblot analysis. Cells were treated with cotransin (30 μm, 17 h) (+) or with DMSO (−). Receptors were precipitated using an anti-GFP antiserum and detected using a monoclonal anti-GFP antibody and horseradish peroxidase-conjugated anti-mouse IgG. Nontransfected HEK 293 cells were used as a control for antibody specificity (Ctrl). Treatment of the receptors with EndoH (EH) or PNGaseF (PF) was used to identify the complex (*) and nonglycosylated (#) receptor forms. An additional faint band (●) represents a degradation product of ETBR.GFP resulting from cleavage by an endogenous metalloprotease (21). The immunoblot is representative for three independent experiments. B, qRT-PCR analysis of ETBR.GFP mRNA expression following cotransin treatment. Cells were treated with DMSO or cotransin (30 μm, 17 h), and mRNA levels were amplified by qRT-PCR. The graphs show the data of one out of three independent experiments. The overlapping solid and dashed lines represent the mRNA amount of ETBR.GFP and GAPDH of DMSO and cotransin-treated cells, respectively. The table shows the corresponding CT values (±S.D.). Nontransfected HEK 293 cells were used as a control yielding a similar curve only in the case of GAPDH (data not shown). C, upper panel, flow cytometry measurements of the GFP fluorescence signals of the ETBR.GFP, ΔSP-ETBR.GFP, μOR.GFP, and SP.ETBR-μOR.GFP following cotransin treatment (30 μm, 17 h; see the text and Fig. 1 for the details of the constructs). Columns represent mean values of the GFP fluorescence signals of 1 × 104 cells of three independent experiments (±S.D.) and are expressed in % of the DMSO control (**, p < 0.01; **, p < 0.01; one-sample two-tailed t test). Lower panel, flow cytometry histograms of a single representative experiment for each construct. The cell counts were plotted against the GFP fluorescence intensities. Gray areas represent the DMSO-treated control cells, the white areas below the black lines represent the cotransin-treated cells. Signal intensities below 10 result from nontransfected or weakly transfected cells.
It has been shown previously that cotransin interferes with the Sec61 translocon complex in a signal sequence-discriminatory mechanism; transcription of the mRNAs of the target proteins was unaffected (6). To study whether this holds true for the ETBR, we isolated mRNA from transiently transfected HEK 293 cells expressing ETBR.GFP following cotransin treatment (30 μm, 17 h) and performed qRT-PCR experiments. Consistent with the above results, no decrease in ETBR transcription was observed (Fig. 4B). To demonstrate that the cotransin effect is mediated by the signal peptide of the ETBR, a signal peptide deletion mutant was constructed (construct ΔSP-ETBR.GFP; see Fig. 1). Moreover, the ETBR signal peptide was fused N-terminally with the μOR, which is normally cotransin-resistant (resulting construct SP.ETBR-μOR.GFP, Fig. 1). HEK 293 cells were transiently transfected with the constructs and treated with cotransin (17 h, 30 μm), and the GFP fluorescence signals were quantified using flow cytometry measurements (Fig. 4C). The wild type construct ETBR.GFP was again cotransin-sensitive. Deletion of the signal peptide (construct ΔSP-ETBR.GFP) leads to a strong decrease in cotransin sensitivity. Moreover, fusion of the signal peptide to the μOR transfers cotransin sensitivity to this signal anchor sequence receptor demonstrating that the cotransin-mediated inhibition of ETBR expression is solely conferred by the signal peptide of the receptor.
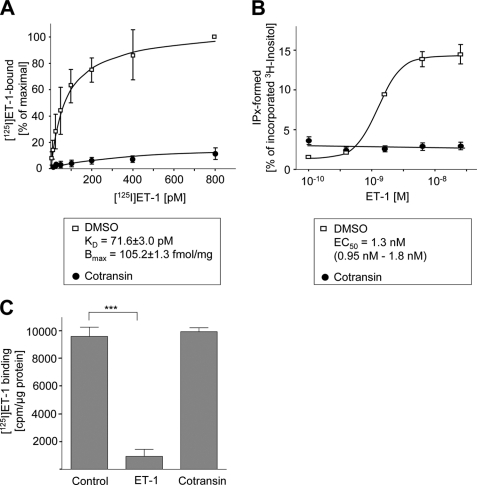
We next analyzed the pharmacological properties of the ETBR.GFP in transiently transfected HEK 293 cells following cotransin treatment. [125I]ET-1 binding profiles of intact cells were recorded using cotransin-treated (30 μm, 17 h) or DMSO-treated cells (Fig. 5A). In the case of the control cells, a typical saturation curve was observed with KD (71.6 ± 3.0 pm) and Bmax (105.2 ± 1.3 fmol/mg) values for [125I]ET-1 binding similar to previously published results (22). Following cotransin treatment, [125I]ET-1-binding sites were only barely detectable, and KD and Bmax values could consequently not be calculated. We next measured inositol phosphate accumulation in transiently transfected HEK 293 cells following ET-1 stimulation (Fig. 5B). In the case of the control cells, a typical concentration-response curve was observed with an EC50 value of 1.3 nm (95% confidence limits, 0.95-1.8 nm) similar to previously published results (22). Following cotransin treatment, however, second messenger formation was completely abolished. To demonstrate that cotransin does not interfere with the ligand binding site of the receptor, a [125I]ET-1 displacement binding assay was performed using unlabeled ET-1 or cotransin (1 μm each) to compete for [125I]ET-1 binding (Fig. 5C). Binding of the radioligand was only impaired by ET-1 demonstrating formally that cotransin is not a competitive antagonist for the ETBR.
FIGURE 5.
Pharmacological properties and subcellular localization of ETBR.GFP in transiently transfected HEK 293 cells following cotransin treatment. A, specific [125I]ET1 binding profiles of crude membranes from cotransin-treated (30 μm, 17 h) or DMSO-treated cells. Data points represent mean values of three independent experiments each performed in triplicate (±S.D.). The calculated KD and Bmax values for DMSO-treated cells are indicated. B, ET-1-mediated inositol phosphate accumulation in intact cotransin-treated (30 μm, 17 h) or DMSO-treated cells. Data points represent mean values of three independent experiments each performed in triplicate (S.D.). The calculated EC50 value for the DMSO-treated cells is indicated. C, [125I]ET-1 displacement assay. A binding assay was performed as described in A using 50 pm of the radioligand (Control). Unlabeled ET-1 or cotransin (1 μm each) was added to compete for [125I]ET-1 binding. Columns represent mean values of specific [125I]ET-1 binding of three independent experiments each performed in triplicate (±S.D.) (***, p < 0.001; one-sample two-tailed t test).
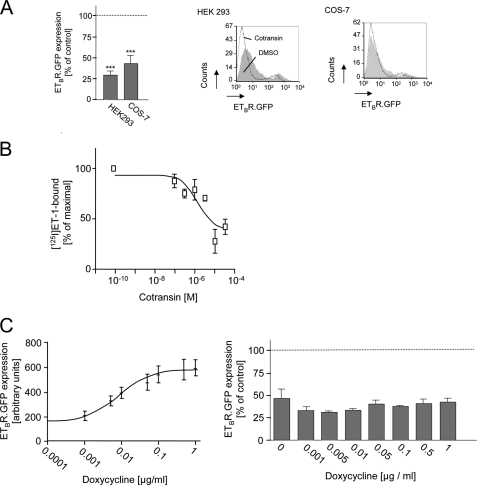
We next studied whether cotransin inhibits ETBR expression in other cell types, too. Transiently transfected HEK 293 or COS-7 cells expressing ETBR.GFP were treated for 17 h with 30 μm cotransin, and receptor expression was quantified by flow cytometry measurements of the GFP fluorescence signals (Fig. 6A). ETBR expression was decreased to a similar extent in both cell types demonstrating that the results are not cell type-specific. Moreover, cotransin treatment (17 h) decreased [125I]ET-1 binding to the endogenous ETBR present in primary cultured astrocytes (23) in a concentration-dependent manner (Fig. 6B) (IC50 = 1.3 ± 0.4 μm). In addition, we studied whether the cotransin effect in HEK 293 cells is influenced by receptor expression levels. HEK 293 Tet-On advanced cells were transiently transfected with ETBR.GFP cloned in the vector plasmid pTRE-Tight, and receptor expression was quantified by flow cytometry measurements of the GFP fluorescence signals. Induction of the cells with doxycycline strongly induced receptor expression (Fig. 6C, left panel). However, cotransin-mediated inhibition of ETBR biosynthesis (30 μm, 17 h) remained constant under these conditions (Fig. 6C, right panel) indicating that receptor expression levels do not influence cotransin sensitivity within the limits of HEK 293 cells.
FIGURE 6.
Influence of cell type and receptor expression levels on the cotransin sensitivity of ETBR.GFP. A, left panel, flow cytometry measurement of the GFP fluorescence signals of ETBR.GFP in cotransin-treated (30 μm, 17 h) HEK 293 or COS-7 cells. Columns represent mean values of three independent experiments (±S.D.) and are expressed in % of the DMSO control (***, p < 0.001; one-sample two-tailed t test). Right panel, flow cytometry histograms of a single representative experiment for each cell type. The cell counts were plotted against the GFP fluorescence intensities. Gray areas represent the DMSO-treated control cells; the white areas below the black lines represent the cotransin-treated cells. Signal intensities below 10 result from nontransfected or weakly transfected cells. B, specific [125I]ET-1 binding profiles of crude membranes from cotransin-treated (30 μm, 17 h) or DMSO-treated primary cultured astrocytes. ETBR-specific binding was guaranteed by subtracting values obtained in the presence of the ETBR-specific agonist IRL 1620 (29). Data points represent mean values of two independent experiments each performed in triplicate (±S.D.). C, influence of receptor expression levels on cotransin sensitivity in transiently transfected HEK 293 Tet-On advanced cells. Left panel, receptor expression following induction with doxycycline. Data points represent mean values of the GFP fluorescence signals (arbitrary units) of 1 × 104 cells of three independent experiments (±S.D.). Right panel, cotransin-mediated inhibition of ETBR biosynthesis (30 μm, 17 h) in doxycycline-induced cells. Columns represent mean values of the GFP fluorescence signals of 1 × 104 cells of three independent experiments (±S.D.) and are expressed in % of the DMSO control (differences were not significant in a one-sample two-tailed t test).
Taken together, our results show that the ETBR can be classified among the small number of cotransin-sensitive proteins. Moreover, complete prevention of ETBR signaling following cotransin treatment indicates that the substance may be of pharmacological significance in the case of this receptor.
In the flow cytometry quantification and immunoprecipitation experiments using HEK 293 cells, ETBR.GFP signals were still detectable following cotransin treatment, albeit in low amounts. The even more sensitive ligand binding and second messenger accumulation experiments in HEK 293 cells indicate, however, that no functional receptors were present under these conditions (Fig. 5, A and B). This phenomenon might be explained by the presence of an intracellular receptor population that does not contribute to functional receptors (e.g. aggregates). Indeed, subcellular localization of the GFP fluorescence signals of ETBR.GFP by confocal LSM revealed the presence of a stable intracellular receptor population following cotransin treatment (30 μm, 17 h; data not shown). Assuming that this receptor population is formed by pre-existing receptors that have a long half-life, their insensitivity to cotransin is easily explicable.
Cotransin-insensitive proteins with a long half-life would represent an experimental problem, in particular in the case of integral membrane proteins because they cannot be washed out like secretory proteins prior to cotransin treatment. Thus, simple immunoprecipitation experiments and flow cytometry measurements of GFP fluorescence signals are not the optimal solution for deriving concentration-response curves for cotransin action, for example. Conventional pulse-chase experiments may help to overcome this problem. They are very extensive, however, when using different cotransin concentrations. Ligand binding displacement assays, as shown in Fig. 6B, are expensive and specific for only one target protein. Taking these considerations into account, we decided to develop biosynthesis assays that could be easily used to measure cotransin effects on different integral membrane proteins independent of long half-life protein populations.
Establishment of Biosynthesis Assays to Quantify Cotransin Action on ETBR Biosynthesis
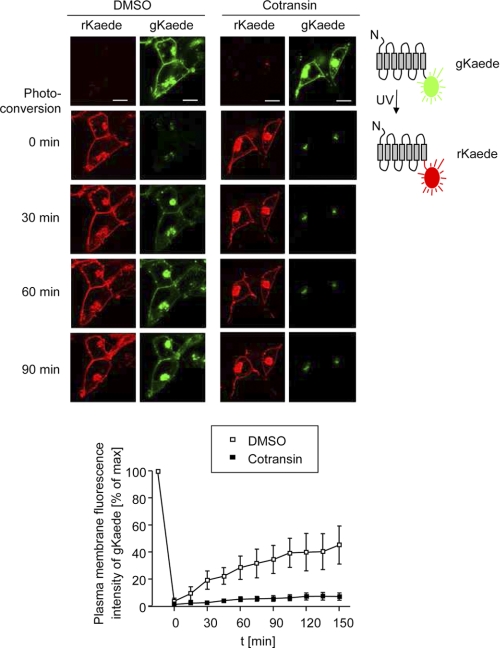
We have previously shown that the nonbioluminescent fluorescent protein Kaede from the stony coral T. geoffroy can be fused with GPCRs and be used to localize receptors within cells (13). In this recent study, C-terminally fused Kaede neither influenced GPCR trafficking nor receptor pharmacology. The advantage of Kaede in comparison with the widely used GFP derivatives is that its fluorescence can be photoconverted from green (gKaede signals) to red (rKaede signals) following UV irradiation, due to rearrangements in its chromophore (24). This property allows us, for example, to visualize trafficking of Kaede fusion proteins between different subcellular compartments using a confocal LSM. Once the Kaede fusion protein has reached a particular region of the cell, its fluorescence can be switched from green to red, and its trafficking to the target compartment can be easily studied by monitoring the new fluorescence (13). We assumed that a modification of this technique would also be suitable to measure biosynthesis of integral membrane proteins or its inhibition. In such an experiment, the gKaede signals of the target proteins are completely photoconverted at time t = 0 to rKaede. Time-dependent reappearance of the gKaede signals by receptor biosynthesis can then be measured microscopically in real time using single cells (“pulse-chase microscopy”). A proof of principle of such a methodology was recently published using the tagged KV1.1 channel (25). A pulse-chase microscopy experiment for transiently transfected HEK 293 cells expressing ETBR.Kaede is shown in Fig. 7; cells were pretreated with cotransin (30 μm, 3 h) or left untreated, and ETBR.Kaede biosynthesis and its inhibition by cotransin could be measured following photoconversion without disturbing signals of pre-existing receptors.
FIGURE 7.
Single cell pulse-chase microscopy assay to quantify the biosynthesis of ETBR.Kaede in transiently transfected HEK 293 cells using confocal LSM. Top, cells were treated with DMSO or cotransin (30 μm, 3 h). The gKaede signals of ETBR.Kaede were completely photoconverted to rKaede by UV irradiation, and reappearance of the gKaede signals was analyzed in a time series (180 min total; one picture every 15 min; only pictures at the indicated times are shown). The horizontal xy scans are representative for five independent experiments. Scale bar, 10 μm. Bottom, quantification of the reappearance of the gKaede signals at the plasma membrane. Data points represent mean values of the signals of five cells (±S.E.) analyzed in a time series (180 min total; one picture every 15 min). Plasma membrane gKaede signals were quantified using an 8-bit gray scale ranging from 0 to 255. The curve is representative for three independent experiments.
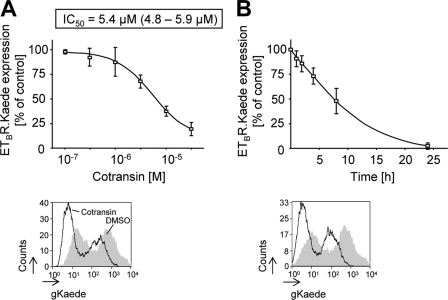
As an alternative to the pulse-chase microscopy working at a single cell level, flow cytometry measurements of reappearing gKaede signals of cell populations are possible. We used such an assay to derive a concentration-response curve for the cotransin action on ETBR.Kaede biosynthesis in transiently transfected HEK 293 cells. The gKaede signals of the cells were photoconverted to rKaede; cells were treated with increasing cotransin concentrations for 17 h, and reappearance of the gKaede signals was measured by flow cytometry (Fig. 8A). From the resulting concentration-response curve, an IC50 value of 5.4 μm (95% confidence limits, 4.8–5.9 μm) could be calculated, which was ∼10-fold higher than that reported for the biosynthesis inhibition of the secretory VCAM1 protein (6).
FIGURE 8.
Flow cytometry assays to quantify the cotransin-mediated biosynthesis inhibition of ETBR.Kaede in transiently transfected HEK 293 cells. A, upper panel, concentration-response curve. The gKaede signals were photoconverted to rKaede; cells were treated with increasing concentrations of cotransin for 17 h or DMSO, and reappearance of the gKaede signals during that time was analyzed by flow cytometry measurements (the fluorescence background of vector-transfected cells and residual rKaede signals present immediately after photoconversion were subtracted). Data points represent mean values of three independent experiments (±S.D.) and are expressed in % of DMSO control. The calculated IC50 value is indicated. Lower panel, flow cytometry histogram of a representative experiment using DMSO-treated cells and cells treated with a single cotransin concentration (30 μm). The cell counts were plotted against the gKaede fluorescence intensities. The gray area represents the DMSO-treated control cells, and the white area below the black line represents the cotransin-treated cells. Signal intensities below 10 result from nontransfected cells. B, time dependence of the cotransin action at a concentration of 30 μm. Upper panel, gKaede fluorescence signals of the cells were photoconverted to rKaede; cells were incubated for different times with cotransin (30 μm) or DMSO, and reappearance of the gKaede signals during these times was analyzed by flow cytometry measurements. Data points represent mean values of three independent experiments (±S.D.) and are expressed in % of the DMSO control. Lower panel, flow cytometry histogram of a representative experiment using DMSO-treated cells and cotransin-treated cells at a single incubation time (24 h). The cell counts were plotted against the gKaede fluorescence intensities. The gray area represents the DMSO-treated control cells, the white area below the black line represents the cotransin-treated cells. Signal intensities below 10 result from nontransfected cells.
We also used these flow cytometry measurements to get more detailed data about the incubation times needed to measure cotransin effects (30 μm) in cells. The gKaede signals of transiently transfected HEK 293 cells expressing ETBR.Kaede were photoconverted to rKaede; cells were treated for different times with cotransin, and reappearance of the gKaede signals was measured by flow cytometry (Fig. 8B). Using 30 μm of cotransin, half of the effect was observed after ∼8 h of treatment.
DISCUSSION
In summary, our results show that the G protein-coupled ETBR belongs to the small group of cotransin-sensitive proteins. Moreover, we have established biosynthesis assays with Kaede fusion proteins allowing the quantification of cotransin effects on membrane protein biosynthesis with minimal problems resulting from long half-life receptor pools.
The group of highly cotransin-sensitive proteins (low micromolar concentrations) includes VCAM1, P-selectin, angiotensinogen, β-lactamase, CRF1R, and ETBR. A comparison of the signal peptides of these proteins with those of various nonsensitive proteins (6) such as Eotaxin-3, monocyte chemotactic protein-1 (MCP-1), decay-accelerating factor (DAF1/CD55), urokinase-type plasminogen activator receptor (uPAR/CD87), and the vascular endothelial growth factor receptor-2 (VEGFR2) reveals no obvious consensus sequence in the signal peptides. This is similar to what was described for the HUN-7293 derivative CAM741 and its VCAM1 and VEGF signal peptide targets (9, 11), where different residues were identified to be critical for compound sensitivity. In the case of the VEGF, it was speculated that helix formation propensity and enhanced flexibility of the signal peptide may be responsible for its sensitivity to CAM741 and that the substance competes with the signal peptide for binding to a specific site within the Sec61 translocon complex (9). The same may hold true for the cotransin-sensitive signal peptides, but the exact mechanism remains elusive as long as the exact binding site(s) of the heptadepsipeptides are unknown. It is noteworthy that all highly cotransin-sensitive proteins described so far possess cleavable signal peptides (three secretory and two integral membrane proteins). The putative signal anchor sequence-containing GPCRs tested in this study were invariantly cotransin-resistant, even to very high cotransin concentrations (see Fig. 3B). Thus, a high hydrophobicity of a signal sequence (as it is frequently observed in the case for signal anchor sequences) may correlate with cotransin resistance.
At the moment, only six highly cotransin-sensitive proteins are known, including the ETBR (see above). Cotransin does not affect overall protein synthesis significantly (Fig. 2A) and is consequently a relatively nontoxic substance (Fig. 2B). These data indicate that cotransin acts relatively selectively and may impair signal sequence-dependent protein synthesis of a relatively limited subset of proteins. It should be noted, however, that the exact amount of cotransin-sensitive proteins is not known at the moment and can not even be estimated as long as proteomic analyses of secreted and membrane proteins of various cell types following cotransin treatment are not available.
The fact that ETBR signaling could be completely abolished by cotransin treatment, at least in HEK 293 cells, indicates that this compound may be significant for the pharmacology of this receptor. In the case of primary cultured astrocytes expressing endogenous ETBRs (Fig. 6B), the observed IC50 value of cotransin action was even 4-fold lower than that observed in HEK 293 cells (Fig. 8A) (1.3 versus 5.4 μm, respectively) confirming a potential role of cotransin for the ETBR pharmacology. The slight difference in the observed IC50 values may be due to the different methodology used (ligand binding displacement versus flow cytometry assay, respectively). A better cotransin permeation in astrocytes or the substantially lower amounts of the endogenous receptor may also play a role. It should be noted, however, that [125I]ET-1 binding could not be reduced in the astrocytes as completely as in the transfected HEK 293 cells. The reason for this phenomenon remains unclear.
One may speculate that derivatization of cotransin may lead to substances that are specific for this receptor or even specific for other target signal sequences. In the case of CAM741, a proof of principle for changing selectivity was published recently (9). Whereas the substance CAM741 inhibits both VCAM1 and VEGF biosynthesis, the derivative NFI028 only inhibits that of VCAM1. In the case of cotransin, the synthesis of such derivatives will be facilitated by the recently established solid phase synthesis protocol for this substance (14, 26).
We have previously shown that fusions of the photoconvertible Kaede protein to GPCRs are useful to study trafficking of the receptors between individual subcellular compartments (13). The prerequisite for using this methodology was that fused Kaede did not influence the pharmacological properties of the receptors and did not oligomerize when fused to integral membrane proteins (13). In these trafficking experiments, the receptor's gKaede fluorescence signals are switched to rKaede in a particular compartment using a confocal LSM, and the fate of the rKaede signals is analyzed independently from the residual receptor pool which is still of the gKaede fluorescence type. Here, we have developed an additional application of the Kaede technology; by complete photoconversion of the gKaede signals at a certain time point and by analysis of their reappearance, protein synthesis could be measured quantitatively either on a single cell level in real time by confocal LSM or by flow cytometry measurements integrating the signals of as many cells as needed for the experimental setup. This technique also avoids problems with receptor pools having a long half-life because they are of rKaede fluorescence type in these measurements. It is conceivable that this type of assay can be easily adapted to high throughput screening, and this may lead to the identification of novel substances having a similar mechanism of action as cotransin.
Acknowledgments
We thank Gunnar Kleinau and Gerd Krause for useful discussions. We thank Erhard Klauschenz from the DNA sequencing service group for contributions. Björn Lamprecht (Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany) was helpful in the establishment of the FACS assays. Primary-cultured mouse astrocytes were a gift from H. Kettenmann (MDC, Berlin, Germany) and were isolated and cultured by Maria Pannell. We also thank Jenny Eichhorst and Angelika Ehrlich for excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft Grant SFB 449 and Grant SCHU 1116/2-1.
C. Westendorf, A. Schmidt, I. Coin, J. Furkert, I. Ridelis, D. Zampatis, C. Rutz, B. Wiesner, W. Rosenthal, M. Beyermann, and R. Schülein, unpublished results.
- ER
- endoplasmic reticulum
- CRF2(a)R
- corticotropin-releasing factor receptor type 2(a)
- ETBR
- endothelin B receptor
- ET-1
- endothelin-1
- EndoH
- endoglycosidase H
- GPCR
- G protein-coupled receptor
- LSM
- laser scanning microscopy
- PEI
- polyethyleneimine
- μOR
- μ opioid receptor
- PNGaseF
- peptide N-glycosidase F
- qRT-PCR
- quantitative real time PCR
- AT2R
- angiotensin 2 receptor
- CRF1R
- corticotropin-releasing factor receptor type 1
- μOR
- μ opioid receptor
- PAR1
- protease-activated receptor 1
- UTR2
- urotensin 2 receptor
- V1aR
- vasopressin V1a receptor
- V2R
- vasopressin V2 receptor.
REFERENCES
- 1. von Heijne G. (1985) J. Mol. Biol. 184, 99–105 [DOI] [PubMed] [Google Scholar]
- 2. von Heijne G. (1990) Curr. Opin. Cell Biol. 2, 604–608 [DOI] [PubMed] [Google Scholar]
- 3. Higy M., Junne T., Spiess M. (2004) Biochemistry 43, 12716–12722 [DOI] [PubMed] [Google Scholar]
- 4. Audigier Y., Friedlander M., Blobel G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5783–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Besemer J., Harant H., Wang S., Oberhauser B., Marquardt K., Foster C. A., Schreiner E. P., de Vries J. E., Dascher-Nadel C., Lindley I. J. (2005) Nature 436, 290–293 [DOI] [PubMed] [Google Scholar]
- 6. Garrison J. L., Kunkel E. J., Hegde R. S., Taunton J. (2005) Nature 436, 285–289 [DOI] [PubMed] [Google Scholar]
- 7. Boger D., L., Keim H., Oberhauser B., Schreiner E. P., Foster C. A. (1999) J. Am. Chem. Soc. 121, 6197–6205 [Google Scholar]
- 8. Schreiner E. P., Kern M., Steck A., Foster C. A. (2004) Bioorg. Med. Chem. Lett. 14, 5003–5006 [DOI] [PubMed] [Google Scholar]
- 9. Harant H., Wolff B., Schreiner E. P., Oberhauser B., Hofer L., Lettner N., Maier S., de Vries J. E., Lindley I. J. (2007) Mol. Pharmacol. 71, 1657–1665 [DOI] [PubMed] [Google Scholar]
- 10. MacKinnon A. L., Garrison J. L., Hegde R. S., Taunton J. (2007) J. Am. Chem. Soc. 129, 14560–14561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harant H., Lettner N., Hofer L., Oberhauser B., de Vries J. E., Lindley I. J. (2006) J. Biol. Chem. 281, 30492–30502 [DOI] [PubMed] [Google Scholar]
- 12. Ando R., Hama H., Yamamoto-Hino M., Mizuno H., Miyawaki A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12651–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt A., Wiesner B., Weisshart K., Schulz K., Furkert J., Lamprecht B., Rosenthal W., Schülein R. (2009) Traffic 10, 2–15 [DOI] [PubMed] [Google Scholar]
- 14. Coin I., Beerbaum M., Schmieder P., Bienert M., Beyermann M. (2008) Org. Lett. 10, 3857–3860 [DOI] [PubMed] [Google Scholar]
- 15. Alken M., Rutz C., Köchl R., Donalies U., Oueslati M., Furkert J., Wietfeld D., Hermosilla R., Scholz A., Beyermann M., Rosenthal W., Schülein R. (2005) Biochem. J. 390, 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sambrook J., Russel D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Wietfeld D., Heinrich N., Furkert J., Fechner K., Beyermann M., Bienert M., Berger H. (2004) J. Biol. Chem. 279, 38386–38394 [DOI] [PubMed] [Google Scholar]
- 18. Kyhse-Andersen J. (1984) J. Biochem. Biophys. Methods 10, 203–209 [DOI] [PubMed] [Google Scholar]
- 19. Köchl R., Alken M., Rutz C., Krause G., Oksche A., Rosenthal W., Schülein R. (2002) J. Biol. Chem. 277, 16131–16138 [DOI] [PubMed] [Google Scholar]
- 20. Rutz C., Renner A., Alken M., Schulz K., Beyermann M., Wiesner B., Rosenthal W., Schülein R. (2006) J. Biol. Chem. 281, 24910–24921 [DOI] [PubMed] [Google Scholar]
- 21. Grantcharova E., Furkert J., Reusch H. P., Krell H. W., Papsdorf G., Beyermann M., Schulein R., Rosenthal W., Oksche A. (2002) J. Biol. Chem. 277, 43933–43941 [DOI] [PubMed] [Google Scholar]
- 22. Alken M., Schmidt A., Rutz C., Furkert J., Kleinau G., Rosenthal W., Schülein R. (2009) Mol. Pharmacol. 75, 801–811 [DOI] [PubMed] [Google Scholar]
- 23. Möller T., Kann O., Prinz M., Kirchhoff F., Verkhratsky A., Kettenmann H. (1997) Neuroreport 8, 2127–2131 [DOI] [PubMed] [Google Scholar]
- 24. Mizuno H., Mal T. K., Tong K. I., Ando R., Furuta T., Ikura M., Miyawaki A. (2003) Mol. Cell 12, 1051–1058 [DOI] [PubMed] [Google Scholar]
- 25. Raab-Graham K. F., Haddick P. C., Jan Y. N., Jan L. Y. (2006) Science 314, 144–148 [DOI] [PubMed] [Google Scholar]
- 26. Coin I., Beyermann M., Bienert M. (2007) Nat. Protoc. 2, 3247–3256 [DOI] [PubMed] [Google Scholar]
- 27. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 28. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 29. Watakabe T., Urade Y., Takai M., Umemura I., Okada T. (1992) Biochem. Biophys. Res. Commun. 185, 867–873 [DOI] [PubMed] [Google Scholar]