Background: Recent studies suggest that deficiency of the antioxidant enzyme glutathione peroxidase-1 (GPx-1) may contribute to cardiovascular disease risk.
Results: In human endothelial cells, GPx-1 deficiency and its accompanying oxidant stress increased adhesion molecule expression by augmenting NFκB and JNK activation.
Conclusion: GPx-1 deficiency promotes a proinflammatory state by redox mechanisms.
Significance: This study provides insights into the mechanisms by which GPx-1 regulates endothelial activation and atherogenesis.
Keywords: Cell Adhesion, Cytokine Action, Dual Specificity Phosphoprotein Phosphatase, Endothelium, Inflammation, MAP Kinases (MAPKs), NFκB, Oxidative Stress, Tumor Necrosis Factor (TNF), Glutathione Peroxidase
Abstract
Glutathione peroxidase-1 (GPx-1) is a crucial antioxidant enzyme, the deficiency of which promotes atherogenesis. Accordingly, we examined the mechanisms by which GPx-1 deficiency enhances endothelial cell activation and inflammation. In human microvascular endothelial cells, we found that GPx-1 deficiency augments intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) expression by redox-dependent mechanisms that involve NFκB. Suppression of GPx-1 enhanced TNF-α-induced ROS production and ICAM-1 expression, whereas overexpression of GPx-1 attenuated these TNF-α-mediated responses. GPx-1 deficiency prolonged TNF-α-induced IκBα degradation and activation of ERK1/2 and JNK. JNK or NFκB inhibition attenuated TNF-α induction of ICAM-1 and VCAM-1 expression in GPx-1-deficient and control cells, whereas ERK1/2 inhibition attenuated only VCAM-1 expression. To analyze further signaling pathways involved in GPx-1-mediated protection from TNF-α-induced ROS, we performed microarray analysis of human microvascular endothelial cells treated with TNF-α in the presence and absence of GPx-1. Among the genes whose expression changed significantly, dual specificity phosphatase 4 (DUSP4), encoding an antagonist of MAPK signaling, was down-regulated by GPx-1 suppression. Targeted DUSP4 knockdown enhanced TNF-α-mediated ERK1/2 pathway activation and resulted in increased adhesion molecule expression, indicating that GPx-1 deficiency may augment TNF-α-mediated events, in part, by regulating DUSP4.
Introduction
To maintain cellular homoeostasis, intracellular levels of reactive oxygen species (ROS)2 are tightly controlled by antioxidant factors, including enzymes such as the glutathione peroxidases (GPxs), superoxide dismutases, catalase, and peroxiredoxins, as well as other redox-active proteins, such as thioredoxin and glutaredoxin, and small redox-active molecules, such as glutathione and NADPH. Among these, the GPxs comprise a group of selenocysteine-containing proteins (1, 2) responsible for reducing various hydroperoxides with reducing equivalents provided by the co-substrate glutathione (3). GPx-1, cellular glutathione peroxidase, is one of the most abundant GPx isoforms within eukaryotic cells, and is important in preventing oxidant-mediated cell death (4).
Studies in knock-out and transgenic mice have shown that GPx-1 modulates vascular function (5, 6). GPx-1-deficient mice have an increase in the oxidant stress marker, F2-isoprostanes (iPF2α-III), manifest endothelial dysfunction, have abnormal cardiac function following ischemia/reperfusion injury, and have increased infarct size and vascular permeability in response to cerebral ischemia/reperfusion injury (5–7). Other studies suggest that overexpression of GPx-1 decreases tissue damage after cerebral or myocardial ischemia/reperfusion injury, and that GPx-1 can protect against angiotensin II-mediated vascular dysfunction (8–10). Recent findings indicate that GPx-1 deficiency contributes to atherosclerosis and cardiovascular disease risk. In the context of ApoE deficiency, GPx-1 deficiency augments atherosclerosis in mice (11, 12) and human subjects with coronary artery disease, GPx-1 activity is inversely correlated with, and an independent risk factor for, future cardiovascular events (13, 14).
Proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α), stimulate endothelial cell expression of adhesion molecules and chemokines, which guide leukocytes to the vascular wall promoting inflammation and atherogenesis. TNF-α activation of these pathways requires generation of ROS (15) that promote kinase activation and phosphatase inactivation (16). NFκB and MAPK are involved in these pathways. MAPK pathways are evolutionarily highly conserved and involved in diverse cellular functions, including cell proliferation, differentiation, and stress responses (17). Activated MAPKs are inactivated through dephosphorylation of threonine and/or tyrosine residues within the activation loop; dephosphorylation can be achieved by serine/threonine phosphatase, tyrosine phosphatase, and dual-specificity phosphatases (DUSP). The regulation and activation of DUSP family members in different cells and tissues control MAPK signaling intensity, duration, and subcellular localization (18).
In the present study, we sought to investigate the molecular mechanism by which GPx-1 regulates TNF-α induction of ICAM-1 and VCAM-1 in endothelial cells. The data presented here suggest that GPx-1 modulates TNF-α-mediated signaling by regulating ROS flux to enhance MAPK pathways, in part, by regulating expression of the dual specificity phosphatase, DUSP4, to stimulate ERK1/2-activation. Furthermore, GPx-1 deficiency simultaneously augments JNK and NFκB activation, contributing to enhanced adhesion molecule expression in the absence of GPx-1.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
N-Acetyl-l-cysteine (NAC) and pyrrolidine dithiocarbamate (PDTC) were purchased from Sigma. U0126, the MEK1/2 inhibitor (Cell Signaling, Danvers, MA), and SP600125, the JNK inhibitor (Assay Design, Plymouth Meeting, PA), were dissolved in dimethyl sulfoxide, which also served as the vehicle control. Other chemicals and reagents were purchased from Bio-Rad unless otherwise specified.
Cell Culture
Primary human microvascular endothelial cells (HMVEC) were maintained in endothelial basal medium-2 supplemented with EGM-2 MV (Lonza, Walkersville, MD) at 37 °C in 5% CO2. Experiments were performed on cells from passage 5 to 8. Cells were treated with TNF-α (R&D Systems, Minneapolis, MN) (5–40 ng/ml) for 1–4 h.
Transfection with Small Interfering RNA (siRNA) and Recombinant Adenoviral Vector Infection
To decrease GPx-1 expression, near confluent HMVEC were transfected with 80 nmol/liter of stealth siRNA to GPx-1 mRNA (310 sequence 5′-GGUUCGAGCCCAACUUCAUGCUCUU-3′) or scrambled control (310 sequence 5′-GGUAGCGCCAAUCCUUACGUCUCUU-3′). Stealth RNAs were transfected as in our previous studies (19), using LipofectamineTM 2000 (Invitrogen) for 3 h in antibiotic-free Opti-MEM ITM (Invitrogen). Transfection medium was replaced with full growth medium, and experiments were performed after 48 h. To decrease DUSP4 mRNA, HMVEC were transfected with 80 nmol/liter of stealth siRNA to DUSP4 mRNA (5′-CCCACCUCGCAGUUCGUCUUCAGCU-3′), for 3 h, and experiments were performed after 24 h. To decrease NADPH oxidase 4 (NOX-4) HMVEC were transfected as above with a stealth siRNA to NOX-4 mRNA (5′-CAGAGUAUCACUACCUCCACCAGAU-3′) and similar methods were used to knockdown mitochondrial complex III subunit 5, the ubiquinol-cytochrome c reductase Rieske iron-sulfate polypeptide 1 (5′-ACACAGACAUCAAGGUGCCUGACUU-3′; ComplxIII).
A recombinant adenoviral vector expressing GPx-1 (AdGPx-1), tagged with a c-Myc epitope at the amino terminus (20), and an empty vector (Ad5Bgl II) were kindly provided by John F. Engelhardt (University of Iowa) through the Gene Transfer Vector Core. Confluent HMVEC were incubated with AdGPx-1 or Ad5Bgl II for 48 h. Conditions that produced >90% transduction with recombinant adenovirus were assessed by immunostaining for N-terminal c-Myc. A recombinant adenovirus vector (Vector Biolabs, Philadelphia, PA) was similarly used to overexpress DUSP4.
Western Blots
Cells were washed in PBS, scraped in PBS, and pelleted at 300 × g for 5 min. Cell pellets were resuspended in ice-cold buffer (50 mm Tris-HCl, pH 7.5, 5 mm EDTA, and 1 mm dithiothreitol (DTT)), and lysed by the freeze-thaw method (21). Alternatively, cells were directly lysed in cell lysis buffer with protease inhibitors, or lysates were prepared using a nuclear and cytoplasmic extraction kit (Active Motif, Carlsbad, CA). Samples were stored at −80 °C. Protein samples (10–40 μg) were separated on 4–15% SDS-polyacrylamide gels (Bio-Rad) and transferred to nitrocellulose membranes (Hybond, Amersham Biosciences). The membranes were incubated with anti-GPx-1 (MBL, Woburn, MA), anti-ICAM-1, anti-VCAM-1, anti-NFκBp50, anti-IκBα, and anti-USF-2 (Santa Cruz, Santa Cruz, CA) antibodies, or with anti-phospho-ERK1/2 or anti-ERK1/2 (Cell Signaling) antibodies overnight at 4 °C, and visualized using the ECL detection system (Amersham Biosciences). The membranes were then stripped and reprobed with a polyclonal rabbit anti-β-actin antibody (Sigma). A VersaDoc scanning system and the accompanying software (Bio-Rad) were used to quantitate band density.
Cellular Glutathione Peroxidase Activity Assay
Cellular GPx activity was determined by an indirect assay that links GPx-1-mediated oxidation of glutathione (GSH) with the recycled reduction of oxidized glutathione (GSSG) to GSH by glutathione reductase using NADPH as a reductant (22). The reaction was carried out in a buffer containing 50 mm Tris-HCl, 5 mm EDTA, 1 mm glutathione, 0.4 units/ml of glutathione reductase, and 0.2 mm NADPH (pH 7.6), and initiated by the addition of tert-butyl-hydroperoxide. Enzyme activity was calculated from the change of absorbance at 340 nm over time, a measure of NADPH oxidation. Enzyme activity was calculated using a molar extinction coefficient for NADPH of 6,220 m−1 cm−1, and normalized to protein concentration.
ROS Measurements
For measurement of the intracellular ROS levels, HMVEC were cultured in 96-well plates, and incubated with 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate ester (DCF) (5 μmol/liter) (Invitrogen), as described previously (23). Adenoviral infection and siRNA transfection were performed 48 h prior to stimulation with TNF-α (20 ng/ml). Cellular ROS accumulation was determined with a microplate fluorometer. Fluorescent DCF accumulation was monitored using an excitation wavelength of 485 nm and recording emission at 518 nm. Readings were calculated as a % change from the initial reading for each well. Each condition was tested in 6–12 wells per experiment.
An Amplex Red hydrogen peroxide detection kit (Invitrogen) was used to measure extracellular hydrogen peroxide. Amplex Red reacts with hydrogen peroxide in the presence of horseradish peroxidase to produce resorufin, a fluorescent compound (24). Briefly, confluent 6- or 12-well plates of transfected cells were rinsed with phosphate-buffered saline (pH 7.4) and then treated with equal volumes of phosphate-buffered saline and Amplex Red/horseradish peroxidase prepared according to the manufacturer's instructions (Invitrogen). Following a 20–30-min incubation in a cell culture incubator, aliquots of supernatant were removed and measured for Amplex Red fluorescence using an excitation wavelength of 530 nm and an emission wavelength of 590 nm. Additives such as TNF-α or PEG catalase were included in the Amplex Red mixture. Hydrogen peroxide (1 μm) and phosphate-buffered saline were used as positive and negative controls for Amplex Red activity, respectively. Values were normalized for protein content.
RNA Isolation and qRT-PCR
Total RNA from HMVEC was extracted with the RNeasy kit (Qiagen, Germantown, MD), incorporating an optional DNase I step to remove residual DNA. Samples were quantitated and checked for purity and quality by A260/A280 measurements and gel electrophoresis. cDNA was synthesized from 0.4 to 1 μg of each total RNA sample with oligo(dT) primers using the Advantage RT-for-PCR kit (Clontech, Mountain View, CA), according to the manufacturer's protocol. Quantitative real-time PCR, including data analysis, were performed on an Applied Biosystems PRISM 7900 HT Sequence Detector (Foster City, CA) (see supplemental Table S1 for specific gene expression primers and reference sequences). PCR products were analyzed by the ΔΔCt method that compares the amount of target gene to an endogenous control (GAPDH), followed by normalization to a control sample to obtain relative levels of gene expression. Cycle parameters were 95 °C for 15 min to activate Taq, followed by 40 cycles of 95 °C for 15 s, 58 °C for 1 min, and 72 °C for 1 min.
Microarray Preparation
HMVEC grown to 90% confluence in a 100-mm culture dish (1 × 107 cells) were transfected with siRNA, treated with 20 ng/ml of TNF-α for 2 h, and gene expression changes were investigated by microarray analysis. Reverse transcription of mRNA, probe labeling, and hybridization were performed using standard Affymetrix protocols, as conducted at the Cambridge HPCGG Gene Chip® Microarray Facility. The DNA hybridization utilized the GeneChip Human Genome U133A 2.0 Array containing >22,000 probe sets and 500,000 distinct oligonucleotide features that analyze the expression level of 18,400 transcripts and variants, including 14,500 well characterized human genes (Affymetrix, Santa Clara, CA). Data were analyzed with GeneChip Operating Software, including the GeneChip Scanner 3000, and image analysis was carried out according to standard Affymetrix protocols.
Data Analysis
All experiments were performed three to seven times, and data were expressed as mean ± S.E. The statistical significance of differences (p < 0.05) was determined by ANOVA followed by Fisher's protected least-square difference (PLSD) pairwise comparisons using STATVIEW 5.0.1 software (Abacus Concepts, Calabasas, CA).
Affymetrix Microarray Suites 5.0 gene expression values were normalized with the median invariant method (25). To filter out significant genes, two approaches were used. The primary filtering method used the standard two-sample t test in pairwise comparisons involving treated groups (NT, siControl, and TNF-α, siGPx-1, and TNF-α). Pairwise comparisons were performed and the gene lists generated. Gene Set Enrichment Analysis (GSEA) was used to identify which gene sets were enriched in each group. Leading edge analysis was then used to determine gene targets that caused the enrichment of each gene set. Expression of gene targets was compared between siGPx-1 plus TNF-α and siControl plus TNF-α to generate a subset of GSEA up- and down-regulated genes. These analyses were performed using the GSEA website.
RESULTS
Tumor Necrosis Factor-α Induces ICAM-1 and VCAM-1 Expression in HMVEC
To determine the effects of TNF-α on ICAM-1 and VCAM-1 expression in HMVEC, cells were exposed to TNF-α in a time- and dose-dependent manner. By 1 h, 20 ng/ml of TNF-α treatment significantly increased ICAM-1 and VCAM-1 mRNA (supplemental Fig. S1A). Total cellular ICAM-1 and VCAM-1 protein levels were increased markedly by 3 h after TNF-α stimulation (p = 0.001, and p < 0.0001, respectively, supplemental Fig. 1B); GPx-1 protein levels did not change with this treatment (supplemental Fig. 1B). Increased expression of ICAM-1 and VCAM-1 was detectable with as little as 5 ng/ml of TNF-α, and persisted at concentrations up to 40 ng/ml (supplemental Fig. 1C).
Knockdown of GPx-1 Augments TNF-α-induced ICAM-1 Expression in HMVEC
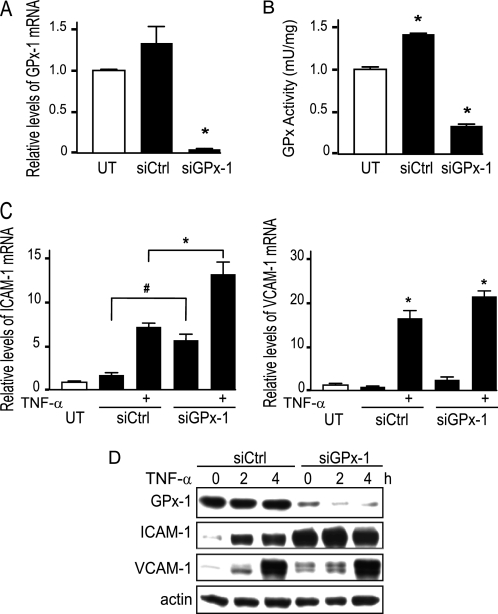
TNF-α signaling is thought to rely, in part, on ROS generation (15). GPx-1 serves an important role in the intracellular defense against oxidant stress. Therefore, to determine the role of GPx-1 in modulating responses to TNF-α, we used siRNA to reduce GPx-1 levels in HMVEC. Targeted knockdown of GPx-1 with siRNA (siGPx-1) decreased GPx-1 mRNA by 96.6% (p < 0.0001) (Fig. 1A) with corresponding reductions in GPx-1 protein expression (91.4%, p < 0.0001) (Fig. 1D) and in GPx-1 activity (78.3%, p < 0.0001) compared with control transfected cells (Fig. 1B). GPx-1 activity was significantly down-regulated in siGPx-1 cells compared with untreated cells (p < 0.0001). In contrast, GPx-1 activity was up-regulated in siControl cells (although GPx-1 protein was not substantially changed, supplemental Fig. 2A). Although the reason for the increase in activity is not apparent from these studies, it has been suggested that GPx-1 may be activated by kinases to augment enzyme activity (26). GPx-1 knockdown plus 2 h of TNF-α treatment further augmented ICAM-1 mRNA (2.1-fold, p < 0.0001) and protein (2.5-fold, p = 0.001) expression compared with TNF-α-treated siControl cells (Fig. 1, C and D), whereas VCAM-1 expression was similarly enhanced in TNF-α-treated siGPx-1 and siControl cells (Fig. 1, C and D). In the absence of TNF-α, GPx-1-deficient cells showed a 3.6-fold increase in ICAM-1 mRNA (p < 0.01) and a trend toward an increase in VCAM-1 mRNA (not significant) (Fig. 1C). ICAM-1 and VCAM-1 proteins were both significantly up-regulated by GPx-1 knockdown (Figs. 1D and 2A). The structural basis for multiple VCAM-1 bands in Western blots is not clear but most likely represents detection of major VCAM-1 isoforms that result from alternative splicing of exon 5, which eliminates one of 6 N-linked glycosylation sites; alternatively, these bands may represent other states of VCAM-1 glycosylation (27).
FIGURE 1.
Knockdown of GPx-1 and TNF-α-induced ICAM-1 and VCAM-1 expression. A, HMVEC were transfected with siGPx-1 or siRNA control (siCtrl) for 48 h and mRNA was measured by qRT-PCR following normalization to GAPDH as endogenous control (n = 4) (p = 0.0003 by ANOVA) and compared with an untransfected sample to obtain relative mRNA levels. B, GPx-1 enzyme activity was determined by an indirect assay (n = 4) (p < 0.001 by ANOVA). UT indicates untransfected cells. Pairwise comparison is shown, *, p < 0.001 versus no treatment. C, transfected cells were treated with 20 ng/ml of TNF-α for 2 h and ICAM-1 and VCAM-1 mRNA were measured by qRT-PCR as in A (n = 5) (#, p < 0.01 and *, p < 0.0001 by Fisher's PLSD). ICAM-1 gene expression was significantly increased over no treatment by GPx-1 knockdown, in the presence or absence of TNF-α and by control transfection plus TNF-α. VCAM-1 gene expression was significantly elevated by TNF-α, compared with all non-TNF-α treated controls (*) and tended to be higher in siGPx-1 cells compared with controls. D, proteins (10 μg) were separated on 4–15% SDS-PAGE gels and transferred to HyBond membrane. Antibodies against GPx-1 (MBL, Woburn, MA), ICAM-1 and VCAM-1 (Santa Cruz Biochemicals), and actin (Sigma) were used to detect the effect of 20 ng/ml of TNF-α in GPx-1-deficient compared with siControl cells for 2 and 4 h. Representative blots are shown.
FIGURE 2.
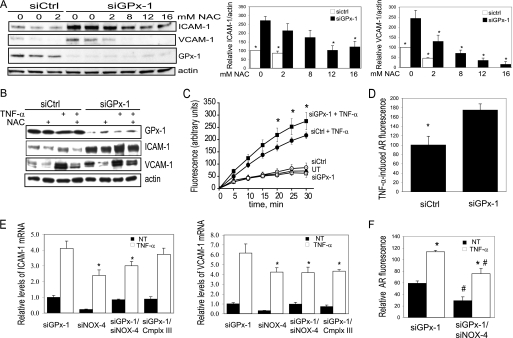
Knockdown of GPx-1 and ROS-mediated effects. A, HMVEC were treated with 0–16 mm NAC immediately following transfection with siGPx-1 or siRNA control (siCtrl). Proteins (10 μg) were harvested 48 h later, separated on 4–15% SDS-PAGE gels, and transferred to HyBond membrane. Western blot was used to detect ICAM-1, VCAM-1, GPx-1, and actin. Plots to the right indicate relative expression of ICAM-1 and VCAM-1 normalized to actin (n = 3). The effects of NAC on ICAM-1 or VCAM-1 were significant by ANOVA (p < 0.005); *, indicates significant pairwise differences with siGPx-1 in the absence of NAC, by post hoc analysis (p < 0.05). B, 48 h after transfection, cells were pretreated 1 h with the antioxidant, NAC, before TNF-α exposure (20 ng/ml). ICAM-1 and VCAM-1 expression was determined by immunoblotting 4 h following TNF-α treatment (n = 4). Representative blots are shown. C, reactive oxygen species accumulation was measured by DCF fluorescence with an excitation wavelength of 485 nm and recording emission at 518 nm. An average of seven experiments with mean ± S.E. is shown (p < 0.0001 by ANOVA and *, p < 0.05 by Fisher's PLSD). Boxes indicate cells treated with siGPx-1, circles indicate cells treated with siControl (siCtrl), filled triangles indicate untransfected (UT) cells, and filled circles and squares represent transfected cells additionally treated with TNF-α. D, relative hydrogen peroxide in media from siGPx-1- and siControl-treated cells was measured 48 h following transfection by monitoring TNF-α-induced Amplex Red (AR) fluorescence in the presence of horseradish peroxidase (excitation 530 nm and emission 590 nm). In the presence of catalase, no AR fluorescence was detected. Fluorescence values are normalized for protein. *, indicates significant differences by t test (p < 0.04, n = 7). E, relative levels of ICAM-1 and VCAM-1 mRNA were measured by qRT-PCR 48 h following transfection with siRNAs; values have been normalized to actin. Values were analyzed by ANOVA followed by pairwise comparisons (p < 0.05 significance). *, significantly different from siGPx-1 treated with TNF-α. All TNF-α values are significantly higher than the no TNF-α (NT) values. F, relative AR fluorescence was measured as in D from cells transfected with siGPx-1 or siGPx-1 together with siNOX-4. The absence of NOX-4 significantly decreased AR fluorescence in the presence or absence of TNF-α. Values were analyzed as in E. *, significantly different from corresponding no TNF-α; #, significantly different from the corresponding siGPx-1 NT or siGPx-1 TNF-α (n = 7).
GPx-1 Modulates Adhesion Molecule Expression
To determine whether accumulation of excess oxidants contributes to the augmented adhesion molecule expression found in GPx-1-deficient cells, we pretreated siGPx-1 and siControl cells with NAC, an antioxidant, immediately after transfection. This treatment significantly attenuated the up-regulation of ICAM-1 and VCAM-1 caused by GPx-1 deficiency alone (Fig. 2A). Similarly, pretreatment with NAC 1 h before TNF-α exposure reduced adhesion molecule expression in the presence and absence of TNF-α (Fig. 2B). Use of diphenyleneiodonium, an NADPH oxidase inhibitor, also attenuated the up-regulation of ICAM-1 and VCAM-1 in the presence and absence of GPx-1 (supplemental Fig. 2B). These findings implicate ROS in the enhanced expression of adhesion molecules caused by GPx-1 deficiency. To determine whether GPx-1-deficient HMVEC display an inability to remove oxidants, we treated siGPx-1 and control cells with TNF-α, incubated these cells with dicholorofluorescein (DCF), and measured the change in DCF fluorescence over time, as a nonspecific indication of intracellular ROS (Fig. 2C). Untransfected HMVEC showed significantly increased intracellular ROS following 30 min of TNF-α treatment (55.0 ± 14.8, untreated, versus 156.5 ± 19.1%, TNF-α treatment, p < 0.0001). Similarly, TNF-α increased ROS levels in siControl and siGPx-1 cells, with enhanced DCF fluorescence detectable in siGPx-1 cells compared with control cells within 20 min of TNF-α treatment (p = 0.01). In the absence of TNF-α, however, there was no detectable difference in DCF fluorescence over time between siGPx-1 and siControl cells. To confirm these findings, we used Amplex Red to measure extracellular oxidant release: Amplex Red reacts with hydrogen peroxide in the presence of horseradish peroxidase to produce resorufin, a fluorescent compound (24). Amplex Red detected similar differences in extracellular ROS: TNF-α-induced a 70% increase (p < 0.04) in measurable hydrogen peroxide in the medium of GPx-1-deficient cells compared with siControl-transfected cells (Fig. 2D). Inclusion of catalase in the Amplex Red reaction mixtures completely eliminated Amplex Red fluorescence, indicating that this signal is due to hydrogen peroxide and not other redox-active compounds. To determine the possible source of the excess ROS generation, we transfected siRNAs specific for NOX-4 and the mitochondrial complex III subunit 5 (ComplxIII), and determined the effect of these siRNAs on TNF-α-mediated up-regulation of ICAM-1 and VCAM-1 gene expression. These siRNAs reduced their respective mRNAs by 89.5 (NOX-4, p < 0.0001) and 92.8% (ComplxIII, p < 0.0001). Only NOX-4 deficiency inhibited TNF-α-mediated up-regulation of both ICAM-1 and VCAM-1 mRNA in the presence and absence of GPx-1 (Fig. 2E). NOX-4 suppression also reduced detectable baseline and TNF-α-mediated extracellular hydrogen peroxide in GPx-1-deficient cultures (Fig. 2F).
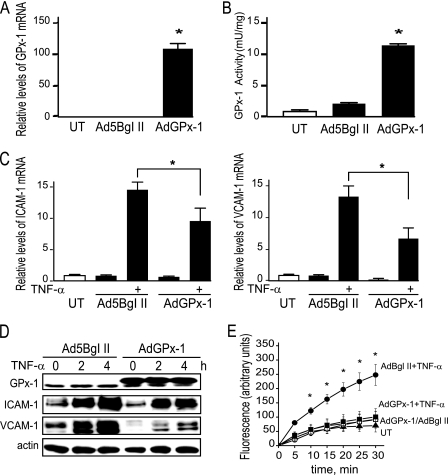
Collectively, these data show that GPx-1-deficient HMVEC have a reduced capacity to neutralize peroxides, suggesting that GPx-1 deficiency impairs the defense of HMVEC against oxidative stress, leading to enhanced endothelial cell activation. We next determined whether increasing oxidant removal by GPx-1 overexpression could reduce TNF-α-induced ICAM-1 and VCAM-1 expression. Adenoviral-mediated overexpression of a myc-tagged GPx-1 significantly increased GPx-1 mRNA 105.5-fold (p < 0.0001) (Fig. 3A) with a 7.5-fold (p < 0.0001) increase in GPx-1 protein expression (Fig. 3D), and 12.6-fold (p < 0.0001) increase in GPx-1 activity (Fig. 3B). Overall, adenoviral overexpression of GPx-1 attenuated TNF-α-mediated responses, reducing ICAM-1 and VCAM-1 mRNA and protein (Fig. 3, C and D), as well as ROS accumulation (Fig. 3E), compared with TNF-α-treated control HMVEC. These data support a role for GPx-1 in mediating the ROS-dependent effects of TNF-α.
FIGURE 3.
Overexpression of GPx-1 and TNF-α-induced ICAM-1 and VCAM-1 expression and ROS accumulation. A, adenovirus treatment for 48 h enhances GPx-1 expression (AdGPx-1) compared with empty vector control (Ad5Bgl II). mRNA was measured by qRT-PCR (n = 5) using GAPDH as an endogenous control, and compared with untransduced cells to obtain relative mRNA levels. B, GPx-1 enzyme activity was measured by an indirect assay (n = 4). Means were significantly different by ANOVA (p < 0.0001), and post hoc comparison found (*) p < 0.00001 compared with no treatment. C, adenovirus treatment of cells to enhance GPx-1 expression resulted in decreased ICAM-1 and VCAM-1 mRNA, relative values of mRNA were calculated as in A. Values were analyzed by ANOVA followed by pairwise comparison, which showed significant differences between empty vector control and GPx-1 overexpressing cells (n = 4–5, *, p < 0.05). Data are presented as mean ± S.E. D, protein expression after 20 ng/ml of TNF-α treatment for 2–4 h in adenovirus-treated cells. Representative blots are shown. Note that the c-Myc-tagged recombinant GPx-1 in the AdGPx-1 lanes migrates more slowly than the endogenous GPx-1. E, DCF fluorescence was measured after cells were treated with adenovirus for 48 h and grown in 96-well plates to near confluence. Prior to treatment with 20 ng/ml of TNF-α, cells were loaded with 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate ester (n = 5) (*, p < 0.05 by Fisher's PLSD). Circles indicate cells treated with control adenovirus (AdBglII), squares represent cells treated with AdGPx-1, and filled circles and squares represent adenovirus-transformed cells additionally treated with TNF-α. Untreated, untransformed cells (UT) are represented by closed triangles.
GPx-1 Mediates the Effects of Redox-sensitive Signaling Pathways in TNF-α-treated HMVEC
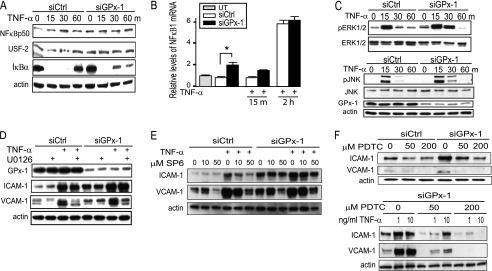
To determine how GPx-1 may modulate TNF-α signaling, we examined its effects on NFκB and MAPK activation. NFκB is viewed as the primary transcription factor that stimulates adhesion molecule gene expression in response to TNF-α (28, 29). In untreated cells, NFκB is complexed with the inhibitory IκB proteins. Therefore, to examine whether GPx-1 influences TNF-α-induced expression of this transcription factor in endothelial cells, we examined IκBα expression as well as NFκB p50 nuclear translocation. In response to stimulation with TNF-α, IκBα was degraded by 15 min, and NFκB translocation to the nucleus was induced (Fig. 4A). GPx-1-deficient cells showed slower recovery of IκBα protein expression following TNF-α treatment (Fig. 4A) compared with TNF-α-treated control cells that had restored IκBα protein levels to those of untreated cells after 60 min. These data suggest prolonged NFκB signaling in GPx-1-deficient cells. After 2 h of TNF-α stimulation, NFκB1 mRNA was increased (5.8-fold, p < 0.0001) in both siGPx-1 and siControl, whereas in siGPx-1-treated cells, NFκB1 mRNA was increased 1.9-fold compared with siControl (p = 0.04) without additional TNF-α treatment (Fig. 4B). Similarly, in the microarray analysis, we also found that NFκB2 gene expression was up-regulated in both siGPx-1 and siControl TNF-α-treated HMVEC (p < 0.0001) (supplemental Table S2).
FIGURE 4.
Knockdown of GPx-1 and NFκB and MAPK signaling pathways in TNF-α-treated cells. A, to determine the effect of TNF-α on NFκB signaling in GPx-1-deficient and control cells by immunoblotting, nuclear extraction was used to detect NFκB p50 and USF-2 protein (a nuclear control) and the cytoplasmic fraction for IκBα and actin protein (cytoplasmic control) (n = 3). B, qRT-PCR was used to measure NFκB1 mRNA after 10 ng/ml of TNF-α for 15 min and 2 h in GPx-1-deficient and control cells. UT indicates untransfected cells. mRNA was measured by qRT-PCR following normalization to GAPDH as endogenous control and compared with an untransfected sample to obtain relative mRNA levels. Significant differences were observed between transfected cells analyzed by pairwise comparison with the Fisher's PLSD test (*, p < 0.05, n = 4). C and D, Western blot analysis: C, the effect of TNF-α on MAPK signaling was tested by analyzing phospho-ERK1/2 protein and phospho-JNK in GPx-1-deficient and control cells (n = 4). D, GPx-1-deficient and control cells were pretreated with MEK1/2 inhibitor (U0126) 1 h prior to TNF-α exposure at 10 ng/ml for 4 h. E, GPx-1-deficient and control cells were pretreated with a JNK inhibitor (SP600125) 1 h prior to TNF-α exposure. F, GPx-1-deficient and control cells were pretreated with PDTC, an NFκB inhibitor, immediately following transfection. Shown is the effect of PDTC on ICAM-1 and VCAM-1 in the absence or presence of TNF-α (1 ng or 10 ng/ml).
We next evaluated the role of MAPK pathways in the TNF-α-induced expression of adhesion molecules in GPx-1-deficient HMVEC. ROS-dependent phospho-ERK1/2 activation in response to TNF-α was prolonged in siGPx-1 compared with siControl cells (Fig. 4C). Similarly, TNF-α activation of JNK was up-regulated by GPx-1 deficiency. Inhibition of ERK1/2 with the specific MEK1/2 inhibitor (U0126) (10 μm) 1 h prior to TNF-α treatment decreased VCAM-1 expression (Fig. 4D), but had little effect on the expression of ICAM-1 in siGPX-1-deficient cells. In contrast, use of SP60125, the JNK inhibitor, reduced TNF-α-mediated ICAM-1 and VCAM-1 expression in siControl- and siGPx-1-treated cells (Fig. 4E). Similarly, use of an NFκB inhibitor, PDTC, attenuated ICAM-1 and VCAM-1 expression, in the presence and absence of TNF-α, in control and GPx-1-deficient cells (Fig. 4F).
GPx-1 Deficiency Alters Gene Expression in Response to TNF-α
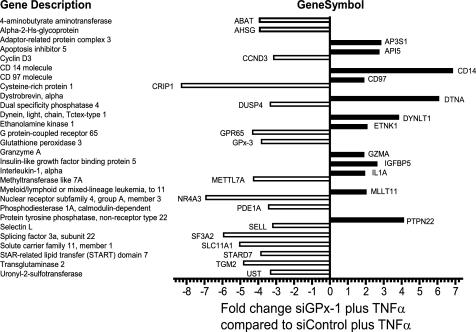
To identify early changes in gene expression induced by ROS generated by inflammatory cytokine stimulation, we compared transcripts from untreated, siControl plus TNF-α-treated, and siGPx-1 plus TNF-α-treated cells using the Affymetrix Human Genome U133A 2.0 gene array. Gene set enrichment analysis was used to filter the expression data, and each TNF-α-treated set was compared with the untreated sample to determine a possible set of TNF-α-responsive genes. Over 430 genes up- or down-regulated by more than 2-fold were identified in siGPx-1 plus TNF-α-treated cells, and 340 genes in siControl plus TNF-α treatment. Using leading edge analysis, a subset of genes differentially regulated in siGPx-1 plus TNF-α-treated cells compared with siControl plus TNF-α-treated cells were identified (Fig. 5, and supplemental Tables S3 and S4). By this analysis, 12 target genes were up-regulated ∼2-fold or more and 71 genes down-regulated ∼2-fold or more by GPx-1 deficiency.
FIGURE 5.
Genes differentially regulated by siGPx-1 deficiency. Genes were selected by leading edge analysis from the top 100 sets identified by Gene Set Enrichment Analysis. Supplemental Table S4 lists additional targets. Positive values indicate increased expression in GPx-1-deficient plus TNF-α-treated cells and negative values indicate increased expression in siControl plus TNF-α treatment.
To analyze specifically TNF-α-mediated pathways, a subset of genes involved in MAPK, NFκB, and other key inflammatory pathways was further confirmed by qRT-PCR (supplemental Table S2). After normalization with GAPDH, the fold-changes determined by qRT-PCR were compared with the fold-values obtained from microarray results. For all genes tested, the directional change (increase/decrease) was consistent between microarray data and qRT-PCR results, demonstrating the fidelity of the microarray analysis in detecting gene expression changes. Additionally, adenoviral-mediated overexpression of GPx-1 attenuated TNF-α-mediated up-regulation of ICAM-1, VCAM-1, and NFκB2 gene expression (supplemental Table S2).
To investigate the role of other antioxidant genes that may function in a compensatory capacity, analysis of a subgroup of genes encoding antioxidant enzymes and redox-active proteins was performed. Among these changes, superoxide dismutase-2 (SOD2) showed a 6.5-fold increase in TNF-α-treated control and TNF-α-treated GPx-1-deficient cells (supplemental Table S5). Of this extensive list, only heme oxygenase-1, GPx-3, GPx-5, and the uncoupling protein homolog were differentially regulated by GPx-1 deficiency (using a stringent cutoff of p < 0.005).
DUSP4 was among the targets that were substantially different between TNF-α-treated siGPX-1 and TNF-α-treated siControl cells, with a significant reduction caused by GPx-1 deficiency (Fig. 5 and supplemental Table S2). DUSP4 is a MAPK phosphatase that has been shown to attenuate both ERK1/2 and JNK signaling in some cell types (18).
Knockdown of DUSP4 Augments TNF-α-induced VCAM-1 Expression in HMVEC
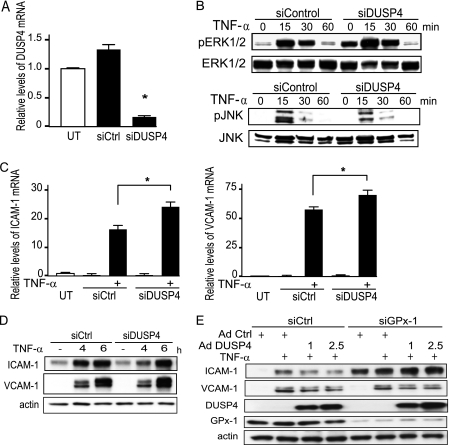
To determine whether down-regulation of DUSP4 could potentiate TNF-α signaling in HMVEC, we used siRNA to knockdown DUSP4 expression. siDUSP4 decreased DUSP4 mRNA by 78.5% (p < 0.0001) (Fig. 6A). DUSP4 knockdown augmented TNF-α-mediated ERK1/2 phosphorylation but not JNK phosphorylation (Fig. 6B). Although DUSP4 deficiency significantly increased TNF-α-induced ICAM-1 and VCAM-1 mRNA expression (p < 0.0005 and p < 0.005, respectively) compared with TNF-α-treated siControl cells (Fig. 6C), only TNF-α induction of VCAM-1 protein was augmented in siDUSP4-transfected cells (Fig. 6D). We next used an adenovirus construct to overexpress DUSP4 in endothelial cells following the suppression of GPx-1. Excess DUSP4 had only a modest effect in decreasing TNF-α-mediated VCAM-1 overexpression (Fig. 6E) that was apparent only with lower doses of TNF-α (5 ng/ml). DUSP4 overexpression had no effect on ICAM-1 expression in GPx-1-deficient cells.
FIGURE 6.
GPx-1 and DUSP4 in TNF-α-treated cells. HMVEC were transfected with siDUSP4 or siRNA control (siCtrl) for 24 h. A, DUSP4 transcripts were measured by qRT-PCR (n = 5) following normalization to GAPDH as endogenous control (n = 4) and compared with an untransfected sample to obtain relative mRNA levels. Using ANOVA (p < 0.0001) and Fisher's PLSD post hoc testing (*, p < 0.05), significant differences were observed. B, siDUSP- and siControl-treated cells were treated with TNF-α and activation of ERK1/2 and JNK was analyzed by Western blot detection of their phosphorylated forms. C and D, the effect of TNF-α treatment on ICAM-1, and VCAM-1 mRNA (C) and protein (D) expression in DUSP4-deficient cells was compared with that in siCtrl cells as determined by qRT-PCR as described in A, and immunoblotting. E, the ability of DUSP4 to modify TNF-α-mediated ICAM-1 and VCAM-1 protein expression was tested in GPx-1-deficient and control transfected cells by using an adenovirus to DUSP4 or a control adenovirus (AdCtrl) and analyzing extracts by Western blot.
DISCUSSION
Recent findings have suggested that deficiency of GPx-1 augments the development of atherosclerosis in susceptible mice (11, 12), and in human subjects with coronary artery disease, GPx-1 activity is inversely correlated with, and an independent risk factor for, future cardiovascular events (13, 14). The molecular basis for these effects, however, has not been completely elucidated. Our study specifically addresses the mechanisms by which GPx-1 deficiency promotes endothelial activation and promotes inflammatory responses to the cytokine, TNF-α. Strikingly, we found that loss of GPx-1 alone causes a proinflammatory phenotype in endothelial cells by increasing ICAM-1 and VCAM-1 expression, events that may facilitate the recruitment of leukocytes to the endothelium. Our findings also show that in the context of GPx-1 deficiency, the proinflammatory effects of TNF-α are augmented. Redox-dependent processes contribute to these proinflammatory responses, as antioxidant treatments attenuate adhesion molecule expression caused by GPx-1 deficiency in the absence or presence of TNF-α. Similarly, the effects of TNF-α on endothelial cells can be decreased with GPx-1 overexpression, consistent with previous studies using GPx-1 mimics (30).
Neither the antioxidant treatments nor GPx-1 overexpression completely block the ability of TNF-α to promote adhesion molecule expression. Possibly these treatments fail to remove all of the TNF-α-mediated ROS flux in cells; however, it is also likely that non-ROS-dependent pathways also contribute to the effects of TNF-α on cellular activation. It is difficult to distinguish between these possibilities as a method such as DCF fluorescence may not detect all essential intracellular ROS flux due to limitations in sensitivity, and subcellular location and duration of oxidant flux. DCF fluorescence is also nonspecific in that it detects several species of ROS (31). In addition, these measurements were normalized to the initial levels of DCF fluorescence in each well to account for potential differences in cell number; however, this method does not allow for a comparison of initial DCF levels. Additionally, DCF measurements may be affected by the formation of a DCF radical that may subsequently cause the generation of superoxide anion and lead to a propagation of the oxidant signal, at least in the presence of heme containing enzymes, such as horseradish peroxidase, as has been shown in in vitro assays (32, 33). DCF fluorescence, however, remains a widely used method to monitor intracellular oxidant production (31, 34) and the dynamics of DCF fluorescence in our study are inversely related to the presence or absence of GPx-1, an important intracellular antioxidant enzyme. Nonetheless, to confirm that loss of GPx-1 augments oxidant levels in endothelial cells, we utilized Amplex Red, which is highly specific for hydrogen peroxide, to monitor extracellular release of oxidants. Consistent with a role for redox-dependent effects mediated by GPx-1 deficiency, these data indicated that loss of GPx-1 significantly increases hydrogen peroxide release to the media following TNF-α treatment.
To confirm further a role for the antioxidant GPx-1 in modulating intracellular ROS, we next used siRNA to suppress the expression of various oxidant generators. Based on our preliminary studies we found that suppression of NOX-4 or mitochondrial complex III subunit expression reduced TNF-α-mediated induction of adhesion molecules to some extent, but neither of these molecules are sufficient to overcome completely the actions of TNF-α. It is likely that in the absence of GPx-1, excess ROS may accumulate due to the actions of multiple ROS systems, including NOX-4, other NOXs, and xanthine oxidase, as well as mitochondrial sources. It is, however, difficult to eliminate all of these sources at once without compromising cell integrity. Our findings indicate that eliminating NOX-4 has a greater effect than eliminating other oxidant generators tested. Furthermore, deficiency of NOX-4 significantly lowered hydrogen peroxide generation in cells lacking GPx-1 in the presence or absence of TNF-α, suggesting a role for NOX-4 in generating intracellular oxidants at baseline. Our findings are consistent with recent findings that suggest a role for NOX-4 in basal hydrogen peroxide generation (35) and with studies suggesting that NOX-4 plays a role in TNF-α-mediated oxidant stress in cerebral microvascular endothelial cells from the pig (36). Interestingly, the latter study implicated NOX-4 in generating superoxide, rather than directly generating hydrogen peroxide. Superoxide, however, is rapidly converted to hydrogen peroxide either spontaneously or via the actions of superoxide dismutases (37), such as SOD2, which is up-regulated by TNF-α. Hydrogen peroxide has an essential role in cell signaling via oxidation of protein thiols, although, in excess, it can contribute to oxidative stress (38).
TNF-α has been previously shown to up-regulate ERK1/2, JNK, and NFκβ pathways by mechanisms involving ROS (15). Our findings identify a crucial role for GPx-1 in modulating activation of these pathways, as loss of GPx-1 augments each of them. Consistent with previous studies indicating that excess GPx-1 or antioxidants can suppress the activation of proinflammatory signaling responses (39, 40), we also found that excess GPx-1 attenuated cytokine action. Similarly, lack of GPx-1, in deficient mice, was found to enhance NFκB activation following cerebral ischemia-reperfusion injury (41), a process that theoretically involves the production of excess oxidants. Consistent with this observation, we found that GPx-1 deficiency in human endothelial cells increased NFκB (p50 subunit) translocation to the nucleus and augmented the degradation of IκBα, prolonging NFκB-dependent signaling. Furthermore, in the context of GPx-1 deficiency, augmented NFκB activation and JNK pathway activation contribute to enhanced endothelial expression of adhesion molecules.
Several studies have found that ICAM-1 and VCAM-1 are differentially regulated, with ICAM-1 transcription primarily responsive to NFκB pathways (28), whereas cytokine-induced transcription of the VCAM-1 gene requires combinatorial interactions of NFκB with other nuclear activators, such as stimulatory protein-1 (42), interferon regulatory factor-1 (43), and activator protein-1 (44). Accordingly, we found that inhibitors directed to ERK1/2 or JNK had a greater effect on VCAM-1 expression than ICAM-1 expression, whereas inhibiting NFκB activation suppressed the expression of both ICAM-1 and VCAM-1. Interestingly, in GPx-1-deficient cells, NFκB inhibition also attenuated the increase of adhesion molecule expression found in the absence of TNF-α, thereby suggesting that tonic activation of NFκB pathways is enhanced by GPx-1 deficiency. In contrast, short-term incubation with ERK1/2 inhibitors had no effect on the enhanced levels of adhesion molecule expression in GPx-1-deficient cells in the absence of TNF-α, whereas short-term incubation with JNK inhibitors decreased somewhat the augmented adhesion molecule expression caused by GPx-1 deficiency alone, suggesting that JNK pathways may also contribute to the basal up-regulation of adhesion molecules in GPx-1-deficient cells. Interestingly, a recent study in endothelial cells found that mechanical stress up-regulated GPx-1 expression, thereby suppressing inflammatory responses in these cells (45). Our findings indicate that lack of GPx-1 alone can promote inflammatory responses in endothelial cells.
To investigate further the role of intracellular signaling pathways in the up-regulation of adhesion molecules under oxidative stress, microarray analysis was performed. Other studies have shown that TNF-α induces gene expression of specific adhesion molecules, chemokines, transcription factors, and signal transduction molecules in endothelial cells (46, 47). Included in these clusters, adhesion molecules ICAM-1 and VCAM-1 (48, 49) were highly up-regulated, as was the antioxidant SOD-2. It is well known that NFκB activates the SOD2 gene and that TNF-α induces SOD-2 (50). In our data set, SOD-2 up-regulation to TNF-α was not altered by GPx-1 deficiency.
Of 14,000 target genes, gene set enrichment analysis followed by leading edge analysis identified 12 target genes up-regulated by ∼2-fold or more in the TNF-α-treated GPx-1-deficient cells and 71 down-regulated compared with the TNF-α-treated control cells. Among these, DUSP4, encoding a MAPK phosphatase, was a novel target down-regulated in TNF-α-treated GPx-1-deficient cells. DUSP4 is one of several dual specificity phosphatases that has been recently described (51), and despite a fairly detailed knowledge of its biochemical properties (52, 53), its physiological function is less completely studied. DUSP4 may play a role in suppression of ERK- and JNK-dependent events (54, 55), and, thus, decreased expression of DUSP may explain, in part, augmented MAPK activation. ROS may also inactivate DUSPs by oxidation of their catalytic cysteine to sulfenic acid (15). Other studies have shown that DUSP4 gene expression may be induced by ROS-mediated p53 activation in some cells (56) or indirectly through ROS-dependent ERK1/2 activation (54), possibly as a feedback mechanism by which to decrease MAPK signaling. In human embryonic kidney 293T cells, hydrogen peroxide or xanthine/xanthine oxidase led to inducible expression of multiple DUSPs, and the suppression of DUSP4 or DUSP16 led to sustained JNK activation following hydrogen peroxide treatment (57). In our studies, DUSP4 expression was decreased in TNF-α-treated GPx-1-deficient cells, even though these cells had elevated accumulation of intracellular ROS. In fact, following a 2-h TNF-α treatment, DUSP4 was modestly reduced (by 15–25%) in control transfectants as well, suggesting that modest suppression of DUSP4 may be a response to TNF-α that is enhanced by GPx-1 deficiency. In our microarray analysis, of all the DUSP family gene targets, only DUSP4 and DUSP9 were differentially regulated by GPx-1 deficiency (supplemental Fig. S3). In contrast, the expression of DUSP5, DUSP6, and DUSP8 was up-regulated to a similar extent in GPx-1-deficient and control cells by TNF-α treatment. Consistent with these results, DUSP5, DUSP6, and DUSP7, which have been designated as ERK-specific phosphatases, were not increased in hydrogen peroxide-treated cells, concomitant with the prolonged phosphorylation of ERKs observed under these conditions in 293T cells (57). Although DUSPs may be post-translationally inactivated to enhance MAPK activation, decreased expression of DUSP4 may also contribute to MAPK stimulation. In support of this view, targeted knockdown of DUSP4 augmented TNF-α-induced phosphorylation of ERK1/2. Consistent with the role of ERK1/2 in VCAM-1 expression and DUSP4 in regulating ERK1/2, lack of DUSP4 enhanced TNF-α-mediated expression of VCAM-1 mRNA and protein. In contrast, lack of DUSP4 did not enhance ICAM-1 expression by Western blot. Furthermore, lack of DUSP4 alone (without cytokine stimulation) had no effect on adhesion molecule expression. DUSP4 knockdown did not augment TNF-α-mediated JNK activation; rather, there was a slight suppression in JNK activation in DUSP4-deficient cells that may relate to compensatory mechanisms by other DUSPs in cells with DUSP4 knockdown. This is in contrast to cells with GPx-1 deficiency, in which lack of GPx-1 augments JNK activation, possibly by ROS-mediated mechanisms. Thus, loss of DUSP4 contributes to enhanced ERK1/2 activation in GPx-1-deficient cells but ERK1/2 pathways do not regulate all of the TNF-α-mediated responses caused by lack of GPx-1.
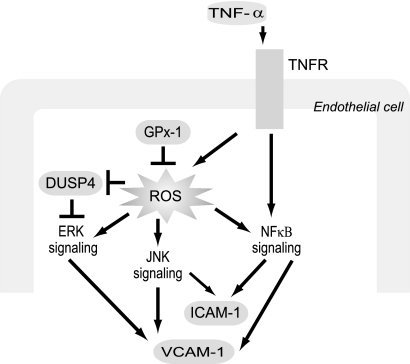
To summarize, our current findings suggest that GPx-1 deficiency and its accompanying oxidant stress increase ICAM-1 and VCAM-1 expression primarily via NFκB and JNK activation (Fig. 7). Additionally, we found that oxidant stress evoked by GPx-1 deficiency was associated with decreased expression of DUSP4, suggesting a mechanism by which to prolong ERK signaling, thereby, enhancing endothelial responses to inflammatory cytokines. This study provides new insights into the mechanisms by which GPx-1 deficiency may contribute to endothelial activation and atherogenesis by contributing to a proinflammatory environment and augmenting the activation of cytokine-stimulated ROS-dependent signaling cascades.
FIGURE 7.
Proposed model illustrating a role for GPx-1 in TNF-α activation of endothelial cells. In response to extracellular (TNF-α) and intracellular (GPx-1 deficiency) oxidative stress, NFκB is activated, increasing ICAM-1 and VCAM-1 expression. In addition, GPx-1 deficiency plus TNF-α treatment augments phospho-ERK1/2 and JNK activation, in part, by decreasing the expression of the gene encoding DUSP4, a MAPK phosphatase. ERK1/2 pathways contribute to VCAM-1 expression and other TNF-α-mediated effects that are not shown.
Supplementary Material
Acknowledgment
We thank Stephanie Tribuna for expert assistance with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants HL 58976, HL 61795, HV 28178, HL 81587, and HL 48743 from the NHLBI and Deutsche Forschungsgemeinschaft Grants LU 1452/1-1 and LU 1452/2-1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1–S5.
- ROS
- reactive oxygen species
- GPx-1
- glutathione peroxidase-1
- HMVEC
- human microvascular endothelial cells
- ICAM-1
- intercellular adhesion molecule-1
- VCAM-1
- vascular cell adhesion molecule-1
- DCF
- dichlorodihydrofluorescein
- PDTC
- pyrrolidine dithiocarbamate
- NAC
- N-acetyl-l-cysteine
- NOX
- NADPH oxidase
- ComplxIII
- mitochondrial complex III subunit 5, the ubiquinol-cytochrome c reductase Rieske iron-sulfate polypeptide 1
- DUSP4
- dual specificity phosphatase 4
- qRT-PCR
- quantitative real-time RT-PCR
- AdGPx-1
- adenovirus overexpressing glutathione peroxidase-1
- Ad5BgI II
- empty adenovirus vector
- PLSD
- protected least-square difference
- ANOVA
- analysis of variance.
REFERENCES
- 1. Raes M., Michiels C., Remacle J. (1987) Free Radic. Biol. Med. 3, 3–7 [DOI] [PubMed] [Google Scholar]
- 2. Ursini F., Maiorino M., Brigelius-Flohé R., Aumann K. D., Roveri A., Schomburg D., Flohé L. (1995) Methods Enzymol. 252, 38–53 [DOI] [PubMed] [Google Scholar]
- 3. Brigelius-Flohé R. (1999) Free Radic. Biol. Med. 27, 951–965 [DOI] [PubMed] [Google Scholar]
- 4. de Haan J. B., Bladier C., Griffiths P., Kelner M., O'Shea R. D., Cheung N. S., Bronson R. T., Silvestro M. J., Wild S., Zheng S. S., Beart P. M., Hertzog P. J., Kola I. (1998) J. Biol. Chem. 273, 22528–22536 [DOI] [PubMed] [Google Scholar]
- 5. Forgione M. A., Cap A., Liao R., Moldovan N. I., Eberhardt R. T., Lim C. C., Jones J., Goldschmidt-Clermont P. J., Loscalzo J. (2002) Circulation 106, 1154–1158 [DOI] [PubMed] [Google Scholar]
- 6. Forgione M. A., Weiss N., Heydrick S., Cap A., Klings E. S., Bierl C., Eberhardt R. T., Farber H. W., Loscalzo J. (2002) Am. J. Physiol. Heart Circ. Physiol. 282, H1255–1261 [DOI] [PubMed] [Google Scholar]
- 7. Wong C. H., Bozinovski S., Hertzog P. J., Hickey M. J., Crack P. J. (2008) J. Neurochem. 107, 241–252 [DOI] [PubMed] [Google Scholar]
- 8. Weisbrot-Lefkowitz M., Reuhl K., Perry B., Chan P. H., Inouye M., Mirochnitchenko O. (1998) Brain Res. Mol. Brain Res. 53, 333–338 [DOI] [PubMed] [Google Scholar]
- 9. Yoshida T., Maulik N., Engelman R. M., Ho Y. S., Magnenat J. L., Rousou J. A., Flack J. E., 3rd, Deaton D., Das D. K. (1997) Circulation 96, II-216–220 [PubMed] [Google Scholar]
- 10. Chrissobolis S., Didion S. P., Kinzenbaw D. A., Schrader L. I., Dayal S., Lentz S. R., Faraci F. M. (2008) Hypertension 51, 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Torzewski M., Ochsenhirt V., Kleschyov A. L., Oelze M., Daiber A., Li H., Rossmann H., Tsimikas S., Reifenberg K., Cheng F., Lehr H. A., Blankenberg S., Förstermann U., Münzel T., Lackner K. J. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 850–857 [DOI] [PubMed] [Google Scholar]
- 12. Lewis P., Stefanovic N., Pete J., Calkin A. C., Giunti S., Thallas-Bonke V., Jandeleit-Dahm K. A., Allen T. J., Kola I., Cooper M. E., de Haan J. B. (2007) Circulation 115, 2178–2187 [DOI] [PubMed] [Google Scholar]
- 13. Blankenberg S., Rupprecht H. J., Bickel C., Torzewski M., Hafner G., Tiret L., Smieja M., Cambien F., Meyer J., Lackner K. J. (2003) N. Engl. J. Med. 349, 1605–1613 [DOI] [PubMed] [Google Scholar]
- 14. Espinola-Klein C., Rupprecht H. J., Bickel C., Schnabel R., Genth-Zotz S., Torzewski M., Lackner K., Munzel T., Blankenberg S. (2007) Am. J. Cardiol. 99, 808–812 [DOI] [PubMed] [Google Scholar]
- 15. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 16. Bowie A., O'Neill L. A. (2000) Biochem. Pharmacol. 59, 13–23 [DOI] [PubMed] [Google Scholar]
- 17. Roux P. P., Blenis J. (2004) Microbiol. Mol. Biol. Rev. 68, 320–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Camps M., Nichols A., Arkinstall S. (2000) FASEB J. 14, 6–16 [PubMed] [Google Scholar]
- 19. Zhang Y., Handy D. E., Loscalzo J. (2005) Circ. Res. 96, 831–837 [DOI] [PubMed] [Google Scholar]
- 20. Li Q., Sanlioglu S., Li S., Ritchie T., Oberley L., Engelhardt J. F. (2001) Antioxid. Redox Signal. 3, 415–432 [DOI] [PubMed] [Google Scholar]
- 21. Murias M., Rachtan M., Jodynis-Liebert J. (2005) J. Pharmacol. Toxicol. Methods 52, 302–305 [DOI] [PubMed] [Google Scholar]
- 22. Flohé L., Günzler W. A. (1984) Methods Enzymol. 105, 114–121 [DOI] [PubMed] [Google Scholar]
- 23. Leopold J. A., Dam A., Maron B. A., Scribner A. W., Liao R., Handy D. E., Stanton R. C., Pitt B., Loscalzo J. (2007) Nat. Med. 13, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou M., Diwu Z., Panchuk-Voloshina N., Haugland R. P. (1997) Anal. Biochem. 253, 162–168 [DOI] [PubMed] [Google Scholar]
- 25. Li C., Wong W. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao C., Leng Y., Huang W., Liu X., Kufe D. (2003) J. Biol. Chem. 278, 39609–39614 [DOI] [PubMed] [Google Scholar]
- 27. Montes-Sánchez D., Ventura J. L., Mitre I., Frías S., Michán L., Espejel-Nuñez A., Vadillo-Ortega F., Zentella A. (2009) BMC Chem. Biol. 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ledebur H. C., Parks T. P. (1995) J. Biol. Chem. 270, 933–943 [DOI] [PubMed] [Google Scholar]
- 29. Lee C. W., Lin W. N., Lin C. C., Luo S. F., Wang J. S., Pouyssegur J., Yang C. M. (2006) J. Cell Physiol. 207, 174–186 [DOI] [PubMed] [Google Scholar]
- 30. d'Alessio P., Moutet M., Coudrier E., Darquenne S., Chaudiere J. (1998) Free Radic. Biol. Med. 24, 979–987 [DOI] [PubMed] [Google Scholar]
- 31. Rhee S. G., Chang T. S., Jeong W., Kang D. (2010) Mol. Cells 29, 539–549 [DOI] [PubMed] [Google Scholar]
- 32. Rota C., Chignell C. F., Mason R. P. (1999) Free Radic. Biol. Med. 27, 873–881 [DOI] [PubMed] [Google Scholar]
- 33. Rota C., Fann Y. C., Mason R. P. (1999) J. Biol. Chem. 274, 28161–28168 [DOI] [PubMed] [Google Scholar]
- 34. Dikalov S., Griendling K. K., Harrison D. G. (2007) Hypertension 49, 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dikalov S. I., Dikalova A. E., Bikineyeva A. T., Schmidt H. H., Harrison D. G., Griendling K. K. (2008) Free Radic. Biol. Med. 45, 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Basuroy S., Bhattacharya S., Leffler C. W., Parfenova H. (2009) Am. J. Physiol. Cell Physiol. 296, C422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liochev S. I., Fridovich I. (2007) Free Radic. Biol. Med. 42, 1465–1469 [DOI] [PubMed] [Google Scholar]
- 38. Forman H. J., Maiorino M., Ursini F. (2010) Biochemistry 49, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kretz-Remy C., Mehlen P., Mirault M. E., Arrigo A. P. (1996) J. Cell Biol. 133, 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garg A. K., Aggarwal B. B. (2002) Mol. Immunol. 39, 509–517 [DOI] [PubMed] [Google Scholar]
- 41. Crack P. J., Taylor J. M., Ali U., Mansell A., Hertzog P. J. (2006) Stroke 37, 1533–1538 [DOI] [PubMed] [Google Scholar]
- 42. Neish A. S., Khachigian L. M., Park A., Baichwal V. R., Collins T. (1995) J. Biol. Chem. 270, 28903–28909 [DOI] [PubMed] [Google Scholar]
- 43. Neish A. S., Read M. A., Thanos D., Pine R., Maniatis T., Collins T. (1995) Mol. Cell Biol. 15, 2558–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ahmad M., Theofanidis P., Medford R. M. (1998) J. Biol. Chem. 273, 4616–4621 [DOI] [PubMed] [Google Scholar]
- 45. Wagner A. H., Kautz O., Fricke K., Zerr-Fouineau M., Demicheva E., Güldenzoph B., Bermejo J. L., Korff T., Hecker M. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 1894–1901 [DOI] [PubMed] [Google Scholar]
- 46. Murakami T., Mataki C., Nagao C., Umetani M., Wada Y., Ishii M., Tsutsumi S., Kohro T., Saiura A., Aburatani H., Hamakubo T., Kodama T. (2000) J. Atheroscler. Thromb. 7, 39–44 [DOI] [PubMed] [Google Scholar]
- 47. Zhou J., Jin Y., Gao Y., Wang H., Hu G., Huang Y., Chen Q., Feng M., Wu C. (2002) Inflamm. Res. 51, 332–341 [DOI] [PubMed] [Google Scholar]
- 48. Sana T. R., Janatpour M. J., Sathe M., McEvoy L. M., McClanahan T. K. (2005) Cytokine 29, 256–269 [DOI] [PubMed] [Google Scholar]
- 49. Viemann D., Goebeler M., Schmid S., Klimmek K., Sorg C., Ludwig S., Roth J. (2004) Blood 103, 3365–3373 [DOI] [PubMed] [Google Scholar]
- 50. Wong G. H., Elwell J. H., Oberley L. W., Goeddel D. V. (1989) Cell 58, 923–931 [DOI] [PubMed] [Google Scholar]
- 51. Misra-Press A., Rim C. S., Yao H., Roberson M. S., Stork P. J. (1995) J. Biol. Chem. 270, 14587–14596 [DOI] [PubMed] [Google Scholar]
- 52. Farooq A., Zhou M. M. (2004) Cell Signal. 16, 769–779 [DOI] [PubMed] [Google Scholar]
- 53. Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 54. Brondello J. M., Brunet A., Pouysségur J., McKenzie F. R. (1997) J. Biol. Chem. 272, 1368–1376 [DOI] [PubMed] [Google Scholar]
- 55. Sieben N. L., Oosting J., Flanagan A. M., Prat J., Roemen G. M., Kolkman-Uljee S. M., van Eijk R., Cornelisse C. J., Fleuren G. J., van Engeland M. (2005) J. Clin. Oncol. 23, 7257–7264 [DOI] [PubMed] [Google Scholar]
- 56. Shen W. H., Wang J., Wu J., Zhurkin V. B., Yin Y. (2006) Cancer Res. 66, 6033–6039 [DOI] [PubMed] [Google Scholar]
- 57. Teng C. H., Huang W. N., Meng T. C. (2007) J. Biol. Chem. 282, 28395–28407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.