Abstract
microRNAs (miRNAs) are 21–23-nucleotide non-coding RNAs. It has become more and more evident that this class of small RNAs plays critical roles in the regulation of gene expression at the post-transcriptional level. MEF2A is a member of the MEF2 (myogenic enhancer factor 2) family of transcription factors. Prior report showed that the 3′-untranslated region (3′-UTR) of the Mef2A gene mediated its repression; however, the molecular mechanism underlying this intriguing observation was unknown. Here, we report that MEF2A is repressed by miRNAs. We identify miR-155 as one of the primary miRNAs that significantly represses the expression of MEF2A. We show that knockdown of the Mef2A gene by siRNA impairs myoblast differentiation. Similarly, overexpression of miR-155 leads to the repression of endogenous MEF2A expression and the inhibition of myoblast differentiation. Most importantly, reintroduction of MEF2A in miR-155 overexpressed myoblasts was able to partially rescue the miR-155-induced myoblast differentiation defect. Our data therefore establish miR-155 as an important regulator of MEF2A expression and uncover its function in muscle gene expression and myogenic differentiation.
Keywords: Cell Differentiation, Gene Expression, MicroRNA, Skeletal Muscle, Transcription Factors, MEF2A, miR-155
Introduction
Determination of the myogenic lineage and differentiation of skeletal muscle cells are precisely orchestrated by the MyoD family of basic helix-loop-helix proteins (1–4). The MyoD family of myogenic transcription factors also activates muscle gene expression in concert with additional transcription cofactors and modifiers. In particular, MyoD and members of the MEF2 family of transcription factors synergistically activate the myogenic program (5). A key advance in understanding activation of myogenic gene expression was the recognition of functional interactions between myogenic regulatory transcription factors and additional regulators that either alter accessibility of chromatin structure or post-transcriptionally regulate gene expression (6).
MicroRNAs (miRNAs)2 are a class of ∼22-nt non-coding RNAs that regulate gene expression at the post-transcriptional level (7). The involvement of miRNAs in muscle biology recently has been reported. miRNAs regulate the expression of transcription factors and signaling mediators important for cardiac and skeletal muscle development and function (8–10). Aberrant miRNA expression has been observed in muscle diseases, including cardiac and skeletal muscle hypertrophy, heart failure, and muscular dystrophy (11–13). We have previously shown that the expression of muscle-specific miR-1 and miR-133 is induced during skeletal muscle differentiation. We further demonstrated that miR-1 and miR-133 play central regulatory roles in myoblast proliferation and differentiation (14). Interestingly, miR-1 and miR-133 are also important regulators of cardiomyocyte differentiation and heart development (15, 16).
MEF2A, a member of the MEF2 (myocyte enhancer factor 2) family of transcription factors, is highly expressed in cardiac and skeletal muscle (17). Interestingly, an earlier study found that the expression of the Mef2A gene is post-transcriptionally repressed by its 3′-UTR (18). However, the “trans-factors” that mediate such repression was unknown. In this study, we hypothesized that the expression and function of MEF2A is repressed by miRNAs post-transcriptionally. We found that miR-155 represses MEF2A expression in skeletal muscle, playing an important role in skeletal muscle myoblast differentiation.
EXPERIMENTAL PROCEDURES
Plasmids and Reporter Genes
The mouse MEF2A-3′-UTR was PCR amplified from a cDNA pool derived from an embryonic day 15.5 mouse embryo and was ligated 3′ to a CMV promoter luciferase reporter (14). The MEF2A 3′-UTR mutation was introduced using the QuikChange kit from Stratagene. The N-FLAG-MEF2A-UTR was ligated into a modified N-FLAG vector (14). The mouse β-globin 3′-UTR (131 bp) was PCR-amplified from a mouse cDNA pool and cloned into the pGL3-luciferase vector. DNA sequences encoding the primary miR-155 transcript were PCR-amplified from a mouse genomic DNA template and ligated into a modified pcDNA3.1 vector. Mutation of miR-155 was introduced by QuikChange kit (Stratagene). All mutations were confirmed by DNA sequencing. miR-155 mimic oligonucleotides and negative control mimic oligonucleotides were purchased from Dharmacon. Ad-siMEF2A and control virus were described previously (19). Ad-MEF2A and control virus were gifts of Dr. Francisco Naya (Boston University).
Cell Culture, Transfection, and Muscle Differentiation Assays
Transfection of 293T, Cos7, and C2C12 myoblasts was performed as described previously (14, 20). Transient transfection for luciferase reporter assays, unless otherwise indicated, used 100 ng of reporter plasmid and 100 ng of each activator or miRNA plasmid. The total amount of DNA per well was kept constant by adding the corresponding amount of expression vector without a cDNA insert. CMV-LacZ or CMV-GFP was included as an internal control for variations in transfection efficiency. All of the transfection experiments were repeated at least twice in duplicate or triplicate.
C2C12 myoblast cells were cultured and myogenic differentiation was induced as described (20) with minor modifications. Briefly, cells were maintained in DMEM with 10% FBS. We plated cells at ∼50–60% confluence and performed the transfection the following day when they reached ∼90–100% confluence. We collected cells on the same day of transfection (∼ 6 h after transfection) and defined it as day 0 (G0). Cells were switched to medium containing 2% horse serum to induce differentiation, and samples were collected at the indicated dates. Myogenesis was monitored by staining cells with myogenic markers. Cells contain two or more nuclei are viewed as myotubes.
siRNA Knockdown
C2C12 myoblasts cultured in growth medium were infected by adenoviral siMEF2A or control virus (19). 24 h later, culture was harvested as a G0 sample or viral infected growth medium was exchanged by differentiation medium and harvested at the indicated dates.
Immunoblotting and Immunostaining
Immunoblotting (Western blot) was performed as described (21) using antibodies against myogenin, MHC (Santa Cruz Biotechnology), MEF2A (a gift of Dr. John McDermott, York University) and β-tubulin (Sigma). Immunostaining was performed as described (14, 22). Briefly, cells cultured in plates were fixed in 4% paraformaldehyde for 10 min, washed with PBS and 0.1% Nonidet P-40, blocked with 5% goat serum in PBS and 0.1% Nonidet P-40 for 1 h at room temperature, incubated with primary antibodies overnight at 4 °C. After washing, cells were incubated with secondary antibodies for 1 h at room temperature and counterstained with DAPI.
All images were acquired at room temperature from cell culture plates by a camera (ORCA-R2, Hamamatsu) mounted on an inverted microscope (TE2000-U, Nikon). Digital fluorescent images were captured at room temperature with a 10× (Plan Fluor, air, numerical aperture, 0.30), 20× (Plan Fluor, air, numerical aperture, 0.45), or 40× (Plan Fluor, air, numerical aperture, 0.60) objective lens using the least possible exposure to minimize bleaching. The images were processed using SPOT software (version 3.5.4 for MacOS; Diagnostic Instruments) and were scaled down and cropped in Photoshop (Adobe Acrobat) to prepare the final figures. The myofiber surface area was measured by Image J.
qRT-PCR
Total RNA was isolated with TRIzol reagent (Invitrogen). After extraction and purification, 1 μg of RNA was used as template for reverse transcription with random hexamer primers (Invitrogen). All PCR products span intron region of the genes. Signal was detected by the 7500 Real-time PCR System with SYBR green qRT-PCR master mix (Applied Biosystems). Data were normalized by the GAPDH signal.
RESULTS
3′-UTR of Mef2A Gene Mediates Its Post-transcriptional Repression
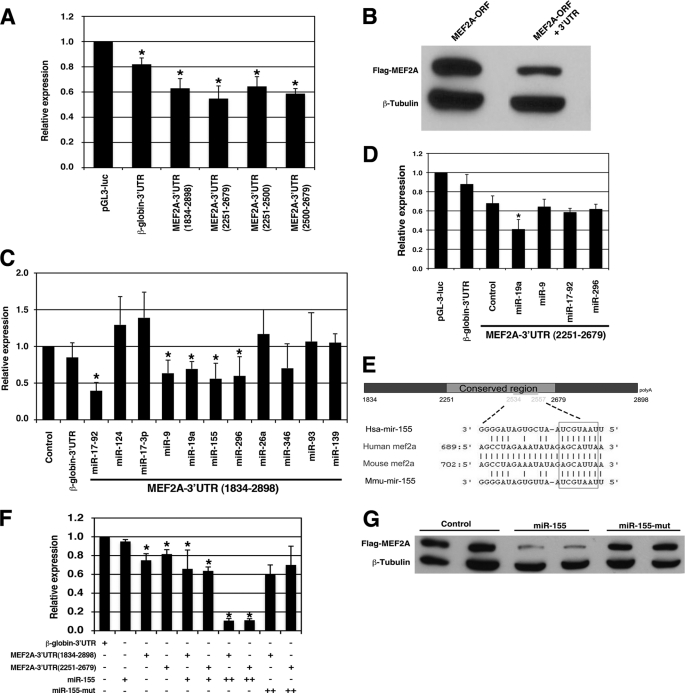
In a prior report, it was shown that the 3′-UTR of the Mef2A transcript functioned as a “cis” element to mediate repression of its expression (18). We cloned the full-length (nt 1834–2898) 3′-UTR of the mouse Mef2A transcript and inserted it downstream of a luciferase reporter. When the reporter was transfected into 293T cells, luciferase activity was significantly repressed when compared with a control construct (Fig. 1A). Similarly, when the Mef2A 3′-UTR full-length construct was transfected into Cos7 cells or C2C12 myoblasts, luciferase activity was repressed (data not shown). When deletion mutagenesis was applied to define regions of the 3′-UTR with the ability to mediate repression of the reporter gene, the region from nt 2251–2679 was able to potently repress luciferase reporter activity (Fig. 1A), consistent with a previous report (18). Further deletion analysis indicated that both the nt 2251–2500 and nt 2500–2679 fragments were also able to mediate the repression of the luciferase reporter gene (Fig. 1A). We included a control reporter in which the 3′-UTR of the mouse β-globin gene was clone downstream of the luciferase reporter gene. We found that this reporter was slightly repressed when transfected into the 293T cells, consistent with a prior report (Fig. 1A) (18). This is not surprising because the β-globin-3′-UTR could be a target for miRNAs to mediate the repression.
FIGURE 1.
The 3′-UTR of the Mef2A gene mediates its repression. A, luciferase reporters containing the 3′-UTRs of the mouse Mef2A gene were transfected into the 293T cells, and luciferase activity was measured 48 h later. Results were presented as relative luciferase activity in which the control was assigned a value of 1. The pGL-3-luciferase and the β-globin-3′-UTR-luciferase reporters were used to serve as controls. Data represent the mean + S.D. from at least three independent experiments in triplicate. *, p < 0.05. B, equal amount of FLAG-tagged MEF2A expression plasmids, which contains or does not contain its 3′-UTR were transfected into 293T cells. Protein expression level was measured by Western blot. β-Tubulin was used as a loading control. C, a luciferase reporter directed by the full-length MEF2A-3′-UTR(1834–2898) was co-transfected with indicated miRNA expression plasmids into the 293T cells, and luciferase activity was measured 48 h later. Results were presented as relative luciferase activity in which the control was assigned a value of 1. The pGL-3-luciferase (control) and the β-globin-3′-UTR-luciferase reporters were used to serve as controls. Data represent the mean + S.D. from at least three independent experiments in duplicate. *, p < 0.05. D, a luciferase reporter directed by a short MEF2A-3′-UTR(2251–2679) was co-transfected with indicated miRNA expression plasmids into the 293T cells, and luciferase activity was measured 48 h later. Results were presented as relative luciferase activity in which the control was assigned a value of 1. The pGL-3-luciferase and the β-globin-3′-UTR-luciferase reporters were used to serve as controls. Data represent the mean + S.D. from at least three independent experiments in duplicate. *, p < 0.05. E, schematic diagram of the mouse Mef2A 3′-UTR and the sequence alignment with miR-155. F, luciferase reporters containing the 3′-UTRs of the mouse Mef2A gene (full-length nt 1834–2898 or the short form nt 2251–2679) were co-transfected with increasing amount of miR-155 or a mutant miR-155 (miR-155-mut) into the 293T cells, and luciferase activity was measured 48 h later. Results were presented as relative luciferase activity in which the control was assigned a value of 1. The β-globin-3′-UTR-luciferase reporter was used to serve as controls. Data represent the mean + S.D. from at least three independent experiments in triplicate. *, p < 0.05. G, FLAG-tagged MEF2A expression plasmids, which contain its 3′-UTR, were co-transfected with miR-155 or a mutant miR-155 into 293T cells. Protein expression level was measured by Western blot using an anti-FLAG antibody. β-Tubulin was used as a loading control.
Next, we transfected 293T cells with FLAG-tagged expression plasmids for MEF2A, with and without 3′-UTR (MEF2A-ORF+3′-UTR or MEF2A-ORF), and then monitored protein expression. MEF2A with the 3′-UTR (MEF2A-ORF+3′-UTR) resulted in a much lower expression level of protein, consistent with the notion that the 3′-UTR mediates its repression (Fig. 1B).
3′-UTR of Mef2A Gene Is Target Site of miRNAs
We hypothesized that the 3′-UTR-dependent repression of the Mef2A gene is mediated by miRNAs. The 3′-UTR of the mouse Mef2A transcript is ∼1054 nt in length. We performed a bioinformatics analysis to identify putative miRNA binding sites (23) and found that multiple miRNAs are predicted to target the Mef2A 3′-UTR. Luciferase reporter assays determined that several miRNAs, including miR-9, miR-17–92, miR-19a, miR-155, and miR-296, repress the activity of the Mef2A 3′-UTR (Fig. 1C).
The nt 2251–2679 region of the Mef2A 3′-UTR was previously reported to mediate its repression most potently (18). We next sought to determine which of the miRNAs predicted to target this region of the Mef2A 3′-UTR sequence could lead to repression of MEF2A expression. Luciferase reporter assay using the MEF2A 3′-UTR(2251–2679) demonstrated that miR-19a (and miR-155) was able to repress MEF2A (Fig. 1D). Sequence alignment showed that miR-155 binds to the Mef2A 3′-UTR at nt 2534–2557 (Fig. 1E), which is within the region previously shown to mediate the repression of MEF2A. Indeed, miR-155 potently represses the MEF2A-3′-UTR luciferase reporters in a dose-dependent manner, but not that of the β-globin 3′-UTR (Fig. 1F). When the seed sequence of miR-155 was mutated, mutant miR-155 was unable to repress the reporter, demonstrating the specificity of such repression (Fig. 1F). Consistent with the results of the luciferase reporter assays, miR-155, but not the miR-155 mutant, significantly represses the expression level of the MEF2A protein when miR-155 and a FLAG-tagged MEF2A expression plasmid containing its 3′-UTR were co-transfected into 293T cells (Fig. 1G). Together, these experimental results indicate that miR-155 represses the Mef2A gene via its 3′-UTR.
miR-155 Represses Myoblast Differentiation by Repressing MEF2A
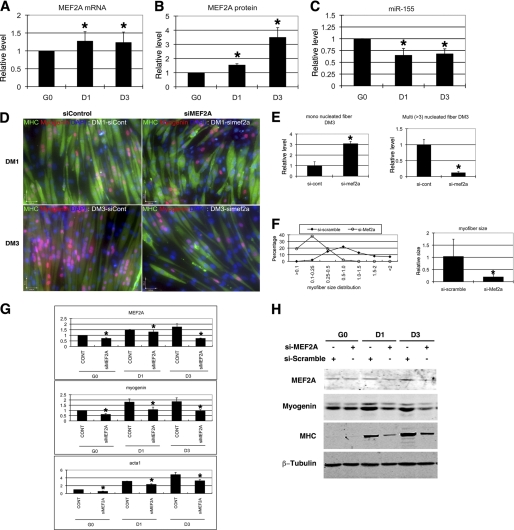
The MEF2 family of transcription factors regulates gene expression in both cardiac and skeletal muscle (17). A gene targeting study demonstrated that MEF2A knock-out mice die post-natally with cardiovascular defects and cardiac failure (24). However, the function of MEF2A in skeletal muscle was not determined. We examined the expression of the Mef2A transcript in C2C12 skeletal muscle myoblasts and found its level was increased when C2C12 myoblasts were induced to differentiate into myotubes (Fig. 2A). Similarly, we observed a significant increase in the expression of MEF2A protein during C2C12 differentiation (Fig. 2B). We investigated the expression of miR-155 in skeletal muscle using Taqman qRT-PCR. Interestingly, the expression level of miR-155 is decreased in differentiating myoblasts, which is inversely correlated with that of MEF2A (Fig. 2C). Note that the decrease of miR-155 expression (a ∼50% decrease) and the increase of MEF2A protein levels (a ∼ 2 fold increase) at Day 3 (D3) is not in a linear inverse correlation, suggesting that additional miRNAs may also contribute to the repression of MEF2A expression. Use of a miR-155 sensor reporter in C2C12 myoblasts further confirmed the above observation (supplemental Fig. S1).
FIGURE 2.
Knockdown of MEF2A impairs myoblast differentiation. A, quantitative real-time PCR measurement of Mef2A transcript expression during C2C12 myoblast differentiation at indicated dates. The result was presented as relative expression level in which the control was assigned a value of 1. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. G0, growth medium day 0; D1, differentiation medium day 1; D3, differentiation medium day 3. B, quantitative measurement of MEF2A protein expression during C2C12 myoblast differentiation at indicated dates. Results from Western blots were quantified by densitometry measurement and presented as relative expression level in which the control was assigned a value of 1. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. C, quantitative Taqman PCR measurement of mature miR-155 expression during C2C12 myoblast differentiation at indicated dates. Result was presented as relative expression level in which the control was assigned a value of 1. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. D, immunohistology of C2C12 cells treated with either control or MEF2A-specific siRNAs. Cells were induced to differentiate at different time courses (differentiation day 1 or day 3) and were stained with antibodies that recognize striate muscle MHC (green) and myogenin (red). DAPI stains nuclei. DM1, differentiation medium day 1; DM3, differentiation medium day 3. E, quantitative analyses of cell numbers of myoblast (MHC-positive, single nucleus) and myotubes (MHC-positive, three or more nuclei) in siMEF2A-treated C2C12 cells when compared with control siRNA treatment. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. F, quantitative analyses of cell size of myoblast and myotubes in siMEF2A-treated C2C12 cells when compared with control siRNA treatment. Left panel, distribution of myoblast and myotube size. Right panel, average size of myoblast and myotube size. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. G, quantitative real-time PCR analyses of gene expression in siMEF2A-treated C2C12 myoblasts and myotubes. Result was presented as relative expression level in which the control was assigned a value of 1. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. H, Western blot analyses of protein expression in siMEF2A-treated C2C12 myoblasts and myotubes, using antibodies recognize indicated proteins.
We asked whether miR-155 mediated repression of MEF2A expression has a functional consequence in skeletal muscle cells. We first tested whether MEF2A is required for myoblast differentiation in cultured C2C12 myoblast cells. Endogenous MEF2A was dramatically knocked down by a MEF2A-specific siRNA but not by a control siRNA. Transfected myoblasts were then switched to differentiation medium to induce myogenic differentiation. We found that myoblast differentiation was significantly impaired, as evidenced by a significant decrease in the number and size of myotubes (Fig. 2D and supplemental Fig. S2). Quantification of mononucleus myoblasts and multinuclei myotubes clearly demonstrated a marked decrease in myotube formation in MEF2A knocked down cells (Fig. 2, E and F). Molecular marker measurements confirmed that the transcripts of myogenic differentiation markers, including myogenin and skeletal muscle α-actin, were all decreased in MEF2A knocked down cells (Fig. 2G). Western blot analysis further supported the notion of decreased myogenic differentiation in MEF2A knocked down myoblasts (Fig. 2H). These data suggest that MEF2A is required for proper differentiation of C2C12 myoblasts. Conversely, adenovirus-based overexpression of MEF2A in C2C12 myoblasts enhances myoblast differentiation (supplemental Fig. S3).
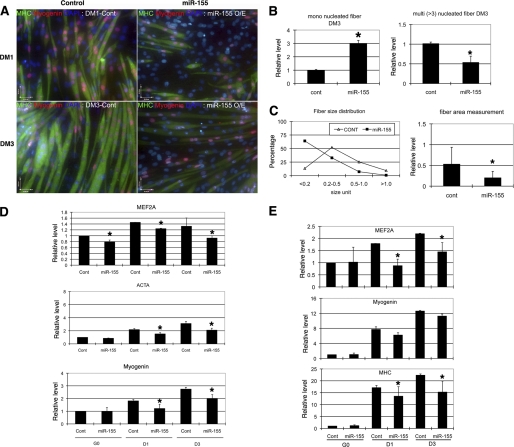
Next, we tested whether miR-155 is involved in the regulation of myoblast differentiation. We transfected a miR-155 expression vector or synthetic miR-155 mimics into undifferentiated C2C12 myoblasts. Overexpression of miR-155 repressed the expression level of endogenous MEF2A. As expected, overexpression of miR-155 inhibited the differentiation of myoblasts. Similar to what we observed in MEF2A knockdown myoblasts, but to a lesser extent, we found less and smaller myotube formation in miR-155 overexpressed myoblasts when they were switched to differential medium to induce differentiation (Fig. 3A and supplemental Fig. S4). Quantitative measurement of the number and size of myoblast and myotube demonstrated that miR-155 decreased the size and number of myotubes (Fig. 3, B and C). Molecular marker analyses, using both quantitative RT-PCR and Western blot, confirm that miR-155 represses myoblast differentiation (Fig. 3, D and E). In addition to miR-155, several other miRNAs also repressed MEF2A-3′-UTR-luc reporter (Fig. 1C). Among them, miR-19a binding site was mapped to the region between nt 2251–2679 of the MEF2A-3′-UTR. We tested whether miR-19a and miR-296 could also repress C2C12 myoblast differentiation. Unlike that of miR-155, miR-19a and miR-296 did not affect myoblast differentiation (supplemental Fig. S5).
FIGURE 3.
MEF2A is repressed by miR-155 via its 3′-UTR. A, immunohistology of C2C12 cells treated with either control or miR-155. Cells were induced to differentiate at indicated dates and were stained with antibodies that recognize striate muscle MHC (green) and myogenin (red). DAPI stains nuclei. DM1, differentiation medium day 1; DM3, differentiation medium day 3. B, quantitative analyses of cell numbers of myoblast (MHC-positive, single nucleus) and myotubes (MHC-positive, three or more nuclei) in miR-155 overexpressed C2C12 cells when compared with control miRNA treatment. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. C, quantitative analyses of cell size of myoblast and myotubes in miR-155 overexpressed C2C12 cells when compared with control miRNA treatment. Left panel, distribution of myoblast and myotube size. Right panel, average size of myoblast and myotube size. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. D, quantitative real-time PCR analyses of gene expression in miR-155 overexpressed C2C12 myoblasts and myotubes. Result was presented as relative expression level in which the control was assigned a value of 1. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. ACTA, skeletal muscle α-actin; G0, growth medium day 0; D1, differentiation medium day 1; D3, differentiation medium day 3. E, quantitative measurement of protein expression in miR-155 overexpressed C2C12 myoblasts, using antibodies recognize indicated proteins. Results from Western blots were quantified by densitometry measurement and presented as relative expression level in which the control was assigned a value of 1. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05.
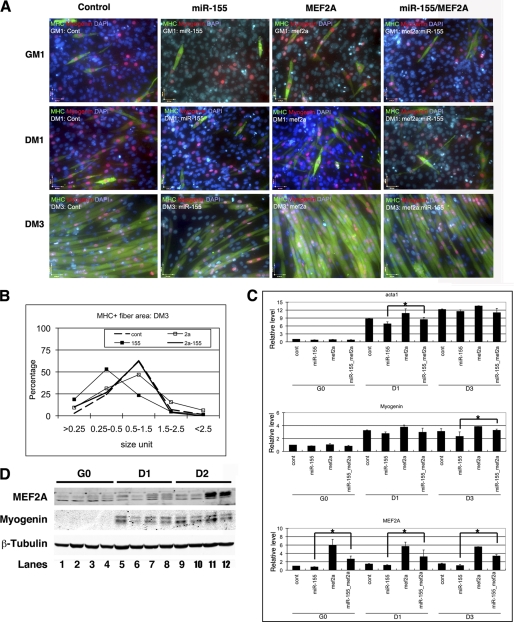
Overexpression of MEF2A Partially Rescues miR-155-mediated Myoblast Differentiation Defects
The above results indicate that either knockdown of MEF2A or overexpression of miR-155 leads to the inhibition of myoblast differentiation. Given that miR-155 inhibits the expression of MEF2A post-transcriptionally, we tested whether overexpression of MEF2A could rescue, at least in part, miR-155-mediated C2C12 myoblast differentiation defects. An adenoviral expression vector containing the Mef2A ORF without the 3′-UTR was used to overexpress the MEF2A protein. As shown in Fig. 4A, whereas miR-155 repressed myoblast differentiation, overexpression of MEF2A partially suppressed miR-155-mediated inhibition of myogenic differentiation. Results from quantitative measurement of myoblast and myotube size supported the view (Fig. 4B). qRT-PCR and Western blot analyses showed that the expression of myogenic markers was partially rescued by overexpression of MEF2A (Fig. 4, C and D).
FIGURE 4.
miR-155 inhibits myoblast differentiation which is suppresses by MEF2A. A, immunohistology of C2C12 cells treated with control, miR-155, Ad-MEF2A, or both miR-155 and Ad-MEF2A. Cells were induced to differentiate at indicated dates and were stained with antibodies that recognize striate muscle MHC (green) and myogenin (red). DAPI stains nuclei. GM1, growth medium day 1; DM1, differentiation medium day 1; DM3, differentiation medium day 3. B, quantitative analyses of cell size of MHC positive myoblast and myotubes in C2C12 cells treated with control, miR-155, MEF2A, or both miR-155 and MEF2A. The distribution of myoblast and myotube size was presented as percentage. C, quantitative real-time PCR analyses of gene expression in C2C12 cells treated with control, miR-155, MEF2A, or both miR-155 and MEF2A. Result was presented as relative expression level in which the control was assigned a value of 1. Data represent the mean + S.D. from at least three independent experiments. *, p < 0.05. G0, growth medium day 0; D1, differentiation medium day 1; D3, differentiation medium day 3. D, Western blot analyses of protein expression levels in C2C12 cells treated with control (contl), miR-155, MEF2A, or both miR-155 and MEF2A. D2, differentiation medium day 2.
DISCUSSION
In this study, we found that miR-155 represses the level of transcription factor MEF2A by binding to the 3′-UTR of Mef2A mRNA. We showed that miR-155-mediated repression of MEF2A plays a role in the regulation of muscle gene expression. Our findings therefore uncover a novel function for miR-155 and MEF2A in regulating the differentiation of skeletal muscle myoblasts.
miRNAs are a class of novel regulators for gene expression, and they have been demonstrated to be involved almost every aspects of biology, from stem cells, cell proliferation, differentiation, apoptosis to related diseases. We and others (14, 25–27) demonstrated previously that miR-1, miR-133, miR-206, and other muscle miRNAs (myomiRs) regulate skeletal muscle proliferation and terminal differentiation. Given the importance of MEF2A in cardiac and skeletal muscle gene expression, it is conceivable that the repression of MEF2A, either by siRNA-dependent repression or miR-155-mediated inhibition, impaired skeletal muscle myoblast differentiation. Indeed, our data demonstrated that inhibition of MEF2A by miR-155 represses myogenic differentiation. Recently, we also found that miR-1 represses MEF2A in cardiomyocytes (28).
Whereas our data clearly established a role for miR-155 in repressing MEF2A expression and myogenic differentiation, we are aware of the observation that miR-155-mediated repression of MEF2A only led to modest repression of myoblast differentiation. Our observation is consistent with many prior reports and current view on how most miRNAs work, in which miRNAs are thought to modestly modulate target gene expression and tips the balance of the biological system (22, 29–32). We noticed that additional cis-element presented in the 3′-UTR of the Mef2A transcript could also mediate its repression, indicating that miR-155 is not the only miRNA that represses the expression and inhibits the function of MEF2A. In fact, our results show that the 3′-UTR of Mef2A mediates its repression by miR-17–92, miR-9, miR-19a, miR-296, and probably other miRNAs (Fig. 1C). Whether such repression mediates MEF2A function in myogenesis remains to be determined.
miR-155 is derived from the non-protein-coding transcript of the BIC locus (33). Recent studies have established miR-155 a critical player in the immunosystem. It also functions as an oncogenic miRNA (34–37). In those studies, miR-155 was demonstrated to repress the expression of different targets, including SMAD2 (38), tumor protein 53-induced nuclear protein 1 (TP53INP1) (39), and Pu.1 (36). However, there was no report regarding to the potential function of this miRNA in cardiac and skeletal muscle. Our current studies, using cultured myoblast cell line in vitro, convincingly demonstrate the function of miR-155 in myoblast differentiation. It will be important for the future to determine whether the repression of MEF2A by miR-155 contributes to skeletal muscle development and function in vivo. It will also be interesting to investigate whether miR-155 and MEF2A participate in skeletal muscle degeneration/regeneration process as well as degeneration-related human muscular diseases, such as muscular dystrophy.
Supplementary Material
Acknowledgments
We thank members of the Wang laboratory for discussion and support. We are grateful to Dr. John McDermott (York University) for generous gift of anti-MEF2A antibody and to Dr. Isidore Rigoutsos (Thomas Jefferson University) for the initial analyses of miRNAs that target MEF2A. We thank Dr. Francisco Naya (Boston University) for sharing unpublished results and reagents.
This work was supported, in whole or in part, by National Institutes of Health. This work was also supported by the March of Dimes Foundation and Muscular Dystrophy Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- miRNA
- microRNA
- nt
- nucleotide(s)
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S. (1991) Science 251, 761–766 [DOI] [PubMed] [Google Scholar]
- 2. Arnold H. H., Winter B. (1998) Curr. Opin. Genet. Dev. 8, 539–544 [DOI] [PubMed] [Google Scholar]
- 3. Molkentin J. D., Olson E. N. (1996) Curr. Opin. Genet. Dev. 6, 445–453 [DOI] [PubMed] [Google Scholar]
- 4. Tapscott S. J. (2005) Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 5. Molkentin J. D., Black B. L., Martin J. F., Olson E. N. (1995) Cell 83, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 6. Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G. J., Parker M. H., MacQuarrie K. L., Davison J., Morgan M. T., Ruzzo W. L., Gentleman R. C., Tapscott S. J. (2010) Dev. Cell 18, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callis T. E., Wang D. Z. (2008) Trends Mol. Med. 14, 254–260 [DOI] [PubMed] [Google Scholar]
- 9. van Rooij E., Liu N., Olson E. N. (2008) Trends Genet. 24, 159–166 [DOI] [PubMed] [Google Scholar]
- 10. Small E. M., Olson E. N. (2011) Nature 469, 336–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., Amato A. A., Lidov H. G., Kang P. B., North K. N., Mitrani-Rosenbaum S., Flanigan K. M., Neely L. A., Whitney D., Beggs A. H., Kohane I. S., Kunkel L. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17016–17021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tatsuguchi M., Seok H. Y., Callis T. E., Thomson J. M., Chen J. F., Newman M., Rojas M., Hammond S. M., Wang D. Z. (2007) J. Mol. Cell. Cardiol. 42, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thum T., Galuppo P., Wolf C., Fiedler J., Kneitz S., van Laake L. W., Doevendans P. A., Mummery C. L., Borlak J., Haverich A., Gross C., Engelhardt S., Ertl G., Bauersachs J. (2007) Circulation 116, 258–267 [DOI] [PubMed] [Google Scholar]
- 14. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y., Ransom J. F., Li A., Vedantham V., von Drehle M., Muth A. N., Tsuchihashi T., McManus M. T., Schwartz R. J., Srivastava D. (2007) Cell 129, 303–317 [DOI] [PubMed] [Google Scholar]
- 16. Ivey K. N., Muth A., Arnold J., King F. W., Yeh R. F., Fish J. E., Hsiao E. C., Schwartz R. J., Conklin B. R., Bernstein H. S., Srivastava D. (2008) Cell stem cell 2, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Black B. L., Olson E. N. (1998) Annu. Rev. Cell Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 18. Black B. L., Lu J., Olson E. N. (1997) Mol. Cell Biol. 17, 2756–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He A., Kong S. W., Ma Q., Pu W. T. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu J., McKinsey T. A., Zhang C. L., Olson E. N. (2000) Mol. Cell 6, 233–244 [DOI] [PubMed] [Google Scholar]
- 21. Cao D., Wang Z., Zhang C. L., Oh J., Xing W., Li S., Richardson J. A., Wang D. Z., Olson E. N. (2005) Mol. Cell Biol. 25, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J. F., Tao Y., Li J., Deng Z., Yan Z., Xiao X., Wang D. Z. (2010) J. Cell Biol. 190, 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. (2006) Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 24. Naya F. J., Black B. L., Wu H., Bassel-Duby R., Richardson J. A., Hill J. A., Olson E. N. (2002) Nat. Med. 8, 1303–1309 [DOI] [PubMed] [Google Scholar]
- 25. Kim H. K., Lee Y. S., Sivaprasad U., Malhotra A., Dutta A. (2006) J. Cell Biol. 174, 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosenberg M. I., Georges S. A., Asawachaicharn A., Analau E., Tapscott S. J. (2006) J. Cell Biol. 175, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams A. H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J. L., Bassel-Duby R., Sanes J. R., Olson E. N. (2009) Science 326, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ikeda S., He A., Kong S. W., Lu J., Bejar R., Bodyak N., Lee K. H., Ma Q., Kang P. M., Golub T. R., Pu W. T. (2009) Mol. Cell Biol. 29, 2193–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sayed D., Abdellatif M. (2011) Physiol. Rev. 91, 827–887 [DOI] [PubMed] [Google Scholar]
- 30. Liu N., Olson E. N. (2010) Dev. Cell 18, 510–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Callis T. E., Deng Z., Chen J. F., Wang D. Z. (2008) Exp. Biol. Med. 233, 131–138 [DOI] [PubMed] [Google Scholar]
- 32. Chen J. F., Callis T. E., Wang D. Z. (2009) J. Cell Sci. 122, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eis P. S., Tam W., Sun L., Chadburn A., Li Z., Gomez M. F., Lund E., Dahlberg J. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tili E., Croce C. M., Michaille J. J. (2009) Int. Rev. Immunol. 28, 264–284 [DOI] [PubMed] [Google Scholar]
- 35. Dorsett Y., McBride K. M., Jankovic M., Gazumyan A., Thai T. H., Robbiani D. F., Di Virgilio M., Reina San-Martin B., Heidkamp G., Schwickert T. A., Eisenreich T., Rajewsky K., Nussenzweig M. C. (2008) Immunity 28, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vigorito E., Perks K. L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P. P., Miska E. A., Rodriguez A., Bradley A., Smith K. G., Rada C., Enright A. J., Toellner K. M., Maclennan I. C., Turner M. (2007) Immunity 27, 847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Science 316, 604–608 [DOI] [PubMed] [Google Scholar]
- 38. Louafi F., Martinez-Nunez R. T., Sanchez-Elsner T. (2010) J. Biol. Chem. 285, 41328–41336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gironella M., Seux M., Xie M. J., Cano C., Tomasini R., Gommeaux J., Garcia S., Nowak J., Yeung M. L., Jeang K. T., Chaix A., Fazli L., Motoo Y., Wang Q., Rocchi P., Russo A., Gleave M., Dagorn J. C., Iovanna J. L., Carrier A., Pébusque M. J., Dusetti N. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16170–16175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.