Background: The transfer of sialic acids is catalyzed by a set of sialyltransferases with defined specificities.
Results: We solved the ternary complex of the sialyltransferase Cst-II and kinetically characterized its mechanism.
Conclusion: Our analysis gives insights into the acceptor specificity and proposes the iso-ordered Bi Bi mechanism.
Significance: This work improves our understanding of sialyltransferase structure/function.
Keywords: Bacteria, Crystal Structure, Enzyme Kinetics, Enzyme Structure, Oligosaccharide, Campylobacter jejuni, Lipo-oligosaccharide, Sialyltransferase
Abstract
Sialic acids play important roles in various biological processes and typically terminate the oligosaccharide chains on the cell surfaces of a wide range of organisms, including mammals and bacteria. Their attachment is catalyzed by a set of sialyltransferases with defined specificities both for their acceptor sugars and the position of attachment. However, little is known of how this specificity is encoded. The structure of the bifunctional sialyltransferase Cst-II of the human pathogen Campylobacter jejuni in complex with CMP and the terminal trisaccharide of its natural acceptor (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc) has been solved at 1.95 Å resolution, and its kinetic mechanism was shown to be iso-ordered Bi Bi, consistent with its dual acceptor substrate specificity. The trisaccharide acceptor is seen to bind to the active site of Cst-II through interactions primarily mediated by Asn-51, Tyr-81, and Arg-129. Kinetic and structural analyses of mutants modified at these positions indicate that these residues are critical for acceptor binding and catalysis, thereby providing significant new insight into the kinetic and catalytic mechanism, and acceptor specificity of this pathogen-encoded bifunctional GT-42 sialyltransferase.
Introduction
Sialic acids (N-acetylneuraminic acid or Neu5Ac) are a family of nine carbon keto sugars, which participate in various biologically important processes, including cellular recognition, microbial infection, and tumor metastasis (7–11). They are widely disseminated molecules not only in mammalian tissues but also in pathogenic bacteria (12). In mammals, sialic acids typically exist as terminal residues on the outermost cell surface glycoconjugates (13, 14). For example, gangliosides located predominantly on vertebrate nerve cell surfaces are complex glycosphingolipids composed of oligosaccharide chains containing one or more terminal sialic acids (15). Sialic acids also have been found as terminal residues on the surface of a number of pathogenic bacteria and shown to play important roles during infection (11, 12, 16). A recent study of the human pathogen Campylobacter jejuni revealed that a sialylated variant of this bacterium showed significantly higher invasion levels of epithelial cells compared with unsialylated versions (17).
C. jejuni is recognized as the leading causative agent of bacterial diarrhea and food-borne gastroenteritis worldwide (1, 2). This organism has been shown to express variable outer core structures of lipo-oligosaccharide (LOS),7 which mimic the carbohydrate moieties of human gangliosides. Because of the molecular mimicry, the LOS of C. jejuni is considered as a major virulence factor, providing a protective barrier to evade the immune system of the host (3, 4). In addition, the carbohydrate mimicry between LOS outer core structures of C. jejuni and human gangliosides has been suggested to trigger the development of autoimmune diseases such as Guillain-Barré syndrome (5, 6). Oligosaccharides found in the outer core region of the LOS of various C. jejuni strains contain terminal sialic acid residues, which provide the diversity in these structures (3).
Sialyltransferases transfer sialic acids from an activated sugar donor (CMP-Neu5Ac) to various acceptor molecules including N-acetylgalactosamine (GalNAc), galactose, or another sialic acid at terminal positions of oligosaccharides of glycoproteins and glycolipids, with distinct linkages based on their specificities (18, 19). They are classified into several glycosyltransferase families in the CAZy data base, according to sequence similarities. Bacterial sialyltransferases are mainly categorized into four distinct families (GT-38, GT-42, GT-52, and GT-80) with polysialyltransferases located in GT-38.
The GT-42 enzyme Cst-II from C. jejuni OH4384 is a bifunctional sialyltransferase, transferring sialic acid to the terminal galactose moiety of the lipo-oligosaccharide with an α-2,3-linkage and subsequently transferring another sialic acid to the initially formed sialoside with an α-2,8-linkage (20). X-ray crystallographic studies of Cst-II in complex with the inert donor analog CMP-3FNeu5Ac7 revealed a structure that was a significant variation of the GT-A fold (21). Structural and mechanistic analyses of Cst-II supported a catalytic mechanism involving a conserved histidine residue as general base (His-188) that deprotonates the acceptor hydroxyl group during its attack on the anomeric carbon (C-2) of the donor substrate (CMP-Neu5Ac) (21, 22). Although the previously reported structure of Cst-II provided valuable information on donor binding, the lack of structural information on the acceptor sugar binding mode has hampered our ability to understand the molecular basis of acceptor binding specificity. Likewise, a detailed kinetic characterization that encompasses the dual acceptor specificity of this important class of glycosyltransferase has not been performed.
Here, we report the structure of Cst-II in complex with the donor analog, CMP, and the terminal trisaccharide of its natural acceptor, Neu5Ac-α-2,3-Gal-β-1,3-GalNAc, along with a detailed analysis of its kinetic mechanism. In addition, the roles of residues that bind to the sialic acid moiety of the acceptor are examined through structural and kinetic analyses of mutants. This work provides a mechanistic and structural basis for understanding acceptor binding during the LOS sialylation of C. jejuni and significantly improves our general understanding of the substrate specificity and mechanistic features of bacterial sialyltransferases.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Cst-IIΔ32 and Its Mutants
All molecular biology procedures were carried out in a similar manner to those described previously. Briefly, the truncated version of Cst-II (32 residue C-terminal deletion) was cloned in pET-28a vector (Novagen) with an N-terminal hexahistidine tag and subsequently transformed into electrocompetent Escherichia coli cells (BL21 λDE3, Novagen) for expression. To produce active site mutants, site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions with the following primers (Integrated DNA Technologies). Cst-II N51A-F (5′-GGTAAAAAATGCAAGGCAGTATTTTACGCTCCTATTCTTTTTTTTGAAC-3′) and Cst-II N51A-R (5′-GTTCAAAAAAAAGAATAGGAGCGTAAAATACTGCCTTGCATTTTTTACC-3′) were used to generate the N51A mutant. Cst-II Y81F-F (5′-CCGAACTAATTATGTGTTCTAATTTCAACCAAGCTCATCTAG-3′) and Cst-II Y81F-R (5′-CTAGATGAGCTTGGTTGAAATTAGAACACATAATTAGTTCGG-3′) were used to generate the Y81F mutant. The Cst-II N51A/Y81F double mutant was constructed by using the Cst-II N51A clone as a template with the forward primer Cst-II Y81F-F and the reverse primer Cst-II Y81F-R. Finally, Cst-II R129A-F and Cst-II R129A-R were used to generate the R129A mutant. All mutations were confirmed by DNA sequencing (Genewiz), and colonies with desired mutations were transformed into competent cells as described above. Expression and purification of wild type and mutant enzymes were performed as described previously (21).
Crystallization and Data Collection
Crystals of wild type Cst-IIΔ32 and its mutants were grown in similar conditions to those described previously (21). Briefly, 8 mg/ml of purified protein mixtures containing 10 mm CMP, MgCl2, and disaccharide (Gal-β-1,3-GalNAc) or trisaccharide (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc) acceptor were crystallized in mother liquor containing 100 mm HEPES, pH 7.5, 8% (w/v) polyethylene glycol 6000, and 5% (v/v) 2-methyl-2,4-pentanediol by the hanging-drop vapor diffusion technique at 21 °C. The resulting well occupied ternary complex of wild type Cst-IIΔ32, CMP, and trisaccharide acceptor belonged to the space group P4 with unit cell dimensions a = 117.67, b = 117.67, c = 46.60 Å, and contained two molecules in the asymmetric unit.
We note that various trials confirmed that additional soaking of Cst-II crystals cocrystallized in the presence of both CMP and the trisaccharide with the donor analog CMP-3FNeu5Ac expel bound trisaccharide acceptor from the active site. At the same time, cocrystallization with both CMP-3FNeu5Ac and the trisaccharide acceptor yielded only a binary donor complex. We also note that no observable density for acceptor were visible in trials utilizing the disaccharide (Gal-β-1,3-GalNAc) as acceptor.
The crystal of the Y81F mutant belonged to the same space group as the wild type enzyme (P4) with similar unit cell dimensions (a = 116.62, b = 116.62, and c = 46.84 Å) and also contained two molecules in the asymmetric unit. The crystal of the N51A mutant belonged to the space group I4 and contained one molecule in the asymmetric unit with unit cell dimensions a = 116.19, b = 116.19, and c = 46.95 Å. The crystals were cryoprotected in mother liquor containing 15% 2-methyl-2,4-pentanediol and directly plunged into liquid nitrogen prior to data collection. X-ray diffraction data were collected at 100 K under a nitrogen stream using an in-house CuKα rotating anode x-ray generator coupled to a Mar345 detector or using a synchrotron source (beamline 08ID-1 of the Canadian Light Source (Saskatoon, Saskatchewan, Canada) coupled to a Mar300CCD detector). Collected data were processed by the HKL2000 or MOSFLM suites (23, 24).
Structure Determination and Refinement
All structures were solved by molecular replacement with PHASER (25) using the monomer of wild type Cst-IIΔ32 (Protein Data Bank accession code 1RO8) as the starting model. Subsequent manual model building was performed by COOT (26), and refinement was carried out by REFMAC5 (27) excluding 5% of reflections for the Rfree calculation. In addition, Translation/Libration/Screw motion determination was incorporated into the procedure for further rounds of crystallographic refinement (28). The trisaccharide ligand files were generated from the Dundee PRODRG server (29). All structural figures were generated using PyMOL (30), and electrostatic surface calculations were performed with the APBS plugin (31). Final models were validated with MOLPROBITY (32).
Synthesis of Acceptor Substrates
Synthetic di- and trisaccharides used in this study (Gal-β-1,3-GalNAc-α-OBn, Neu5Ac-α-2,3-Gal-β-1,3-GalNAc-α-OBn, and Neu5Ac-α-2,3-Gal-β-1,3-GalNAc-β-pNP) were prepared in parallel from the corresponding monosaccharides (GalNAc-α-OBn and GalNAc-β-pNP) by glycosyltransferase-catalyzed addition of galactose and sialic acid as described below. GalNAc-α-OBn or GalNAc-β-pNP was dissolved at final concentrations in the reaction of 15 and 4 mm, respectively, in 50 mm NaOAc, pH 6.0, containing 10 mm MnCl2, 1 mm DTT, UDP-galactopyranose (1.1 eq), alkaline phosphatase (Sigma, 10–15 units/ml), and recombinant β-1,3-galactosyltransferase (CgtBOH4384ΔC30-C-term MalE (33), 0.1 mg/ml in reaction). The mixtures were monitored by TLC (mobile phase 7:2:1, ethyl acetate/methanol/water) until the starting material was completely consumed. The reaction mixture was diluted with an equal volume of cold methanol before centrifugation (2 min, 10,000 × g) to remove solids. The supernatant was concentrated, redissolved in water, and applied to a tC18 SepPak cartridge (Waters). After washing the cartridge with water and 5% methanol, products were eluted with higher concentrations of methanol (10–20%), concentrated, re-dissolved in water, and freeze dried. To obtain pure disaccharides, the tC18 SepPak purification was repeated several times. To obtain trisaccharide, the partially purified material was dissolved in 50 mm HEPES, pH 7.5, containing 10 mm MnCl2, 10 mm MgCl2, 4 mm DTT, CMP-Neu5Ac (1.0 eq based on monosaccharide starting material), and alkaline phosphatase (Sigma, 10–15 units/ml) at an estimated acceptor concentration of 2.5 mg/ml. Reaction was initiated by addition of the sialyltransferase Cst-I (34) (<0.1 mg/ml in reaction mixture). After standing overnight, the reaction mixture was applied to a tC18 SepPak cartridge. The cartridge was then washed with water, and the impure trisaccharide was eluted with 10% methanol. Fractions containing the target were pooled and concentrated before further purification in water on a column of BioGel P-2 (1.5 × 75 cm). Combined yields for the two glycosylation reactions were 30–50%.
Characterization of New Compounds
4-Nitrophenyl (5-acetamido-3,5-dideoxy-d-glycero-α-d-galacto-non-2-ulopyranosylonic acid)-(2→3)-β-d-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-β-d-galactopyranoside (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc-β-pNP) is characterized as follows: 1H NMR (600 MHz, D2O, δ) 8.24 (d, 2H, Ar, J = 9.4 Hz), 7.19 (d, 2H, Ar, J = 9.4 Hz), 5.30 (d, 1H, H-1, J1,2 = 8.7 Hz), 4.55 (d, 1H, H-1′, J1′,2′ = 7.7 Hz), 4.34 (dd, 1H, H-2, J2,3 = 10.8 Hz), 4.27 (d, 1H, H-4, J4,3 = 3.0 Hz), 4.07 (dd, 1H, H-3′, J3′,2′ = 9.7 Hz, J3′,4′ = 3.1 Hz), 4.01 (dd, 1H, H-3), 3.95–3.92 (m, 2H, H-4′), 3.88–3.77 (m, 5H, H-8″, H-5″, H-6′a, CH2), 3.75–3.70 (m, 2H, CH2), 3.70–3.60 (m, 4H, H-4″, H-6″, H6′b), 3.60–3.55 (m, 2H, H-5′, H-2′), 2.74 (dd, 1H, H-3eq″, J3eq″,4″ = 4.6 Hz, J3eq″,3ax″ = 12.3 Hz), 2.01 (s, 3H, NHAc), 1.98 (s, 3H, NHAc), 1.78 (dd, 1H, H-3ax″, J3ax″,4″ ∼12 Hz)). 13C NMR (100 MHz, D2O, δ) 175.1, 174.9, 173.9, 161.8, 142.6, 126.1, 116.5, 104.6, 99.6, 98.8, 79.5, 75.5, 75.3, 74.8, 72.8, 71.8, 69.0, 68.4, 68.0, 67.6, 67.6, 62.4, 61.0, 60.7, 51.6, 50.9, 39.7, 22.2, and 22.0. High resolution mass spectrometry calculated for C31H44N3O21 was 794.2473 and found was 794.2476.
Synthesis of Donor Analog CMP-3F-Neu5Ac
Cytidine-5′-monophospho-5-acetamido-3,5-dideoxy-3-fluoro-d-erythro-l- manno-2-nonulopyranosonic acid was synthesized from 3-fluorosialic acid and CTP using CMP-NeuAc synthetase according to a modified version of a previously published protocol (35, 36) 3-Fluorosialic acid (20 mg, 61 μmol) and CTP (50 mg, 92 μmol) were incubated with CMP-NeuAc synthetase (1 mg/ml) and inorganic pyrophosphatase (50 units) in 200 mm Tris, pH 8.5, containing 20 mm MgCl2 and 0.2 mm DTT. Prior to the addition of enzymes, the pH was adjusted to 8.5 by the addition of NaOH. The solution was incubated at room temperature for ∼18 h until reaction was complete (as determined by negative ion mode ESI-MS). Following completion of the reaction, enzyme was removed using a 5-kDa molecular mass cutoff Centricon filter. The retained solution was diluted with water and further spun twice. The combined aqueous solution containing the desired product was lyophilized, dissolved in 2 ml of water, filtered with a 0.2-μm syringe filter, and loaded onto a preparative HPLC TSKgel Amide-80 (Tosoh Bioscience) column. The desired product was purified using a flow rate of 6 ml/min and a gradient of acetonitrile (A)/water (B) (80:20 A/B holding for 20 min, linear gradient from 80:20 A/B to 65:35 A/B over 40 min, and holding 65:35 A/B for 15 min). The 6-ml fractions containing pure CMP-3FNeuAc (as determined by negative ion mode ESI-MS) were combined; solvent was removed under reduced pressure, and the resulting aqueous solution was lyophilized to yield a white fluffy solid (29.7 mg, 77%).
Enzyme Kinetics
Michaelis-Menten kinetic parameters were determined for Cst-II and mutants through variations of the concentration of acceptor at a fixed concentration of CMP-Neu5Ac. Enzyme concentrations were calculated using the method of Bradford (37). Reaction rates were determined using a continuous coupled spectrophotometric assay, as described previously (21, 38). Briefly, the release of CMP was coupled to the oxidation of NADH (λ = 340 nm, ϵ = 6.22 mm−1 cm−1) through the action of nucleoside monophosphate kinase, pyruvate kinase (PK), and lactate dehydrogenase. Rates of both the enzymatic and the spontaneous hydrolysis of CMP-Neu5Ac were measured and subtracted from enzymatic rate measurements. Assays were conducted at 37 °C in a 200-μl volume containing 20 mm HEPES, pH 7.5, 50 mm KCl, 1% (w/v) BSA, 10 mm MnCl2, 10 mm MgCl2, 0.7 mm phosphoenolpyruvate, 0.35 mm NADH, 2 mm ATP, 5.9 units of PK, 8.4 units of lactate dehydrogenase, and 7.9 units of nucleoside monophosphate kinase, 0.75 mm CMP-Neu5Ac, and varying concentrations of acceptor. The assay was initiated after the rate of NADH oxidation had stabilized by addition of enzyme with thorough mixing. Initial rates were plotted against acceptor substrate concentration, and Km and Vmax values were calculated from the best fit of the data to the Michaelis-Menten equation using nonlinear regression analysis with GraFit 5.0 (Erithacus software).
Capillary Electrophoresis-based Stopped Assay for Cst-II Kinetic Characterization
A stopped assay similar to that described previously (39) was utilized to determine inhibition kinetic parameters for Cst-II in the presence of the product inhibitor CMP. The assay mixture (10 μl final volume in triplicate) was composed of 50 mm HEPES, pH 7.5, 10 mm MgCl2, and 24 μg/ml Cst-II. When varying the concentration of CMP-Neu5Ac (50–500 μm), FCHASE-3′-sialyl-lactose acceptor was held at a fixed concentration of 500 μm. When varying the concentration of FCHASE-3′-sialyl-lactose (0.25–1 mm), CMP-Neu5Ac donor was held at a fixed concentration of 500 μm. CMP concentration was varied from 0.1 to 1 mm. Assays were carried out for 10 min, during which time the rate of product formation was linear over the range of substrates used and then quenched by the addition of an equal volume of stop solution consisting of 2% (w/v) SDS, 10 mm EDTA, and 50% (v/v) MeOH and stored at −20 °C prior to analysis by capillary electrophoresis. Reaction mixtures were analyzed by capillary electrophoresis using a Beckman Instruments PACE 5510 equipped with a 3-milliwatt argon ion laser-induced fluorescence detector (λexc = 488 nm, λemi = 520 nm). The capillary was of bare silica (75 μm × 57 cm) with the detector at 50 cm. Following capillary conditioning (washing with 0.2 m NaOH for 2 min, water for 2 min, and then equilibration with 20 mm sodium phosphate, pH 7.5), samples were injected under pressure for 2–5 s, and separation was performed at 18 kV, 75 μA. Peak integration was performed using the Beckman PACE station software and data were analyzed using GraFit 4.0 (Erithacus software).
RESULTS
Overall Structure
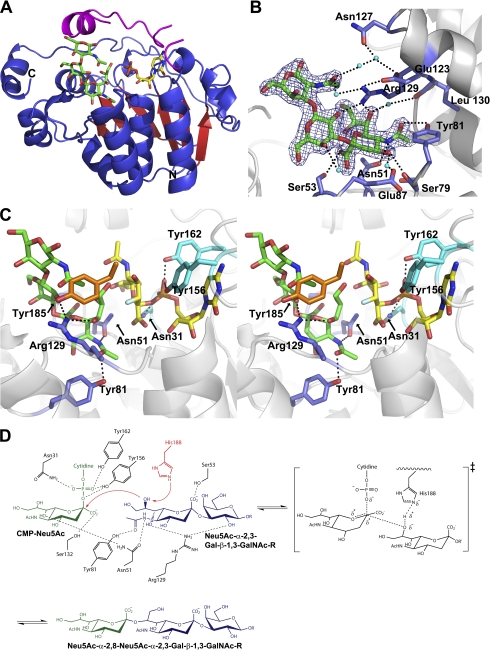
Structures of the ternary complex of wild type Cst-IIΔ32 and various mutants in complex with the donor analog CMP and the trisaccharide acceptor were solved via molecular replacement using the previously determined structure of native Cst-II (21) as a search model. The high resolution edge of the resulting diffraction data varied from 1.95 to 2.20 Å, and the resulting structures were well refined, with Rwork/Rfree values ranging from 17/20 to 18/23%. Full details of the data collection and refinement statistics are summarized in Table 1. The crystal structure of the newly solved Cst-IIΔ32 in complex with the donor analog (CMP) and the trisaccharide acceptor (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc) reveals a highly similar overall architecture to the previously reported Cst-II structure bound to the nonhydrolyzable donor analog (CMP-3FNeu5Ac) (21), with a root mean square deviation of 0.37–0.43 Å on 241 C-α atoms. This earlier structure was shown to form an intimately associated homotetramer (space group P21), with all four expected membrane-associating C-terminal domains aligned on the same face of the tetramer, a likely physiological requirement supported by static light scattering analysis in solution. The structures of the Cst-IIΔ32 ternary complex and Y81F mutant presented in this work belong to the space group P4, containing two molecules in the asymmetric unit, and the N51A mutant belongs to the space group I4, containing one molecule in the asymmetric unit, such that identically arranged tetrameric structures, as observed in the earlier monoclinic space group, can be generated around the 4-fold crystallographic axis of these tetragonal space groups.
TABLE 1.
Data collection and refinement statistics
| Cst-II + CMP + trisaccharide | N51A + CMP | Y81F + CMP | |
|---|---|---|---|
| Data collection | CLS 08ID | CuKα | CuKα |
| Wavelength | 0.9793 Å | 1.5418 Å | 1.5418 Å |
| Resolutiona | 40 to 1.95 Å (2.02–1.95) | 35 to 2.00 Å (2.11–2.00) | 35 to 2.20 Å (2.28–2.20) |
| Space group | P4 | I4 | P4 |
| a | 117.67 Å | 116.19 Å | 116.62 Å |
| b | 117.67 Å | 116.19 Å | 116.62 Å |
| c | 46.60 Å | 46.95 Å | 46.84 Å |
| α, β, γ | 90, 90, 90° | 90, 90, 90° | 90, 90, 90° |
| Rsymb | 8.1% (36.8%) | 8.4% (40.8%) | 4.6% (25.3%) |
| I/σ(I)a | 38.3 (5.0) | 16.6 (3.3) | 17.7 (2.7) |
| Completeness | 99.6% (96.9%) | 100% (100%) | 94.2% (87.7%) |
| Unique reflections | 46,782 (4,468) | 21,401 (3,099) | 30,500 (2,798) |
| Redundancy | 6.3 (5.6) | 4.4 (4.3) | 2.3 (1.8) |
| Refinement | |||
| Rworkc | 16.4% | 18.6% | 17.7% |
| Rfree | 19.8% | 23.0% | 22.8% |
| r.m.s.d bonds | 0.008 Å | 0.007 Å | 0.011 Å |
| r.m.s. angles | 1.068° | 0.986° | 1.228° |
| Average B-factor | |||
| Protein | 22.4 Å2 | 13.3 Å2 | 23.3 Å2 |
| Ligand | 42.8 Å2 | 22.8 Å2 | 40.3 Å2 |
| Water | 32.9 Å2 | 23.7 Å2 | 28.7 Å2 |
| Ramachandran plot | |||
| Most favored | 98.7% | 98.8% | 97.6% |
| Allowed | 1.3% | 1.1% | 2.4% |
a Values in parentheses represent the highest resolution shell.
b Rsym = Σ|I(hkl) − 〈I〉|/ΣI(hkl), where I(hkl) is the measured intensity of a given reflection and 〈I〉 is the average intensity of all symmetry equivalent measurements.
c Rwork = Σ|Fo − Fc|/Σ Fo, where Fo and Fc are observed and calculated structure factors, respectively. 5% of total reflections were excluded from the refinement to calculate Rfree.
d r.m.s., root mean square.
Donor-binding Site
The ternary complex structure of Cst-IIΔ32 shows clear electron density for both CMP in the donor-binding site and the trisaccharide acceptor. The amino acids that line the donor-binding site of the complex adopt a similar conformation to that observed in the previously reported Cst-II structure in complex with the inert donor sugar analog CMP-3FNeu5Ac. All key intermolecular contacts are maintained, including the stabilization of the phosphate moiety of CMP by the conserved Asn-31, Tyr-156, and Tyr-162 residues. As observed in the previous Cst-II-CMP-3FNeu5Ac binary complex (Protein Data Bank code 1RO7, monomer A), the flexible lid domain (residues 155–188) adopts a highly ordered, completely closed form in the new structure of the N51A (CMP) mutant. However, this domain is partially disordered in structures of the wild type ternary complex (Cst-II, CMP, and Neu5Ac-α-2,3-Gal-β-1,3-GalNAc acceptor) and the Y81F (CMP) mutant presented here, whether due to crystal packing restraints or otherwise remaining unclear. As such, support for a potential role of the donor sugar Neu5Ac in stabilizing the lid will require future analysis of other donor-acceptor complexes.
Acceptor-binding Site
The physiological trisaccharide acceptor (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc) binds to the open cleft of Cst-II at the C-terminal half of the Rossmann domain adjacent to the donor-binding site (Fig. 1A). The acceptor substrate binds to the enzyme through a number of hydrogen bonds to the N-acetamide group of the terminal sialic acid moiety. The N-acetamide group interacts with the side chains of both the conserved Asn-51 and Tyr-81. The guanidinium side chain of Arg-129 establishes hydrogen bonds to the hydroxyl group at C-7 of the sialic acid moiety and the hydroxyl group at C-2 of the galactose moiety of the trisaccharide acceptor. In addition, the carboxyl group of the acceptor sialic acid interacts with Ser-53 and the C-4 hydroxyl group with the carbonyl of Ser-79 (Fig. 1B). There are also water-mediated hydrogen bonds between the sialic acid moiety of the trisaccharide and residues lining the acceptor-binding site. For example, the hydroxyl groups at C-7 and C-4 interact with the main chain carbonyl of Ile-130 and the side chain of Glu-87, respectively. Similarly, the carboxyl group of the sialic acid moiety and the hydroxyl group at C-4 of the galactose moiety are involved in water-mediated contacts, interacting with the main chain carbonyl of Leu-86. In addition, the N-acetylgalactosamine moiety of the acceptor interacts with various residues, including Glu-123, Asn-127, and Arg-129 through bound water to further stabilize the acceptor binding.
FIGURE 1.
Structural analysis of the ternary complex of Cst-II and the mechanistic scheme. A, overall structure of Cst-II in complex with donor analog (CMP) and the trisaccharide acceptor (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc). The flexible lid domain (residues 155–188) is shown in magenta. B, interactions between the trisaccharide acceptor and active site residues. Electron density of the trisaccharide acceptor in a refined 2Fo − Fc map contoured at 1.5σ. Dotted lines indicate hydrogen bonding. Residues 82–86 have been omitted for clarity. C, active site of Cst-II with the trisaccharide acceptor and modeled donor (CMP-3FNeu5Ac). The structure is shown in divergent (wall-eyed) stereo. Residues interacting with donor and acceptor are shown in cyan and blue, respectively. Tyr-185 is shown in orange. Carbon atoms are shown in green and yellow for the trisaccharide acceptor and CMP (or CMP-3FNeu5Ac), respectively. The label of Ser-53 has been omitted for clarity. Non-carbon atoms are colored according to atom type (nitrogen, blue; oxygen, red; phosphorus, orange; fluorine, silver). D, proposed mechanism of Cst-II showing all interacting residues for donor and acceptor substrates. Donor substrate (CMP-Neu5Ac) is shown in green, and acceptor substrate is shown in blue. Catalytic base (His-188) is shown in red.
Mutant Structures
Two essential residues (Asn-51 and Tyr-81) interact with the N-acetamide group of the sialic acid moiety of the trisaccharide acceptor. Crystal structures of mutants modified at these positions, Cst-IIΔ32 N51A and Y81F, in the presence of CMP show the same overall tertiary structures as observed in the wild type (root mean square deviation of ∼0.4 Å on 241 C-α atoms). Mutant crystals were grown under the same conditions containing both CMP and the trisaccharide acceptor, as used for the crystallization of the wild type enzyme. However, the structures of these mutants show only electron density corresponding to CMP in the donor-binding site with no visible density corresponding to the trisaccharide acceptor.
Kinetic Analysis of Mutants
Kinetic analyses of wild type Cst-IIΔ32 and mutants were performed to characterize the consequences of these changes upon catalysis (Table 2). All kinetic experiments were carried out at 37 °C, and data were fitted to the Michaelis-Menten equation. For the wild type enzyme, both disaccharide (Gal-β-1,3-GalNAc-α-OBn) and trisaccharide (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc-α-OBn) acceptor substrates were analyzed to compare kinetic parameters for the two distinct acceptors. Kinetic characterizations for mutants (N51A, Y81F, and R129A) were performed only with the trisaccharide acceptor. The trisaccharide is a significantly better acceptor for Cst-IIΔ32 than the disaccharide, with a Km value of 6.9 mm, which is 5-fold lower than that of the disaccharide, and specificity constants (kcat/Km) for the trisaccharide and the disaccharide acceptors of 8.9 and 0.2 min−1mm−1, respectively (Table 2). Kinetic analysis of the mutants reveals only relatively small effects (3-fold decrease in kcat/Km) due to the removal of the hydroxyl group of Tyr-81. However, much larger rate decreases (45-fold decrease in kcat/Km) accompany the removal of the side chains of Asn-51 and Arg-129. Interestingly these effects are expressed almost entirely in reduced kcat values, with relatively little effect upon Km, indicating that interactions that are crucial to transition state binding exclusively are formed at these positions.
TABLE 2.
Kinetic parameters for acceptor substrates with wild type Cst-IIΔ32 and mutants
| Cst-II variant | Acceptor | Km | kcat | kcat/Km |
|---|---|---|---|---|
| mm | min−1 | mm−1min−1 | ||
| WT (Δ32) | Galβ1,3GalNAcαOBn | 34.7 ± 4.0 | 7.9 | 0.2 |
| WT (Δ32) | Neu5Acα2,3Galβ1,3GalNAcαOBn | 6.9 ± 0.4 | 61.1 | 8.9 |
| Y81F | Neu5Acα2,3Galβ1,3GalNAcαOBn | 13.7 ± 1.1 | 39.5 | 2.9 |
| N51A | Neu5Acα2,3Galβ1,3GalNAcαOBn | 5.9 ± 0.7 | 1.1 | 0.2 |
| R129A | Neu5Acα2,3Galβ1,3GalNAcαOBn | 18.6±.3 | 2.8 | 0.2 |
| N51A/Y81Fa | Neu5Acα2,3Galβ1,3GalNAcαOBn | – | – | – |
a Kinetic constants for N51A/Y81F double mutant could not be obtained since it was completely inactive.
Potential Kinetic Mechanisms
In both of the nonhydrolytic reactions catalyzed by Cst-II, two substrates were converted to two products. As such, Cst-II is designated a Bi Bi enzyme according to the notation developed by Cleland for multisubstrate enzymes (40). Such Bi Bi reactions can occur by any one of four possible basic kinetic mechanisms as follows: ping-pong, random sequential, ordered sequential, or Theorell-Chance, as shown in supplemental Fig. S1. Reactions are further termed rapid equilibrium if chemical steps are slower than substrate associations and product release or steady-state if this is not the case. Background on mechanistic dissection is provided in supplemental material.
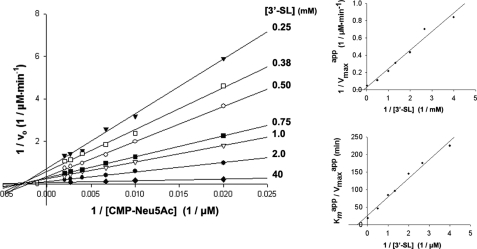
To distinguish between a ping-pong and a sequential mechanism, initial rates were measured with increasing concentrations of CMP-Neu5Ac at various constant concentrations of 3′-sialyl-lactose using the enzyme-coupled continuous assay (Fig. 2). The resulting family of straight lines is clearly not parallel, as would have been expected for a ping-pong mechanism, but instead converges to the left of the 1/vo axis and approximately on the 1/[CMP-Neu5Ac] axis. A ping-pong mechanism for Cst-II can therefore be ruled out, as indeed would be expected for an inverting mechanism for which no stable intermediate species is formed. For a Bi Bi mechanism, these data alone cannot be used to distinguish between the various types of sequential mechanisms, and thus inhibition studies are required.
FIGURE 2.
Double-reciprocal and secondary plots of initial rate data with respect to CMP-Neu5Ac at various constant concentrations of 3′-sialyl-lactose (3′-SL).
Product Inhibition Studies with Cst-II
Product inhibition studies can be used to distinguish whether substrates bind to and products are released from a sequential Bi Bi enzyme in an ordered or random fashion (41–43). For ordered reactions, the patterns of product inhibition can also identify the order of substrate association and product release. The product inhibition patterns for several sequential Bi Bi mechanisms are shown in supplemental Table S1. For a more complete list of inhibition patterns, the reader is directed to the detailed analysis of Segel (41).
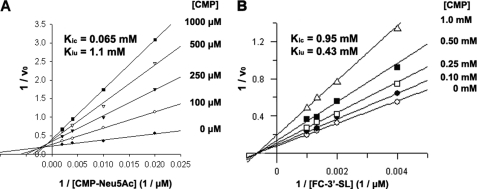
Because CMP release is the mode of detecting transferase activity in the enzyme-coupled continuous assay, to determine patterns of inhibition with respect to the product CMP, a stopped capillary electrophoresis-based assay was used, in which product formation was detected using fluorescein 3′-sialyl-lactose conjugate (FCHASE 3′-SL) as the acceptor substrate. Representative capillary electrophoresis traces for product formation at various concentrations of CMP-Neu5Ac and CMP are shown in supplemental Fig. S2. Because the presence of the fluorescein substituent limits the aqueous solubility of the conjugated acceptor, initial rate measurements could only be obtained using nonsaturating concentrations of the acceptor substrate. In addition, because of the instability of CMP-Neu5Ac and resulting difficulty in obtaining donor substrate free of contaminating CMP (∼10% contamination following HPLC purification), initial rates could also only be obtained under nonsaturating concentrations of the donor substrate. Using CMP as a product inhibitor of the Cst-II reaction, mixed inhibition patterns were observed with respect to both the donor substrate CMP-Neu5Ac (Fig. 3A) and the acceptor substrate FCHASE 3′-SL (Fig. 3B) when the nonvaried substrate was held at a fixed nonsaturating concentration. Competitive (Kic) and uncompetitive (Kiu) inhibition constants were determined by direct fit of the data to the equation for mixed inhibition using the program GraFit 4.0, and these are shown on the double-reciprocal plots (Fig. 3). Somewhat surprisingly, these modes of inhibition are not consistent with either of the expected steady-state ordered or Theorell-Chance Bi Bi mechanism (supplemental Table 1). Mixed product inhibition patterns for both substrates in the presence of fixed nonsaturating concentrations of the other substrate are consistent with either a steady-state random Bi Bi or an iso-ordered Bi Bi mechanism.
FIGURE 3.
Inhibition of Cst-II-catalyzed sialyl transfer to FCHASE 3′-sialyl-lactose (3′-SL) by the product CMP with respect to donor (A) and acceptor (B) substrates.
Inhibition Studies of a Dead-end Donor Substrate Analog
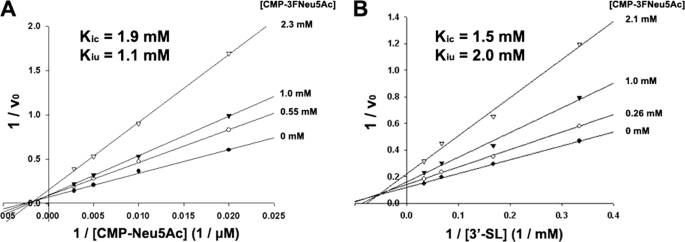
Further insight into the mechanisms of bisubstrate enzyme systems can be obtained from inhibition studies using dead-end substrate analogs (41, 44). Inhibition by the dead-end donor substrate analog CMP-3FNeu5Ac was therefore subjected to detailed kinetic characterization. Earlier kinetic studies with limited quantities of CMP-3FNeu5Ac had suggested that it functioned as a competitive inhibitor of Cst-II with respect to CMP-Neu5Ac (21). However, subsequent more rigorous analysis reported here clearly indicates that using CMP-3F Neu5Ac as a dead-end substrate analog leads to mixed inhibition patterns with respect to both CMP-Neu5Ac (Fig. 4A) and 3′-sialyl-lactose (Fig. 4B) when the nonvaried substrate is held at a fixed nonsaturating concentration. Competitive (Kic) and uncompetitive (Kiu) inhibition constants were determined by direct fit of the data to the equation for mixed inhibition using the program GraFit 4.0, and these values are shown on the double-reciprocal plots (Fig. 4).
FIGURE 4.
Inhibition of Cst-II-catalyzed sialyl transfer to 3′-sialyl-lactose (3′-SL) by the dead-end substrate analog CMP-3FNeu5Ac with respect to donor (A) and acceptor (B) substrates.
As was observed with CMP product inhibition studies, these patterns of inhibition are not consistent with either the ordered or random Bi Bi kinetic mechanism. For both of these mechanisms, competitive inhibition behavior would be expected for CMP-3FNeu5Ac with respect to CMP-Neu5Ac (44). However, the observed modes of inhibition for CMP-3FNeu5Ac are understandable in light of the bifunctional nature of Cst-II. An ability to associate with two distinct stable forms of the enzyme would make CMP-3FNeu5Ac a mixed inhibitor with respect to CMP-Neu5Ac. This mode of inhibition is therefore consistent with a kinetic mechanism that is more complex than that of the steady-state ordered Bi Bi, and although by no means conclusive, an iso-ordered Bi Bi mechanism seems the most probable (Fig. 5).
FIGURE 5.
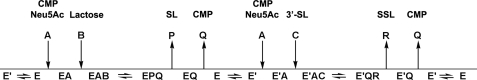
Cleland notation schematic representation of the steady-state iso-ordered Bi Bi mechanism. In the case of Cst-II, E and E′ represent stable enzyme forms that catalyze sialyl transfer to lactose and 3′-sialyl-lactose (SL), respectively. A represents CMP-Neu5Ac; B represents lactose, and C represents 3′-sialyl-lactose (as substrate). P represents 3′-sialyl-lactose (as product); Q represents CMP, and R represents the disialylated product (SSL). Product Q can associate with either stable form of the enzyme E or E′, which would make it a mixed inhibitor with respect to A. The dead-end analog of A could associate with both stable forms of the enzyme E and E′, which would make it a mixed inhibitor with respect to substrate A.
DISCUSSION
Of the few glycosyltransferases that have been subjected to detailed kinetic analysis, to date the majority have been shown to catalyze their respective reactions using an ordered Bi Bi (GnTII (45); XylT (46); FucT (47); MurG (48); LgtC (49); TagA (50); lactose synthase (51); α3GalT (52)) or Theorell-Chance (human blood group B glycosyltransferase (53)) mechanism involving the obligate binding of the nucleotide sugar donor substrate prior to the association of the acceptor. This finding has been rationalized on the basis of obtained three-dimensional x-ray crystal structures of glycosyltransferases with bound donor substrates. Upon binding the donor substrate, a conformational change occurs in which a flexible loop closes over the donor substrate and forms a significant portion of the acceptor substrate binding site, consistent with the observed ordered kinetic mechanisms. An exception to this general observation is the case of a polypeptide GalNAc transferase, which was found to undergo a rapid equilibrium random Bi Bi mechanism using erythropoietin and erythropoietin-like peptides as acceptors and dead-end acceptor analogs (54). Interestingly, unlike most glycosyltransferases that demonstrate a much higher affinity for their donor substrates than their acceptors, the apparent binding affinities of the donor and acceptor substrates for polypeptide GalNAc transferase were found to be comparable.
In the steady-state random Bi Bi mechanism, binding of substrates and release of products does not occur in a defined order but does contribute to the overall observed rate of reaction. Very few examples of the steady-state random mechanism have been reported. The most frequently quoted example, yeast hexokinase, required years of argument and reports of seemingly contrasting data before an authoritative study by Rudolf and Fromm (55) provided significant irrefutable evidence. This mechanism is very difficult to unambiguously identify for several reasons. The rate equation is painfully complex, making the derivation of expected modes of inhibition very difficult to predict. The initial rate data for this mechanism should in theory be nonhyperbolic, although there are several reasons why obtained data can appear hyperbolic (41), which can lead to erroneous interpretations of data. Unfortunately, for reasons described above, in the case of Cst-II initial rates cannot be measured under truly saturating concentrations of donor substrate, and thus curve fitting to nonhyperbolic equations cannot be used to support this mechanism. An additional complexity of the steady-state random mechanism comes from the fact that certain portions of the mechanism can be in rapid equilibrium, although others contribute to the overall rate, further complicating the derivation of the actual rate equation and the prediction of modes of inhibition.
The steady-state iso-ordered Bi Bi mechanism requires an obligate order of substrate binding and product release but also involves an isomerization between two stable enzyme forms that contributes to the overall observed rate of reaction (41). In such a mechanism, product Q will be a mixed inhibitor with respect to substrate A, as was found to be the case with Cst-II, if the two can bind to different stable forms of the enzyme (Fig. 5).
Although we cannot rule out the steady-state random Bi Bi, we favor the steady-state iso-ordered Bi Bi mechanism. Indeed, as Cst-II is a bifunctional enzyme catalyzing two distinct sialyl transfer reactions, it seems quite reasonable that the enzyme could exist in two distinct stable forms, one in which the acceptor site binds sialyl-lactose or equivalents (e.g. Neu5Ac-α-2,3-Gal-β-1,3-GalNAc) and one in which it binds lactose or an equivalent (e.g. Gal-β-1,3-GalNAc). Product CMP could bind to either of these enzyme forms during the product inhibition study, giving the observed mixed inhibition pattern with respect to CMP-Neu5Ac. The existence of two stable enzyme forms of Cst-II is also consistent with the fact that CMP-3FNeu5Ac acts as a mixed inhibitor with respect to CMP-Neu5Ac.
It was of considerable interest to observe these two separate binding modes structurally, hence efforts to derive ternary complex structures with CMP in the donor site and disaccharide (Gal-β-1,3-GalNAc) or (sialyl)trisaccharide (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc) in the acceptor site. Earlier attempts to obtain the acceptor complex of Cst-IIΔ32 by using both cocrystallization and soaking with various acceptor analogs, including lactose, 3′-deoxylactose, and sialyllactose in the presence and absence of the donor analog CMP-3FNeu5Ac, were unsuccessful (21). Here, in our hands, we find that the ternary complex structure can only be obtained by cocrystallization with CMP and at least the terminal trisaccharide of its natural acceptor Neu5Ac-α-2,3-Gal-β-1,3-GalNAc.
Inspection of the resulting structure reveals that interactions between the N-acetamide group of the sialic acid moiety of the trisaccharide acceptor and several conserved residues (Asn-51, Tyr-81, and Arg-129) appear crucial for proper binding. Furthermore, our ensemble of structures suggests the GalNAc moiety of the natural acceptor used here is poised by the nature of its β1,3-linkage to interact with the flexible lid domain (residues 155–188) for proper orientation of the acceptor substrate during the catalytic reaction. Structural superimposition of the current ternary complex and the previously reported donor complex suggests that the side chain of Tyr-185 (Fig. 1C), in the lid domain, is a feasible candidate to interact with the GalNAc N-acetamide group of our bound acceptor. These results also help to explain the inability to trap previously attempted acceptor analogs such as lactose and 3′-deoxylactose analogs (in isolation and in longer variants with β1,4-linkages to downstream galactose or glucose sugars), which would lack the analogous N-acetamide moieties that we observe to be so critical to binding of the physiological acceptor used here.
The importance of residues Asn-51, Tyr-81, and Arg-129 localized in the observed acceptor-binding site was further validated by structural and kinetic analysis of mutants, which showed a complete absence of the trisaccharide acceptor in those active sites as well as a dramatic reduction of the specificity constants (Table 2). Interestingly, kinetic analysis of the Y81F mutant revealed rate constants comparable with the wild type enzyme (Table 2). Structural analysis of the acceptor-binding site identifies that the carbonyl of the N-acetamide group of the sialic acid moiety in the trisaccharide acceptor interacts with Tyr-81 as well as an ordered water molecule. This water in turn directly interacts with the carbonyl of Ile-130 in the enzyme and the C-7 hydroxyl group of the acceptor to further stabilize acceptor binding, which we believe potentially minimizes the adverse effects on acceptor binding imposed by loss of hydrogen bonding in the Y81F mutant form of Cst-II.
Extensive attempts to obtain the complex structure with the disaccharide acceptor (Gal-β-1,3-GalNAc) have been unsuccessful probably due to its weak binding affinity. Previously reported kinetic parameters of wild type Cst-IIΔ32 with lactose-based acceptor analogs demonstrated that lactose, which mimics the disaccharide acceptor, showed a 10-fold higher Km value than that of the 3′-sialyllactose, an analog of the trisaccharide acceptor substrate (21). Similar kinetic behavior regarding acceptor binding was observed here, where the disaccharide acceptor shows 5-fold lower binding affinity compared with that of the trisaccharide.
The superimposition of the acceptor-bound ternary complex of Cst-IIΔ32 coupled with the previous binary complex (Protein Data Bank code 1RO7) provides valuable insights into the catalytic mechanism of α-2,8-sialyl transfer catalyzed by Cst-II. The closure of the flexible lid protects the active site of the enzyme from bulk solvent, favoring electrostatic interactions in the complex and minimizing undesirable hydrolysis of the donor substrate. Our data show that the C-8 carbon of the acceptor sialic acid is ∼4 Å away from both the imidazole side chain of His-188 and the anomeric carbon (C-2) of the sialic acid moiety of the donor substrate. Furthermore, analysis of our structure indicates that the acceptor O-8 hydroxyl group is free to rotate even closer toward the His-188 side chain (∼3 Å). As such, His-188 is suitably located to deprotonate the O-8 hydroxyl group of the sialic acid moiety of the trisaccharide acceptor substrate. At the pH optimum of Cst-II activity, pH 8 (22), His-188 is deprotonated, facilitating this general base role. The activated acceptor sugar in turn is localized for direct attack on the anomeric carbon (C-2) of the sialic acid of the donor substrate, generating the oxocarbenium ion-like transition state aided by interactions with Tyr-156 and Tyr-162 (Fig. 1D).
Further features of the additional α-2,3-sialyl transfer of the bifunctional Cst-II can be rationally proposed by structural comparisons of the ternary complex we present here with the structure of Cst-I, a monofunctional α-2,3-sialyltransferase (34). These sialyltransferases share significant sequence and structural similarities (50% sequence identity, with a root mean square deviation of 0.77 Å on 235 C-α atoms) (supplemental Figs. S3 and S4A). In particular, essential residues in the acceptor binding site of the bifunctional Cst-II show high structural and sequence similarity to corresponding residues of the monofunctional Cst-I. For example, Asn-51, Ser-79, and Arg-129 residues in Cst-II are conserved in Cst-I as Asn-66, Ser-94, and Arg-144, respectively (supplemental Fig. S3). Interestingly, Arg-144 is oriented ∼1.4 Å closer to the acceptor-binding site in the Cst-I structure as compared with its Cst-II Arg-129 counterpart, effectively creating a smaller active site pocket to accommodate its smaller disaccharide acceptor (Gal-β-1,3-GalNAc) (supplemental Fig. S4B). We suggest this arginine residue also plays a role to ensure the proper orientation of the disaccharide acceptor for the α-2,3-sialyl transfer reaction catalyzed by Cst-I and by analogy may play a similar role in acceptor site modulation in accommodating the smaller disaccharide acceptor in the α-2,3-sialyl transfer step of the bifunctional Cst-II. In Cst-I, the disaccharide acceptor can be further stabilized by a hydrogen bonding network involving Asn-66 and the carbonyl of Ser-94, and again by analogy via residues Asn-51 and Ser-79 in Cst-II. Conversely, in the trisaccharide acceptor complex of Cst-II, Tyr-81 specifically interacts with the N-acetamide group of the sialic acid moiety. The corresponding amino acid (Phe-96) of Cst-I is unable to form an analogous direct interaction with the acceptor substrate suggesting that this position is not critical to α-2,3-sialyl transfer in either Cst-I or by analogy, Cst-II.
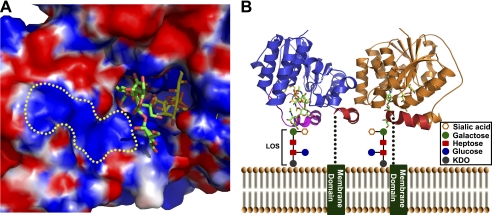
The electrostatic surface map of Cst-II clearly shows the charge complementarity between substrates and the active site of the enzyme (Fig. 6A). In the structure of bacterial LOS, the terminal oligosaccharide consists of a number of negatively charged sugar residues, including sialic acid. The positively charged cluster we observe near the identified acceptor-binding site of Cst-II highlights the potential of the enzyme in recognizing this negatively charged inner core oligosaccharide region of the LOS. Our structural analysis also suggests how the physiological tetrameric form of Cst-II (observed in our structures and in solution) binds to core oligosaccharides of membrane-localized C. jejuni LOS to allow access and transfer of sialic acid to specific acceptor saccharide units therein. The truncated C-terminal region of Cst-II is composed of 32 amino acids, which likely forms a membrane-spanning helix as predicted strongly by various sequence-based algorithms (56). This hydrophobic C-terminal region acts as a membrane anchor to appropriately localize and orient the positively charged Cst-II acceptor sites (which all align on one face of the tetramer) to access the growing membrane-anchored lipo-oligosaccharide chains for the sialyl transfer reaction (Fig. 6B).
FIGURE 6.
Electrostatic surface map of the active site of Cst-II and proposed membrane-associated model of Cst-II tetramer. A, electrostatic molecular surface is shown in red for negatively charged regions and blue for positively charged regions. Both substrates (acceptor and donor) bind to the positively charged active site pocket of the enzyme. The yellow dotted region indicates the potential binding site for inner core oligosaccharide units of the lipo-oligosaccharide. B, membrane-associated anchor domains are shown as green bars. Only two monomeric subunits of the Cst-II tetramer are represented for clarity (in blue and orange). Green dotted lines indicate connecting loops between Cst-IIΔ32 and membrane-anchored domains. C-terminal helices of the Cst-IIΔ32 are shown in red. The flexible lid domain (residues 155–188) of the monomer is shown in magenta. Inner core oligosaccharide units of C. jejuni LOS are represented as schematic diagrams. Carbon atoms of the trisaccharide acceptor (Neu5Ac-α-2,3-Gal-β-1,3-GalNAc) and the nonhydrolyzable donor (CMP-3FNeu5Ac) are shown in green and yellow, respectively. Non-carbon atoms are colored according to atom type (nitrogen, blue; oxygen, red; phosphorus, orange; fluorine, silver).
Our structural and kinetic analysis of the ternary complex of sialyltransferase Cst-II with the donor analog and natural trisaccharide acceptor provides detailed acceptor binding features and further insights into the catalytic mechanism of this class of enzyme, thereby assisting the design of novel therapeutic agents to inhibit the sialylation of pathogenic bacteria.
Supplementary Material
Acknowledgments
We thank the X-ray Crystallography Hub at the Centre for Blood Research (University of British Columbia) and the Canadian Light Source (Saskatoon, Saskatchewan, Canada) for data collection. We also thank Dr. Andrew L. Lovering for synchrotron data collection, and Drs. Leo Yen-Cheng Lin and Francesco V. Rao for fruitful discussions.
This work was supported in part by operating grants from the Canadian Institutes of Health Research (to N. C. J. S., W. W. W., and S. G. W.) and the Howard Hughes Medical Institute International Scholar Program (to N. C. J. S.), with infrastructure funding from the Michael Smith Foundation for Health Research, the Canada Foundation for Innovation, and the B.C. Knowledge Development Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Table S1, and additional references.
The atomic coordinates and structure factors (codes 2X61, 2X62, and 2X63) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LOS
- lipo-oligosaccharide
- CMP-3FNeu5Ac
- CMP-3-fluoro-N-acetylneuraminic acid
- FCHASE
- 6-(fluorescein-5-carbaxamido)hexanoic acid.
REFERENCES
- 1. Nachamkin I., Allos B. M., Ho T. (1998) Clin. Microbiol. Rev. 11, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mead P. S., Slutsker L., Dietz V., McCaig L. F., Bresee J. S., Shapiro C., Griffin P. M., Tauxe R. V. (1999) Emerg. Infect. Dis. 5, 607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilbert M., Karwaski M. F., Bernatchez S., Young N. M., Taboada E., Michniewicz J., Cunningham A. M., Wakarchuk W. W. (2002) J. Biol. Chem. 277, 327–337 [DOI] [PubMed] [Google Scholar]
- 4. Guerry P., Szymanski C. M., Prendergast M. M., Hickey T. E., Ewing C. P., Pattarini D. L., Moran A. P. (2002) Infect. Immun. 70, 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Endtz H. P., Ang C. W., van Den Braak N., Duim B., Rigter A., Price L. J., Woodward D. L., Rodgers F. G., Johnson W. M., Wagenaar J. A., Jacobs B. C., Verbrugh H. A., van Belkum A. (2000) J. Clin. Microbiol. 38, 2297–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuki N., Susuki K., Koga M., Nishimoto Y., Odaka M., Hirata K., Taguchi K., Miyatake T., Furukawa K., Kobata T., Yamada M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11404–11409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Angata T., Varki A. (2002) Chem. Rev. 102, 439–469 [DOI] [PubMed] [Google Scholar]
- 8. Hakomori S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10231–10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schauer R. (2004) Zoology 107, 49–64 [DOI] [PubMed] [Google Scholar]
- 10. Olofsson S., Bergström T. (2005) Ann. Med. 37, 154–172 [DOI] [PubMed] [Google Scholar]
- 11. Sáez-Llorens X., McCracken G. H., Jr. (2003) Lancet 361, 2139–2148 [DOI] [PubMed] [Google Scholar]
- 12. Severi E., Hood D. W., Thomas G. H. (2007) Microbiology 153, 2817–2822 [DOI] [PubMed] [Google Scholar]
- 13. Varki A. (1997) FASEB J. 11, 248–255 [DOI] [PubMed] [Google Scholar]
- 14. Traving C., Schauer R. (1998) Cell. Mol. Life Sci. 54, 1330–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vyas A. A., Schnaar R. L. (2001) Biochimie 83, 677–682 [DOI] [PubMed] [Google Scholar]
- 16. Preston A., Mandrell R. E., Gibson B. W., Apicella M. A. (1996) Crit. Rev. Microbiol. 22, 139–180 [DOI] [PubMed] [Google Scholar]
- 17. Louwen R., Heikema A., van Belkum A., Ott A., Gilbert M., Ang W., Endtz H. P., Bergman M. P., Nieuwenhuis E. E. (2008) Infect. Immun. 76, 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harduin-Lepers A., Recchi M. A., Delannoy P. (1995) Glycobiology 5, 741–758 [DOI] [PubMed] [Google Scholar]
- 20. Gilbert M., Brisson J. R., Karwaski M. F., Michniewicz J., Cunningham A. M., Wu Y., Young N. M., Wakarchuk W. W. (2000) J. Biol. Chem. 275, 3896–3906 [DOI] [PubMed] [Google Scholar]
- 21. Chiu C. P., Watts A. G., Lairson L. L., Gilbert M., Lim D., Wakarchuk W. W., Withers S. G., Strynadka N. C. (2004) Nat. Struct. Mol. Biol. 11, 163–170 [DOI] [PubMed] [Google Scholar]
- 22. Chan P. H., Lairson L. L., Lee H. J., Wakarchuk W. W., Strynadka N. C., Withers S. G., McIntosh L. P. (2009) Biochemistry 48, 11220–11230 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Leslie A. (1992) Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 25. McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28. Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 29. Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 30. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 31. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bernatchez S., Gilbert M., Blanchard M. C., Karwaski M. F., Li J., Defrees S., Wakarchuk W. W. (2007) Glycobiology 17, 1333–1343 [DOI] [PubMed] [Google Scholar]
- 34. Chiu C. P., Lairson L. L., Gilbert M., Wakarchuk W. W., Withers S. G., Strynadka N. C. (2007) Biochemistry 46, 7196–7204 [DOI] [PubMed] [Google Scholar]
- 35. Simon E. S., Bednarski M. D., Whitesides G. M. (1988) J. Am. Chem. Soc. 110, 7159–7163 [Google Scholar]
- 36. Ohrui H., Meguro H., Ioa T. (December 14, 1988) European Patent EP0281067A3
- 37. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 38. Gosselin S., Alhussaini M., Streiff M. B., Takabayashi K., Palcic M. M. (1994) Anal. Biochem. 220, 92–97 [DOI] [PubMed] [Google Scholar]
- 39. Gilbert M., Cunningham A. M., Watson D. C., Martin A., Richards J. C., Wakarchuk W. W. (1997) Eur. J. Biochem. 249, 187–194 [DOI] [PubMed] [Google Scholar]
- 40. Cleland W. W. (1963) Biochim. Biophys. Acta 67, 104–137 [DOI] [PubMed] [Google Scholar]
- 41. Segel I. H. (1975) Enzyme Kinetics: Behaviour and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, pp. 560–656, John Wiley & Sons, Inc., New York [Google Scholar]
- 42. Fromm H. J. (1975) Initial Rate Enzyme Kinetics, pp. 121–144, Springer-Verlag, New York [Google Scholar]
- 43. Cornish-Bowden A. (2004) Fundamentals of Enzyme Kinetics, 3rd Ed., pp. 99–119, Portland Press, London [Google Scholar]
- 44. Piszkiewicz D. (1977) Kinetics of Chemical and Enzyme-catalyzed Reactions, pp. 131–140, Oxford University Press, New York [Google Scholar]
- 45. Bendiak B., Schachter H. (1987) J. Biol. Chem. 262, 5784–5790 [PubMed] [Google Scholar]
- 46. Kearns A. E., Campbell S. C., Westley J., Schwartz N. B. (1991) Biochemistry 30, 7477–7483 [DOI] [PubMed] [Google Scholar]
- 47. Murray B. W., Takayama S., Schultz J., Wong C. H. (1996) Biochemistry 35, 11183–11195 [DOI] [PubMed] [Google Scholar]
- 48. Chen L., Men H., Ha S., Ye X. Y., Brunner L., Hu Y., Walker S. (2002) Biochemistry 41, 6824–6833 [DOI] [PubMed] [Google Scholar]
- 49. Ly H. D., Lougheed B., Wakarchuk W. W., Withers S. G. (2002) Biochemistry 41, 5075–5085 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y. H., Ginsberg C., Yuan Y., Walker S. (2006) Biochemistry 45, 10895–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morrison J. F., Ebner K. E. (1971) J. Biol. Chem. 246, 3977–3984 [PubMed] [Google Scholar]
- 52. Boix E., Zhang Y., Swaminathan G. J., Brew K., Acharya K. R. (2002) J. Biol. Chem. 277, 28310–28318 [DOI] [PubMed] [Google Scholar]
- 53. Kamath V. P., Seto N. O., Compston C. A., Hindsgaul O., Palcic M. M. (1999) Glycoconj. J. 16, 599–606 [DOI] [PubMed] [Google Scholar]
- 54. Wragg S., Hagen F. K., Tabak L. A. (1995) J. Biol. Chem. 270, 16947–16954 [DOI] [PubMed] [Google Scholar]
- 55. Rudolph F. B., Fromm H. J. (1971) J. Biol. Chem. 246, 6611–6619 [PubMed] [Google Scholar]
- 56. Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.